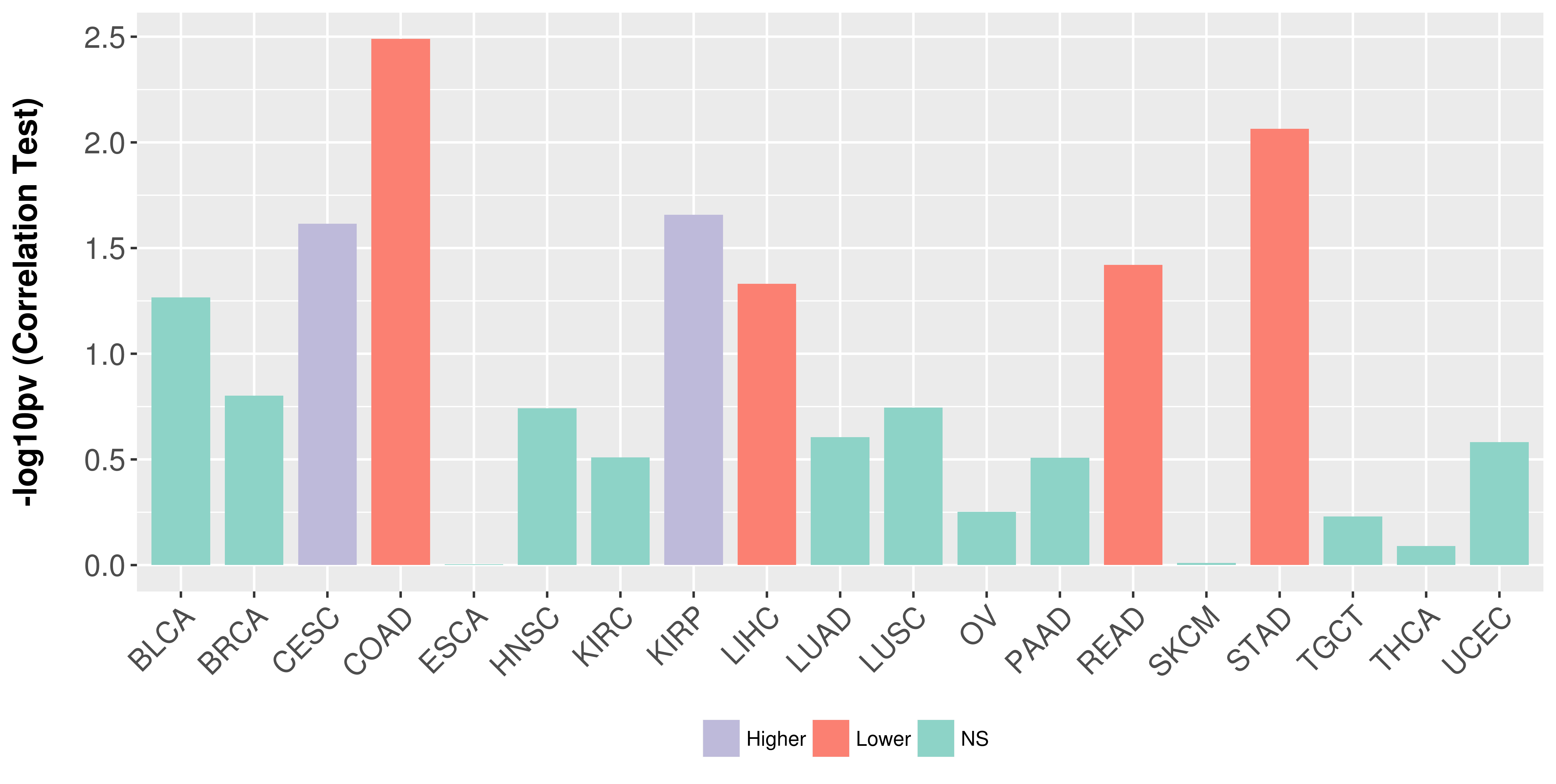
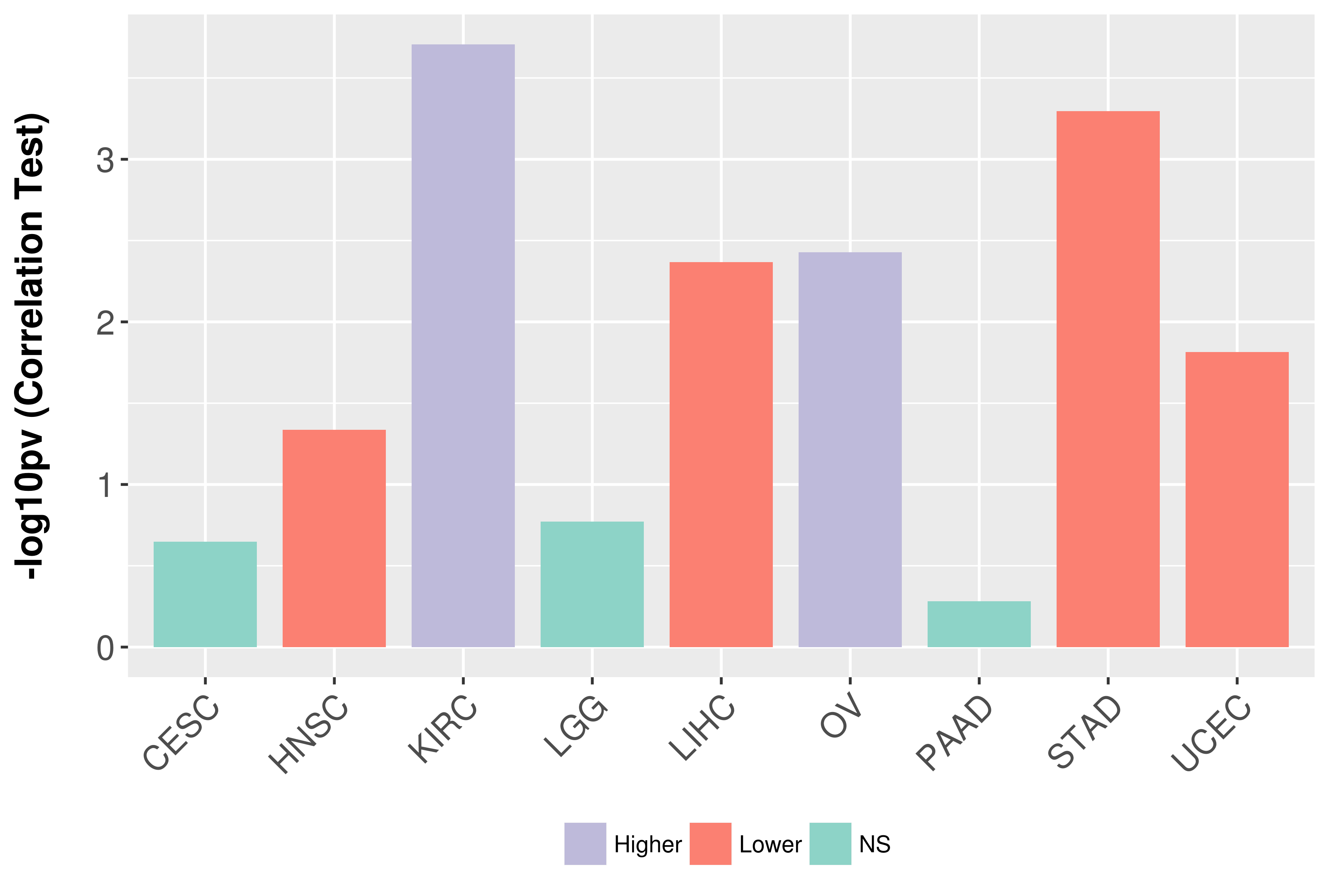
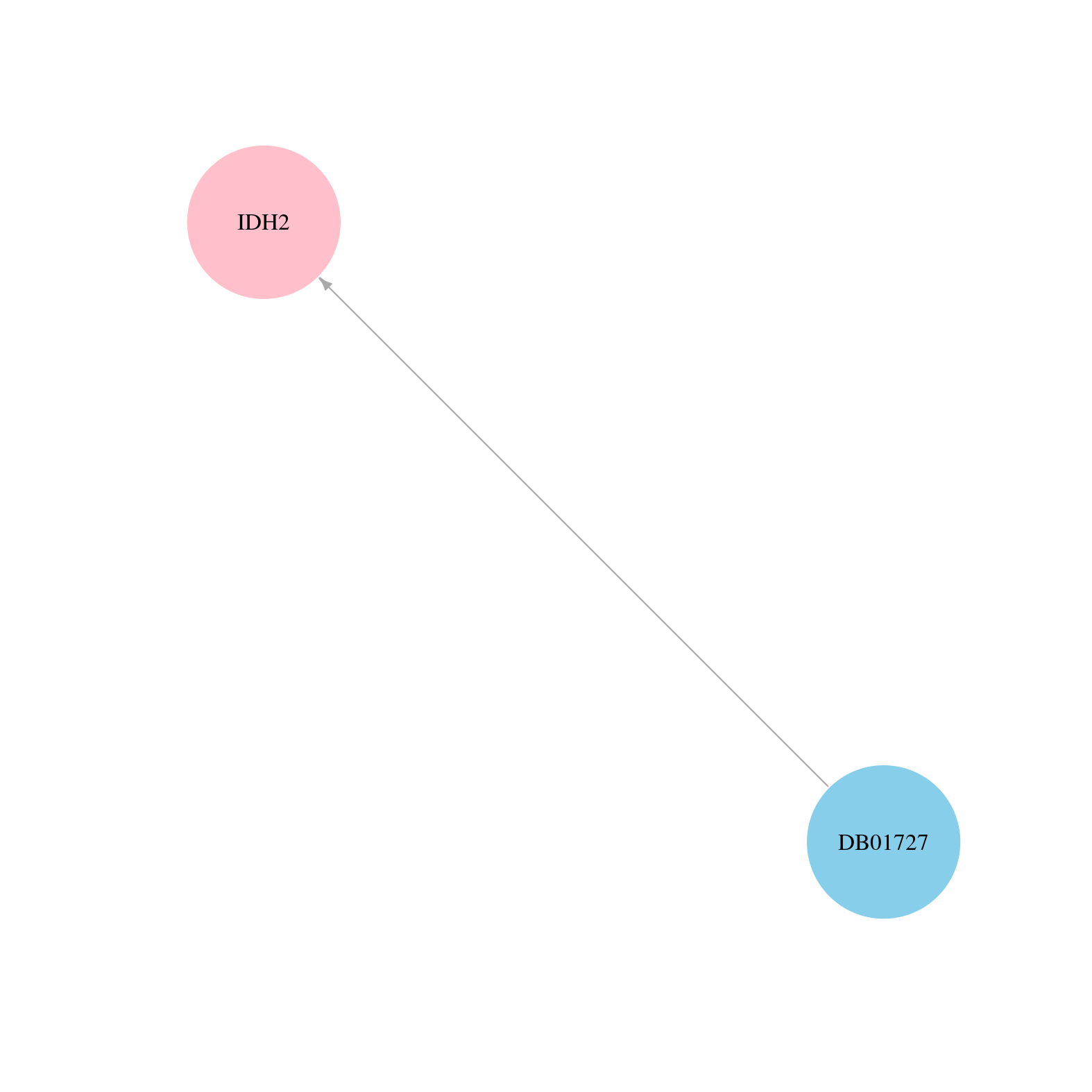
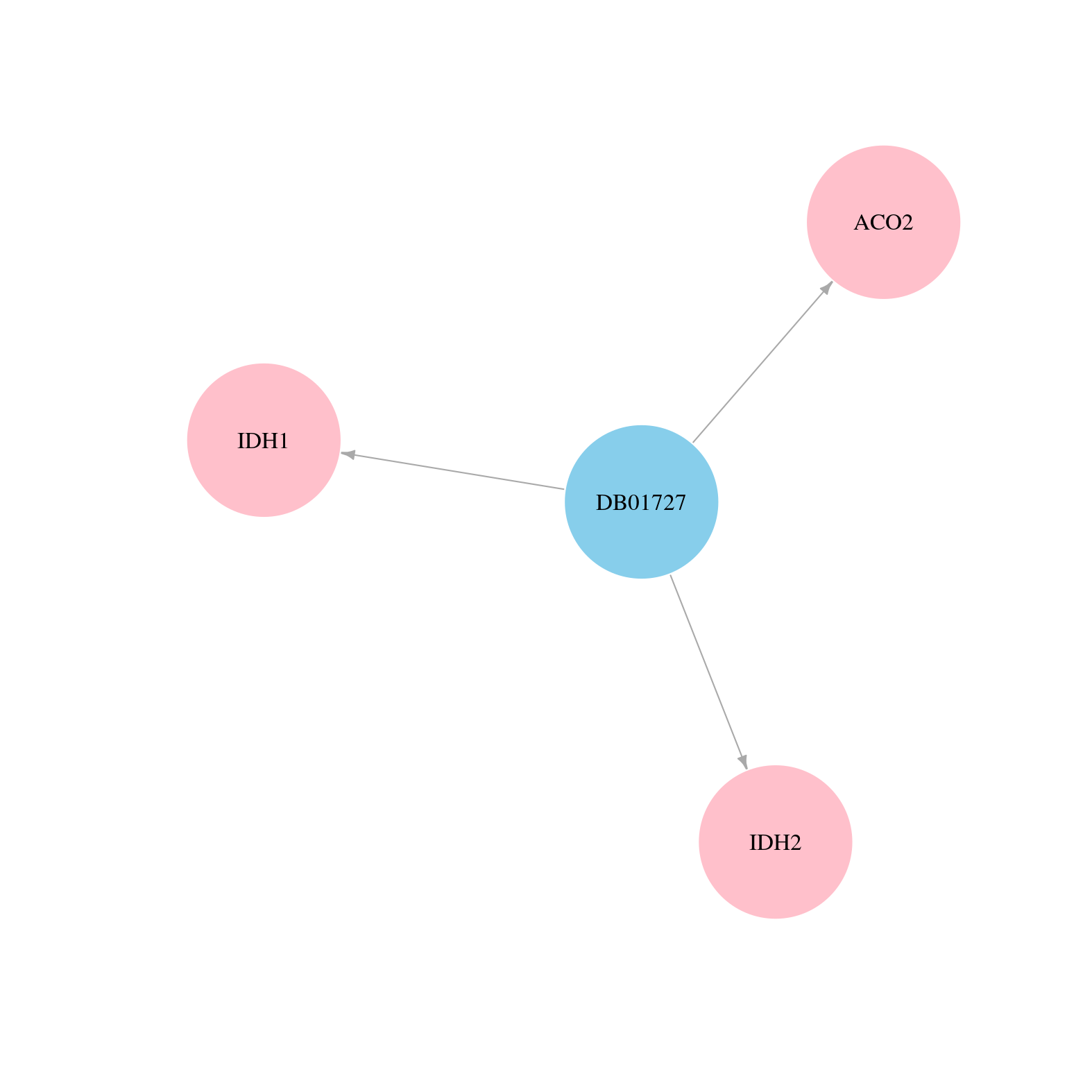
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4506463 | c.720C>T | p.I240I | Substitution - coding silent | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4925898 | c.1084C>T | p.R362W | Substitution - Missense | Liver
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385675 | c.428_429insG | p.T146fs*126 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM223811 | c.731G>A | p.W244* | Substitution - Nonsense | Skin
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Kidney
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Biliary_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5574262 | c.? | p.G176D | Substitution - Missense | Bone
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4990809 | c.148G>A | p.V50M | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4514509 | c.977C>T | p.S326F | Substitution - Missense | Skin
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM379071 | c.562C>T | p.R188W | Substitution - Missense | Lung
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM133169 | c.? | p.D177A | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Prostate
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM3999682 | c.517C>G | p.H173D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5384104 | c.1249G>A | p.G417S | Substitution - Missense | Skin
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM86077 | c.? | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1636997 | c.515G>C | p.R172T | Substitution - Missense | Bone
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4619428 | c.636C>T | p.F212F | Substitution - coding silent | Large_intestine
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM1636997 | c.515G>C | p.R172T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM5417541 | c.?_?insC | p.?fs*? | Unknown | Endometrium
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1737874 | c.418C>G | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4384862 | c.475C>G | p.R159G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5740454 | c.761T>C | p.I254T | Substitution - Missense | Small_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Breast
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM2139728 | c.1129C>T | p.R377C | Substitution - Missense | Large_intestine
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1749373 | c.622G>T | p.E208* | Substitution - Nonsense | Urinary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1737874 | c.418C>G | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5778128 | c.136G>A | p.V46M | Substitution - Missense | Breast
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1375397 | c.829G>A | p.D277N | Substitution - Missense | Large_intestine
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM3355752 | c.429G>C | p.L143L | Substitution - coding silent | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5885267 | c.515_516GG>AA | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3988210 | c.207+2T>C | p.? | Unknown | Kidney
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1749374 | c.144G>T | p.K48N | Substitution - Missense | Urinary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM966441 | c.804G>T | p.E268D | Substitution - Missense | Endometrium
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Large_intestine
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4777517 | c.1256T>C | p.I419T | Substitution - Missense | Liver
|
| COSM144497 | c.? | p.R172T | Substitution - Missense | Soft_tissue
|
| COSM5557399 | c.181A>G | p.I61V | Substitution - Missense | Prostate
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4578611 | c.1156G>C | p.D386H | Substitution - Missense | Bone
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM86077 | c.? | p.R172M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM5945312 | c.518A>T | p.H173L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3957058 | c.526G>T | p.G176C | Substitution - Missense | Lung
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM4767390 | c.? | p.A416fs*? | Frameshift | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4422879 | c.23T>C | p.V8A | Substitution - Missense | Kidney
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1493548 | c.641C>T | p.A214V | Substitution - Missense | Kidney
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM966442 | c.682A>C | p.I228L | Substitution - Missense | Endometrium
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53310 | c.? | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3420710 | c.1261G>A | p.G421S | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3665108 | c.81A>T | p.T27T | Substitution - coding silent | Oesophagus
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Soft_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Soft_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM966444 | c.248A>T | p.D83V | Substitution - Missense | Endometrium
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1636997 | c.515G>C | p.R172T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1749374 | c.144G>T | p.K48N | Substitution - Missense | Urinary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM340426 | c.745A>G | p.S249G | Substitution - Missense | Lung
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3420710 | c.1261G>A | p.G421S | Substitution - Missense | Ovary
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM4990809 | c.148G>A | p.V50M | Substitution - Missense | Kidney
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM3377698 | c.696G>A | p.A232A | Substitution - coding silent | Pancreas
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Bone
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4883838 | c.649G>T | p.V217L | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM6001688 | c.184A>G | p.I62V | Substitution - Missense | Prostate
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4612047 | c.435_436insG | p.T146fs*126 | Insertion - Frameshift | Large_intestine
|
| COSM86077 | c.? | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM966440 | c.1108G>A | p.A370T | Substitution - Missense | Endometrium
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5054592 | c.945G>A | p.V315V | Substitution - coding silent | Stomach
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4652657 | c.1086G>A | p.R362R | Substitution - coding silent | Large_intestine
|
| COSM4803661 | c.467A>G | p.N156S | Substitution - Missense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM86077 | c.? | p.R172M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM86960 | c.512G>A | p.G171D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Soft_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4803661 | c.467A>G | p.N156S | Substitution - Missense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4300420 | c.386A>G | p.K129R | Substitution - Missense | Central_nervous_system
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1737874 | c.418C>G | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Biliary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1375403 | c.435delG | p.T146fs*15 | Deletion - Frameshift | Stomach
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM966443 | c.476G>A | p.R159H | Substitution - Missense | Endometrium
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Biliary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM96477 | c.? | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1235293 | c.? | p.I168I | Substitution - coding silent | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1375398 | c.560A>T | p.D187V | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5650061 | c.1065_1066insA | p.R356fs*>98 | Insertion - Frameshift | Oesophagus
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM5751175 | c.445_446insC | p.R149fs*123 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1651582 | c.? | p.P167S | Substitution - Missense | Soft_tissue
|
| COSM966439 | c.1206C>T | p.C402C | Substitution - coding silent | Endometrium
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5445517 | c.645C>A | p.G215G | Substitution - coding silent | Oesophagus
|
| COSM5991573 | c.409G>A | p.G137R | Substitution - Missense | Skin
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM86077 | c.? | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM4472550 | c.178C>T | p.R60C | Substitution - Missense | Skin
|
| COSM5784561 | c.1325A>G | p.K442R | Substitution - Missense | Breast
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5417540 | c.118G>T | p.A40S | Substitution - Missense | Endometrium
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM86960 | c.512G>A | p.G171D | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144497 | c.? | p.R172T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1375403 | c.435delG | p.T146fs*15 | Deletion - Frameshift | Prostate
|
| COSM4777517 | c.1256T>C | p.I419T | Substitution - Missense | Breast
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53310 | c.? | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4802448 | c.809T>C | p.F270S | Substitution - Missense | Liver
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Bone
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1658762 | c.1054A>C | p.T352P | Substitution - Missense | Breast
|
| COSM4767390 | c.? | p.A416fs*? | Frameshift | Large_intestine
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5991574 | c.433G>A | p.G145R | Substitution - Missense | Skin
|
| COSM5384105 | c.163G>A | p.G55S | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM5043002 | c.189G>A | p.W63* | Substitution - Nonsense | Liver
|
| COSM4169399 | c.? | p.G144R | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Liver
|
| COSM2139728 | c.1129C>T | p.R377C | Substitution - Missense | Large_intestine
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4691742 | c.1324A>G | p.K442E | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1737874 | c.418C>G | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM1749373 | c.622G>T | p.E208* | Substitution - Nonsense | Urinary_tract
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4774901 | c.488G>A | p.G163D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM53310 | c.? | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5436639 | c.583G>A | p.V195I | Substitution - Missense | Oesophagus
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM269346 | c.1322T>C | p.I441T | Substitution - Missense | Large_intestine
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86959 | c.472C>A | p.P158T | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4802448 | c.809T>C | p.F270S | Substitution - Missense | Liver
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3505366 | c.410G>A | p.G137E | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1375401 | c.473C>T | p.P158L | Substitution - Missense | Central_nervous_system
|
| COSM1629738 | c.154G>T | p.E52* | Substitution - Nonsense | Liver
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1638389 | c.514A>C | p.R172R | Substitution - coding silent | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM256046 | c.515_516GG>AT | p.R172N | Substitution - Missense | Biliary_tract
|
| COSM5715080 | c.535-8C>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM4634034 | c.963C>A | p.A321A | Substitution - coding silent | Large_intestine
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Bone
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM6001592 | c.828C>T | p.T276T | Substitution - coding silent | Prostate
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM1736930 | c.484C>T | p.P162S | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM271310 | c.140C>T | p.A47V | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1736930 | c.484C>T | p.P162S | Substitution - Missense | Central_nervous_system
|
| COSM4128570 | c.996C>T | p.S332S | Substitution - coding silent | Thyroid
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144497 | c.? | p.R172T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM96477 | c.? | p.R140G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1236441 | c.1127C>A | p.T376K | Substitution - Missense | Autonomic_ganglia
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5750057 | c.1214C>T | p.T405M | Substitution - Missense | Stomach
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4384860 | c.1354C>T | p.Q452* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5929583 | c.253G>A | p.G85R | Substitution - Missense | Skin
|
| COSM5929582 | c.254G>A | p.G85E | Substitution - Missense | Skin
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4767390 | c.? | p.A416fs*? | Frameshift | Large_intestine
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Urinary_tract
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM41875 | c.419G>T | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1629739 | c.153G>T | p.V51V | Substitution - coding silent | Liver
|
| COSM5609104 | c.523C>T | p.H175Y | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4057844 | c.1124G>A | p.W375* | Substitution - Nonsense | Stomach
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM97048 | c.? | p.R140? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Liver
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM2139738 | c.528C>T | p.G176G | Substitution - coding silent | Large_intestine
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM227366 | c.415A>T | p.I139F | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM5751937 | c.222_223insA | p.V75fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM86960 | c.512G>A | p.G171D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4128570 | c.996C>T | p.S332S | Substitution - coding silent | Soft_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Bone
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM268311 | c.950C>G | p.S317* | Substitution - Nonsense | Large_intestine
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM523140 | c.? | p.R172L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53310 | c.? | p.R140L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1736930 | c.484C>T | p.P162S | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Biliary_tract
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5985374 | c.568G>A | p.G190S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM2139737 | c.569G>A | p.G190D | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945313 | c.407A>G | p.N136S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM99760 | c.516G>A | p.R172R | Substitution - coding silent | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Bone
|
| COSM4384861 | c.676G>A | p.E226K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5575751 | c.374-1G>C | p.? | Unknown | Stomach
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Bone
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1658762 | c.1054A>C | p.T352P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4169399 | c.? | p.G144R | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM86960 | c.512G>A | p.G171D | Substitution - Missense | Skin
|
| COSM4954396 | c.178C>G | p.R60G | Substitution - Missense | Liver
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41877 | c.418C>T | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1210232 | c.743T>C | p.M248T | Substitution - Missense | Large_intestine
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5945714 | c.424A>C | p.I142L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Liver
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Bone
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM86960 | c.512G>A | p.G171D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM131566 | c.? | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM53311 | c.? | p.R140W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM133672 | c.516G>C | p.R172S | Substitution - Missense | Bone
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM52947 | c.? | p.R172? | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4954396 | c.178C>G | p.R60G | Substitution - Missense | Liver
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3999681 | c.518A>C | p.H173P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4254911 | c.409G>T | p.G137* | Substitution - Nonsense | Lung
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM99759 | c.492G>A | p.W164* | Substitution - Nonsense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM24361 | c.880G>A | p.V294M | Substitution - Missense | Skin
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144496 | c.? | p.R172S | Substitution - Missense | Central_nervous_system
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41296 | c.? | p.R172W | Substitution - Missense | Biliary_tract
|
| COSM33731 | c.514A>G | p.R172G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34039 | c.514A>T | p.R172W | Substitution - Missense | Central_nervous_system
|
| COSM966438 | c.1239G>A | p.K413K | Substitution - coding silent | Endometrium
|
| COSM6005781 | c.1303A>T | p.T435S | Substitution - Missense | Skin
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM34090 | c.516G>T | p.R172S | Substitution - Missense | Bone
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Central_nervous_system
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41295 | c.? | p.R172K | Substitution - Missense | Oesophagus
|
| COSM1685352 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33733 | c.515G>A | p.R172K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM49103 | c.? | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM41590 | c.419G>A | p.R140Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM33732 | c.515G>T | p.R172M | Substitution - Missense | Central_nervous_system
|