Browse ING2 in pancancer
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF12998 Inhibitor of growth proteins N-terminal histone-binding |
||||||||||
| Function |
Seems to be involved in p53/TP53 activation and p53/TP53-dependent apoptotic pathways, probably by enhancing acetylation of p53/TP53. Component of a mSin3A-like corepressor complex, which is probably involved in deacetylation of nucleosomal histones. ING2 activity seems to be modulated by binding to phosphoinositides (PtdInsPs). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006476 protein deacetylation GO:0007126 meiotic nuclear division GO:0007127 meiosis I GO:0007140 male meiosis GO:0007141 male meiosis I GO:0007178 transmembrane receptor protein serine/threonine kinase signaling pathway GO:0007179 transforming growth factor beta receptor signaling pathway GO:0007281 germ cell development GO:0007283 spermatogenesis GO:0007286 spermatid development GO:0007548 sex differentiation GO:0007568 aging GO:0007569 cell aging GO:0008356 asymmetric cell division GO:0008406 gonad development GO:0008584 male gonad development GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0016570 histone modification GO:0016575 histone deacetylation GO:0017015 regulation of transforming growth factor beta receptor signaling pathway GO:0017145 stem cell division GO:0022412 cellular process involved in reproduction in multicellular organism GO:0030317 flagellated sperm motility GO:0030511 positive regulation of transforming growth factor beta receptor signaling pathway GO:0031056 regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031063 regulation of histone deacetylation GO:0031065 positive regulation of histone deacetylation GO:0035601 protein deacylation GO:0042078 germ-line stem cell division GO:0042771 intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:0045137 development of primary sexual characteristics GO:0046546 development of primary male sexual characteristics GO:0046661 male sex differentiation GO:0048133 male germ-line stem cell asymmetric division GO:0048232 male gamete generation GO:0048515 spermatid differentiation GO:0048608 reproductive structure development GO:0051321 meiotic cell cycle GO:0061458 reproductive system development GO:0071559 response to transforming growth factor beta GO:0071560 cellular response to transforming growth factor beta stimulus GO:0072331 signal transduction by p53 class mediator GO:0072332 intrinsic apoptotic signaling pathway by p53 class mediator GO:0072520 seminiferous tubule development GO:0090092 regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090100 positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090287 regulation of cellular response to growth factor stimulus GO:0090311 regulation of protein deacetylation GO:0090312 positive regulation of protein deacetylation GO:0090342 regulation of cell aging GO:0090398 cellular senescence GO:0097193 intrinsic apoptotic signaling pathway GO:0097722 sperm motility GO:0098722 asymmetric stem cell division GO:0098728 germline stem cell asymmetric division GO:0098732 macromolecule deacylation GO:1901796 regulation of signal transduction by p53 class mediator GO:1901797 negative regulation of signal transduction by p53 class mediator GO:1902165 regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902166 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902229 regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902230 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902253 regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902254 negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902275 regulation of chromatin organization GO:1902532 negative regulation of intracellular signal transduction GO:1903046 meiotic cell cycle process GO:1903844 regulation of cellular response to transforming growth factor beta stimulus GO:1903846 positive regulation of cellular response to transforming growth factor beta stimulus GO:1905269 positive regulation of chromatin organization GO:2000772 regulation of cellular senescence GO:2001020 regulation of response to DNA damage stimulus GO:2001021 negative regulation of response to DNA damage stimulus GO:2001233 regulation of apoptotic signaling pathway GO:2001234 negative regulation of apoptotic signaling pathway GO:2001242 regulation of intrinsic apoptotic signaling pathway GO:2001243 negative regulation of intrinsic apoptotic signaling pathway |
| Molecular Function |
GO:0003682 chromatin binding GO:0005543 phospholipid binding GO:0035064 methylated histone binding GO:0035091 phosphatidylinositol binding GO:0042393 histone binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0005667 transcription factor complex GO:0016580 Sin3 complex GO:0016602 CCAAT-binding factor complex GO:0044454 nuclear chromosome part GO:0044798 nuclear transcription factor complex GO:0070822 Sin3-type complex GO:0090575 RNA polymerase II transcription factor complex |
| KEGG | - |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-6811555: PI5P Regulates TP53 Acetylation R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804758: Regulation of TP53 Activity through Acetylation R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
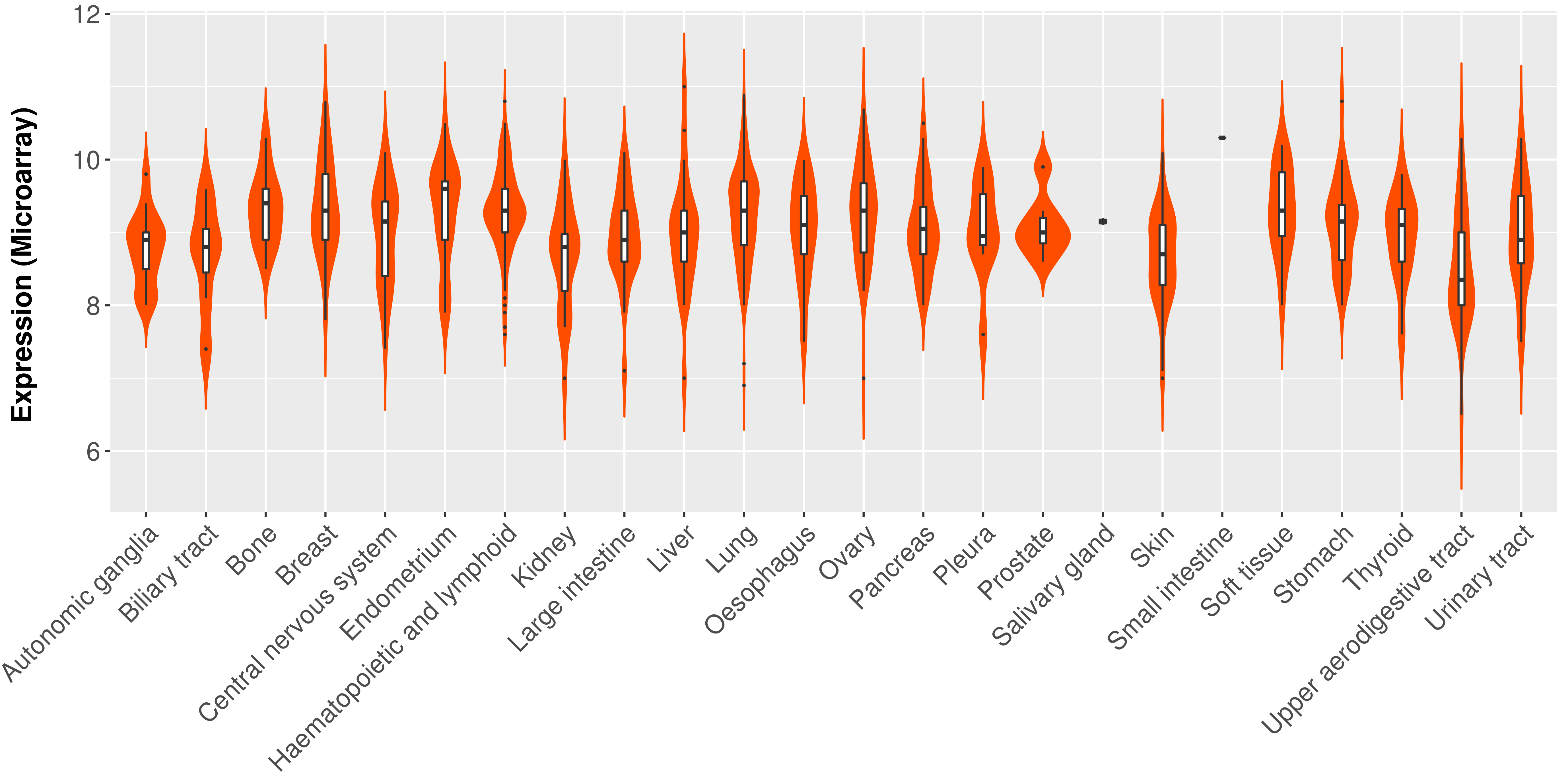
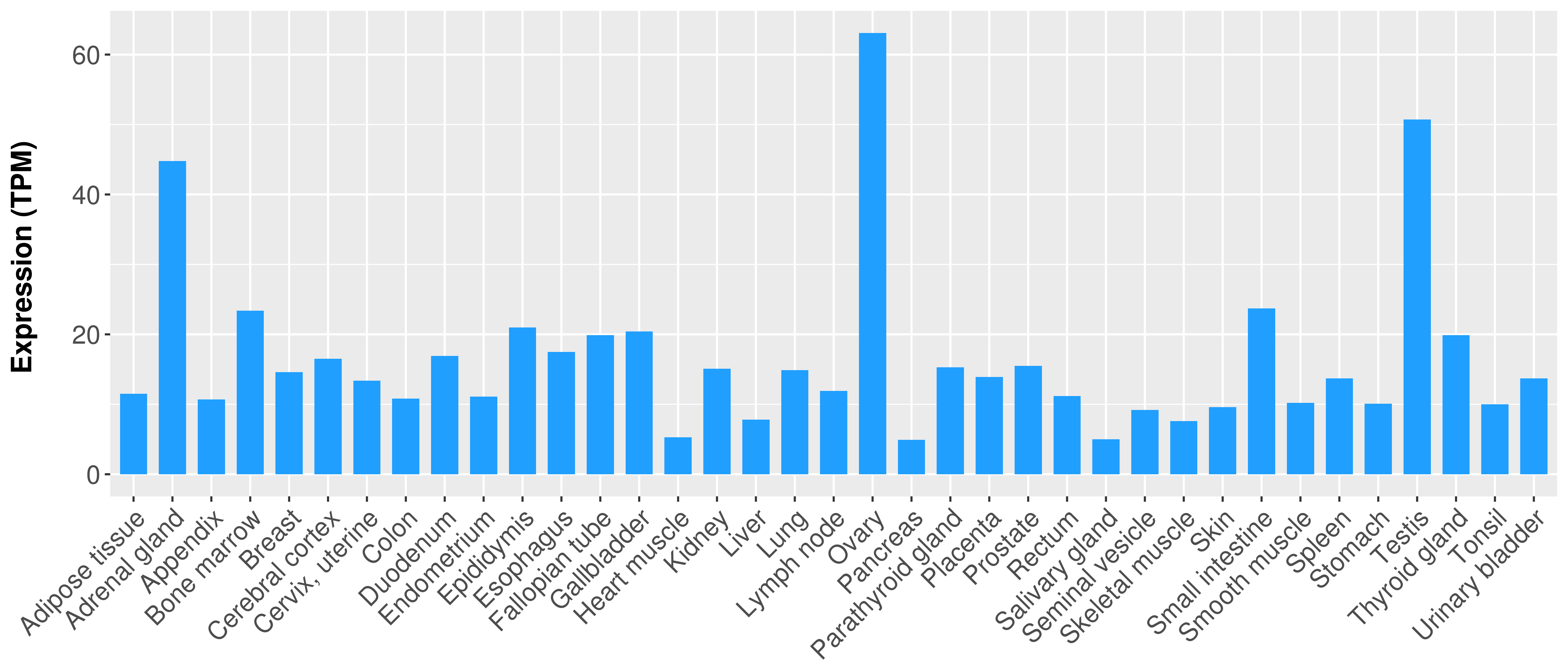
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
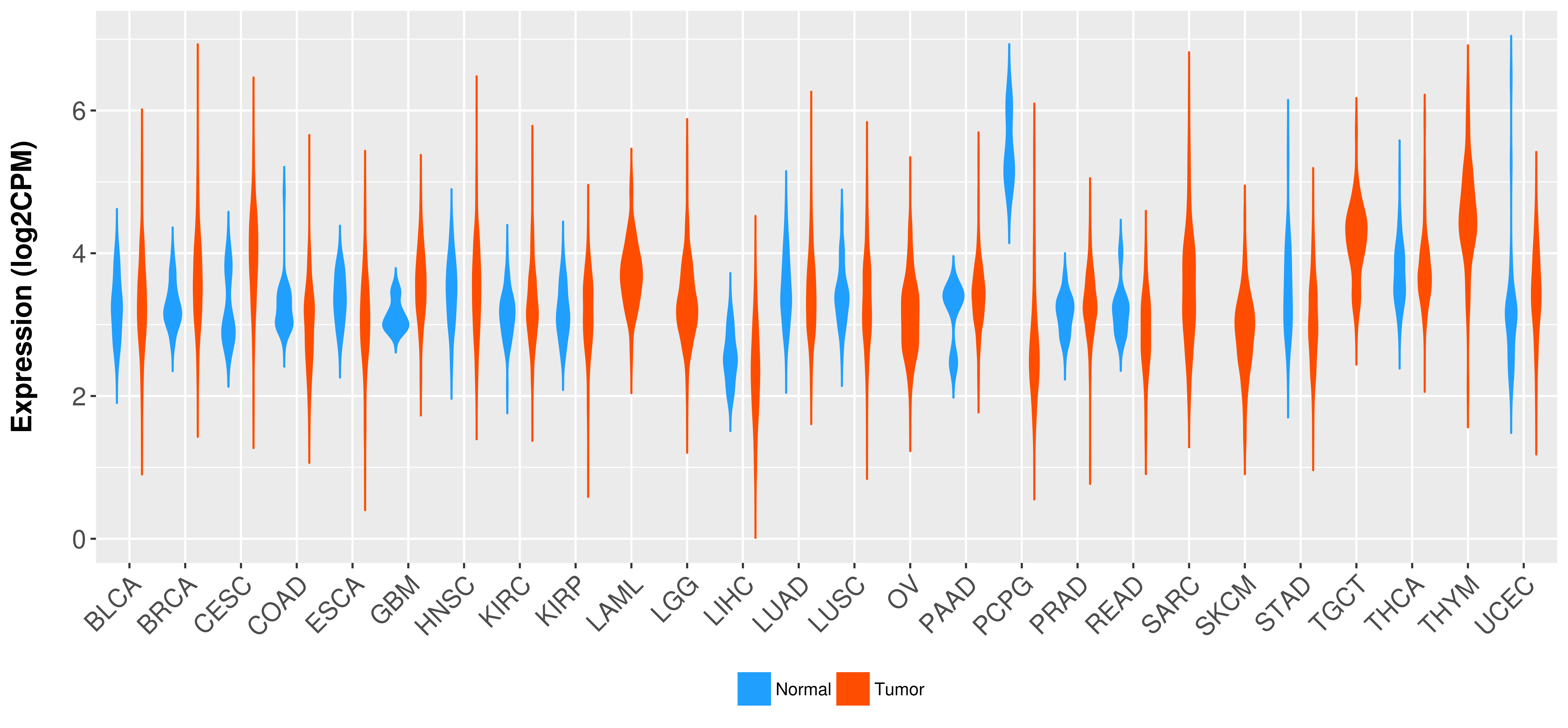
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
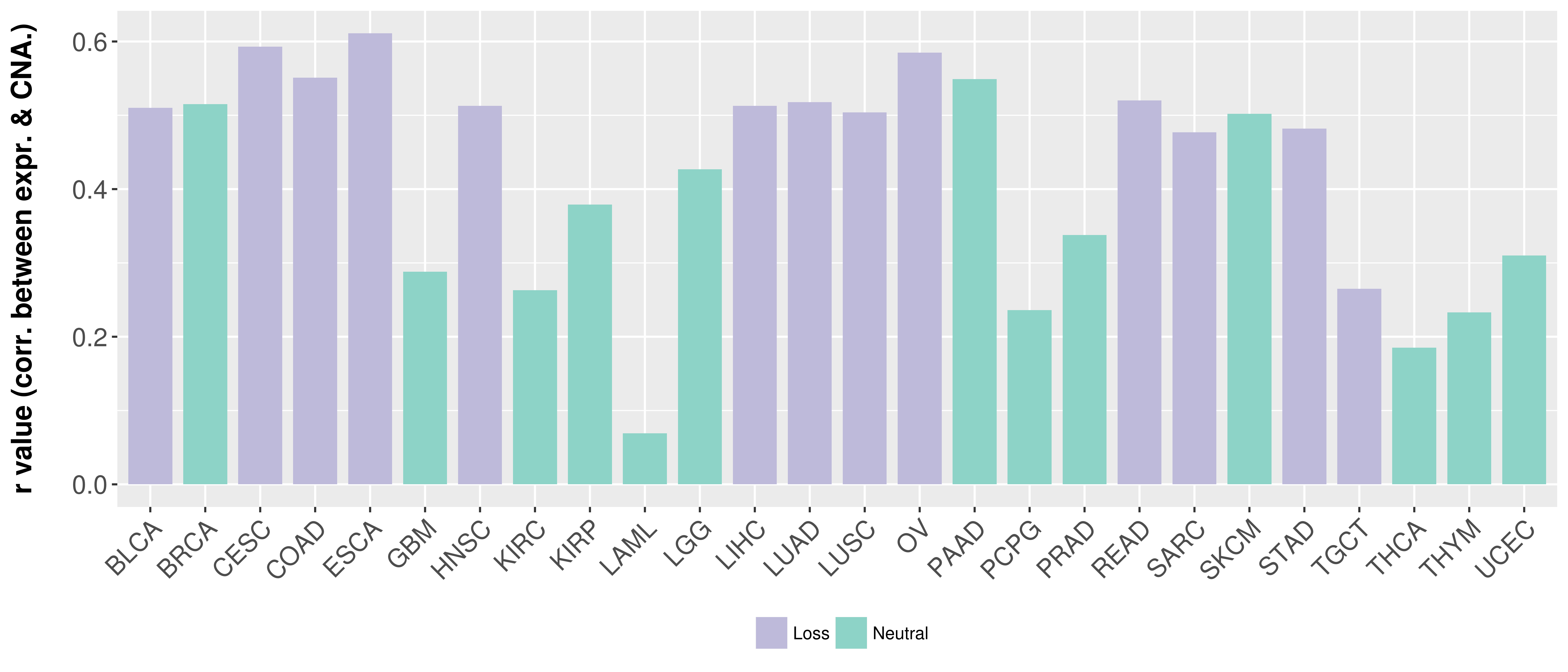
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
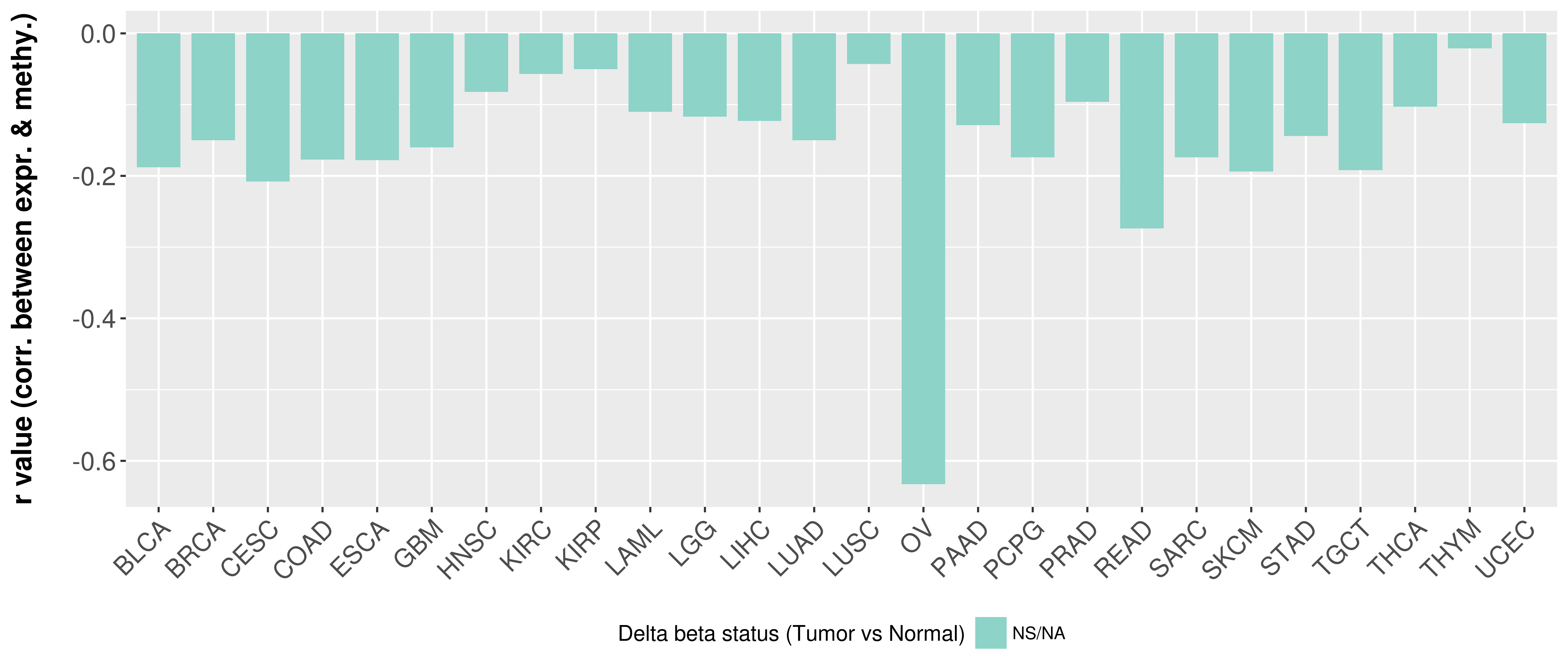
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
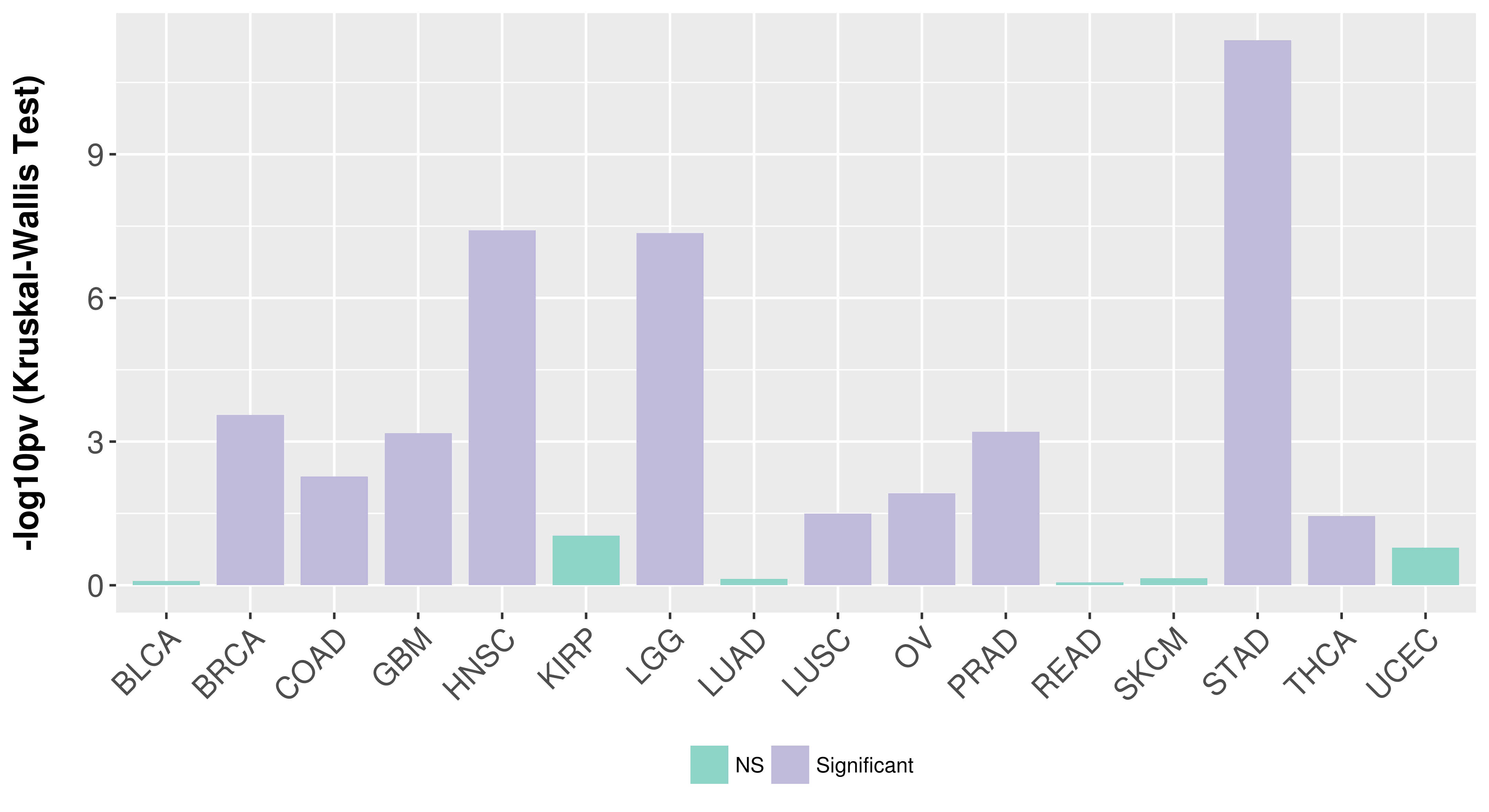
Association between expresson and subtype.
|
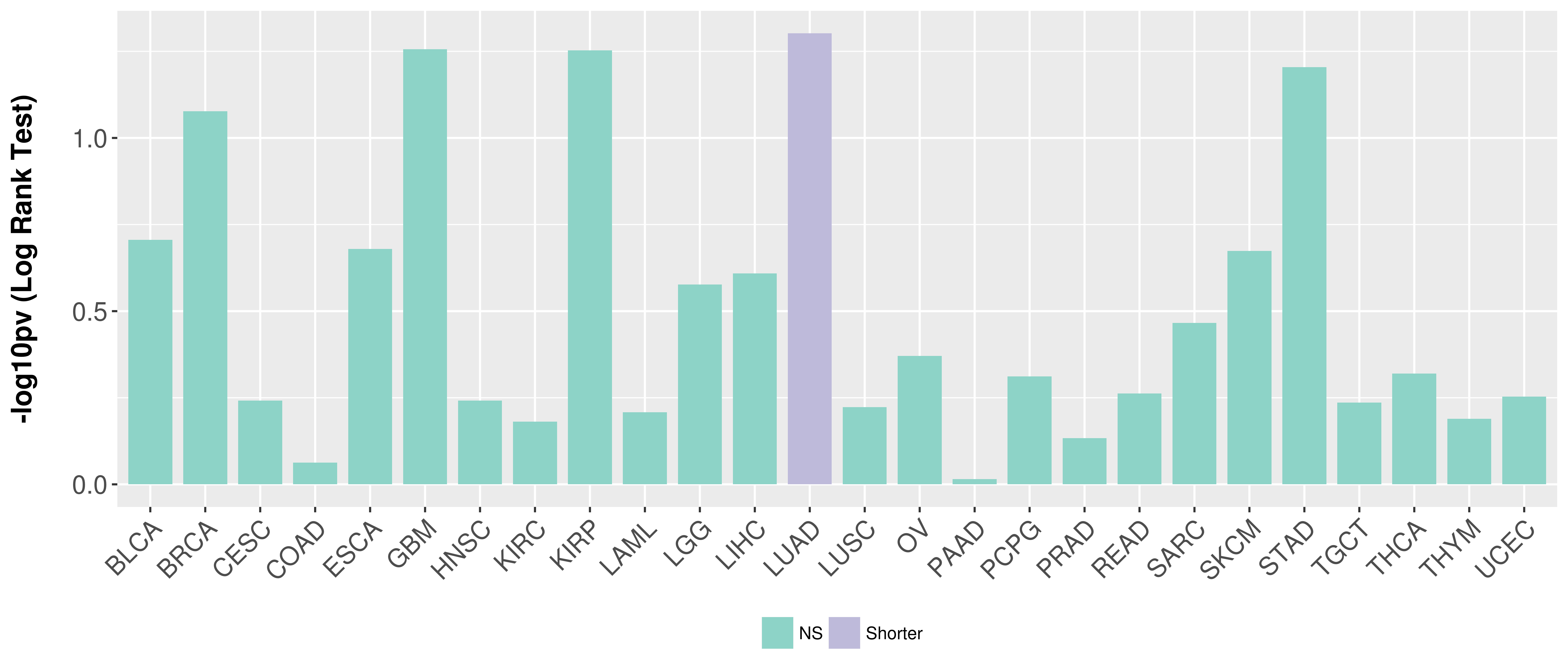
Overall survival analysis based on expression.
|
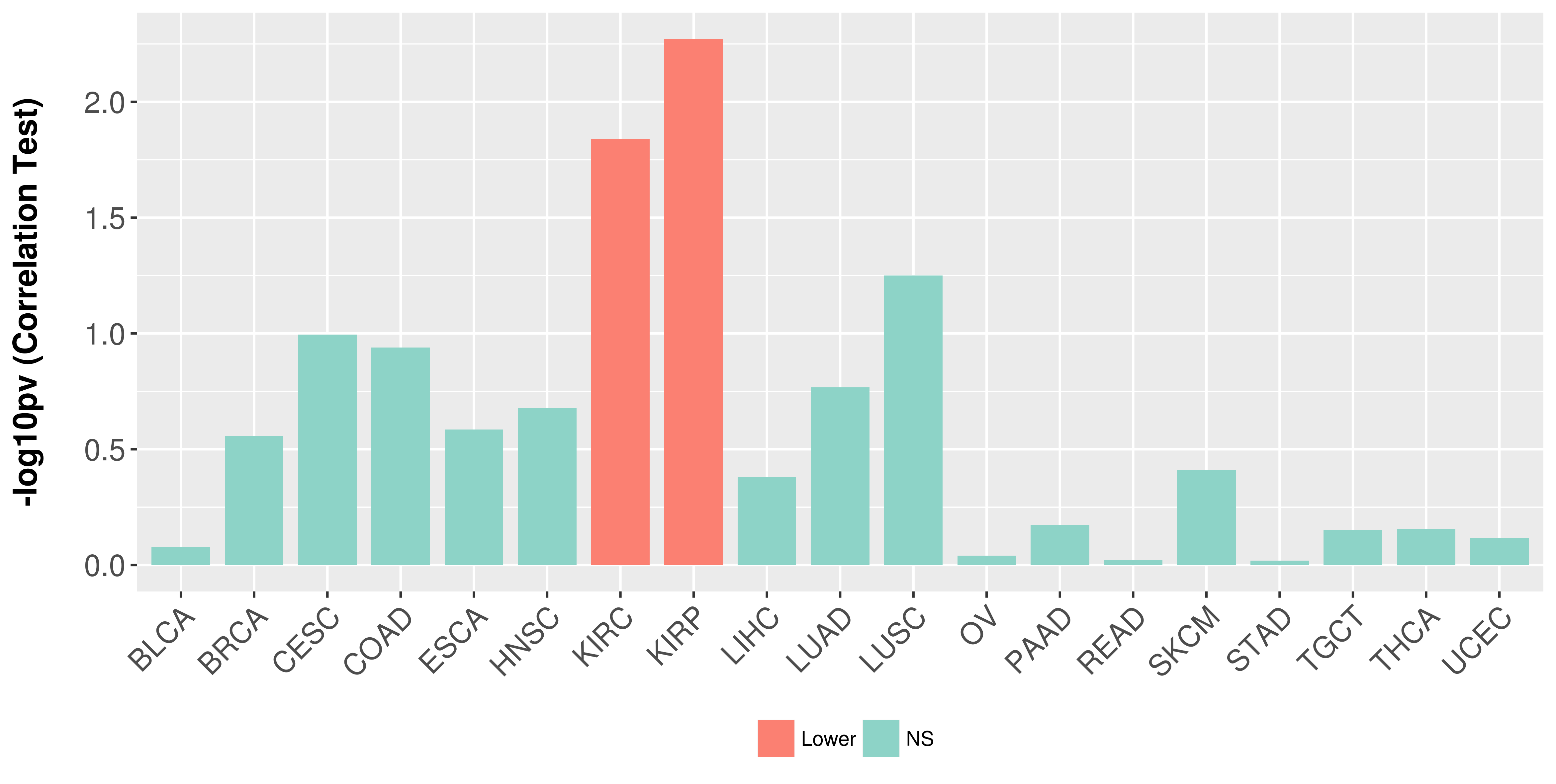
Association between expresson and stage.
|
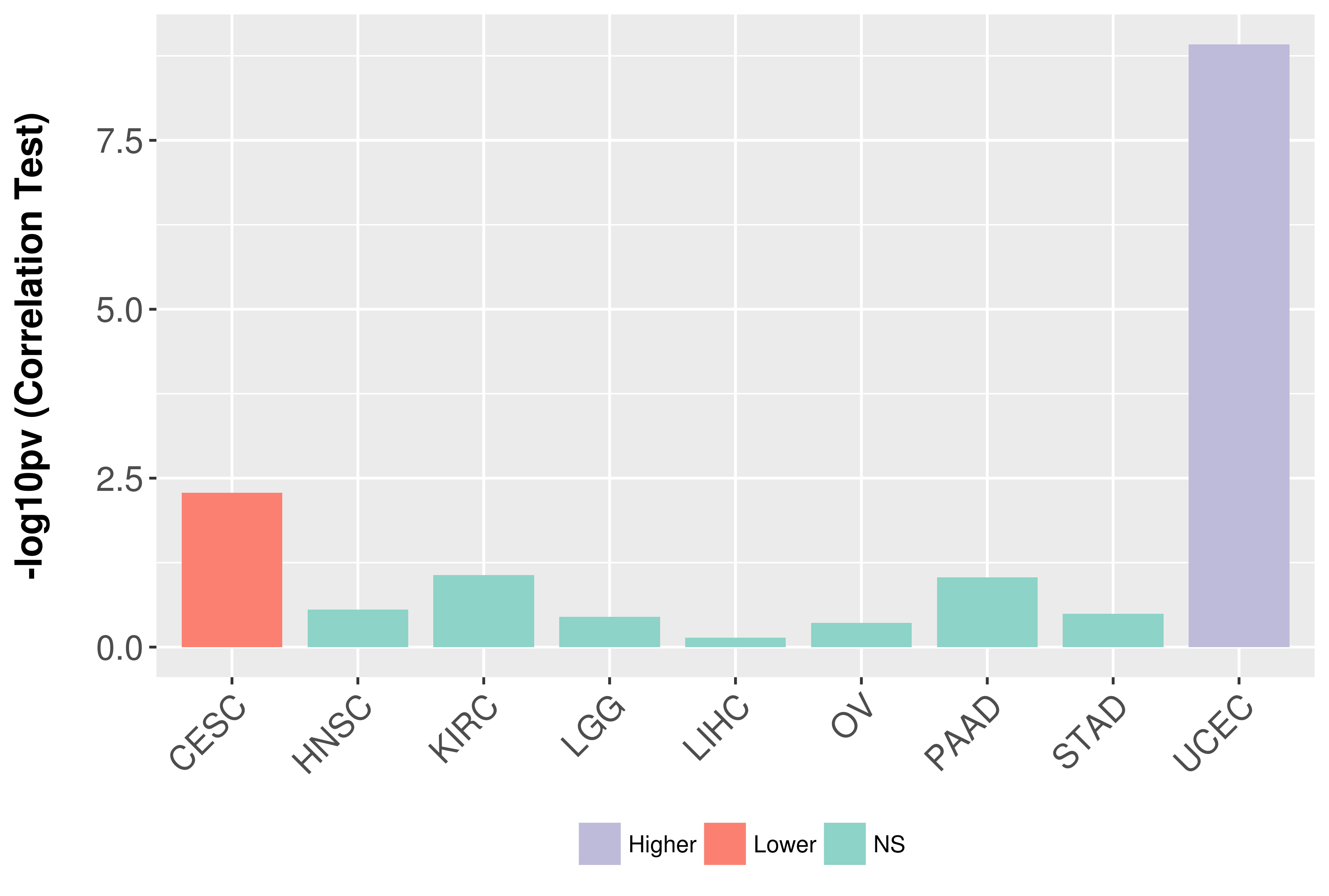
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ING2. |
| Summary | |
|---|---|
| Symbol | ING2 |
| Name | inhibitor of growth family member 2 |
| Aliases | p33ING2; ING1L; inhibitor of growth family, member 1-like; ING1Lp; inhibitor of growth 1-like protein; Inhib ...... |
| Location | 4q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|