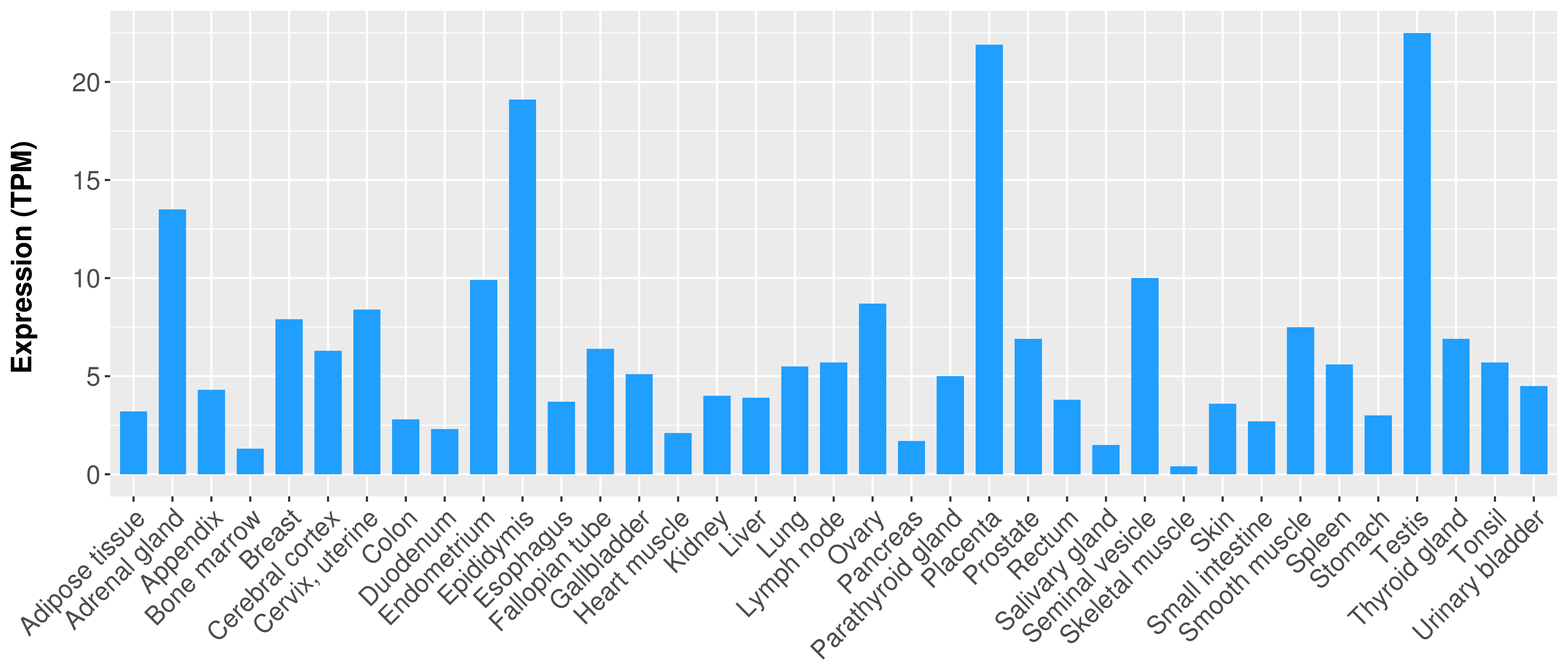
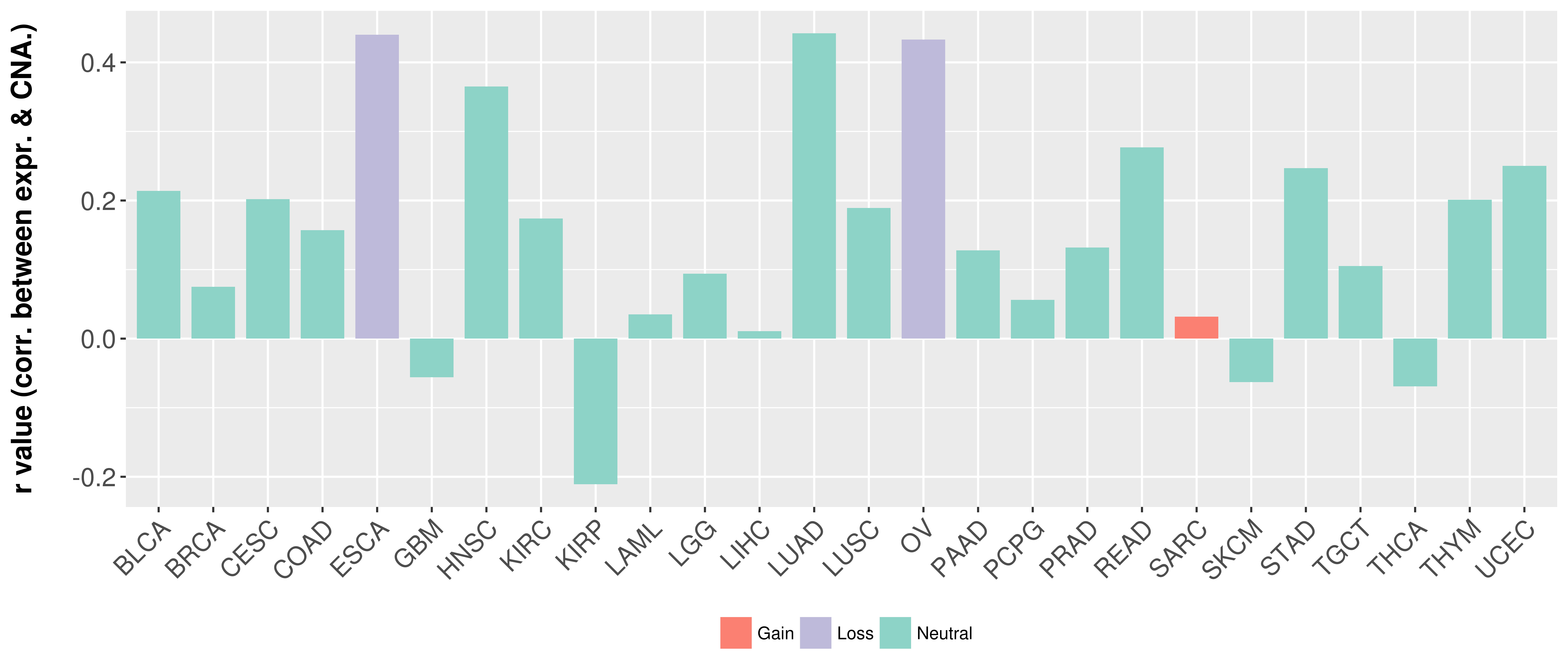
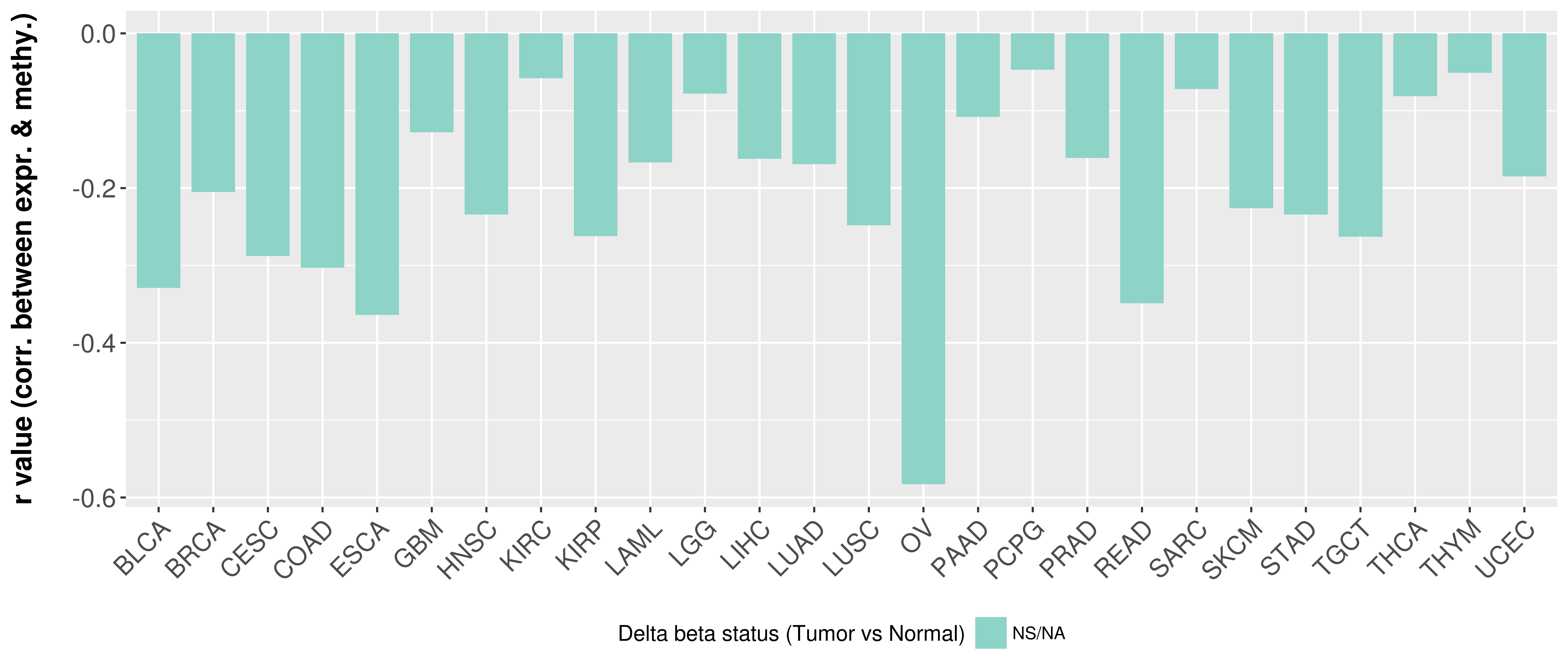
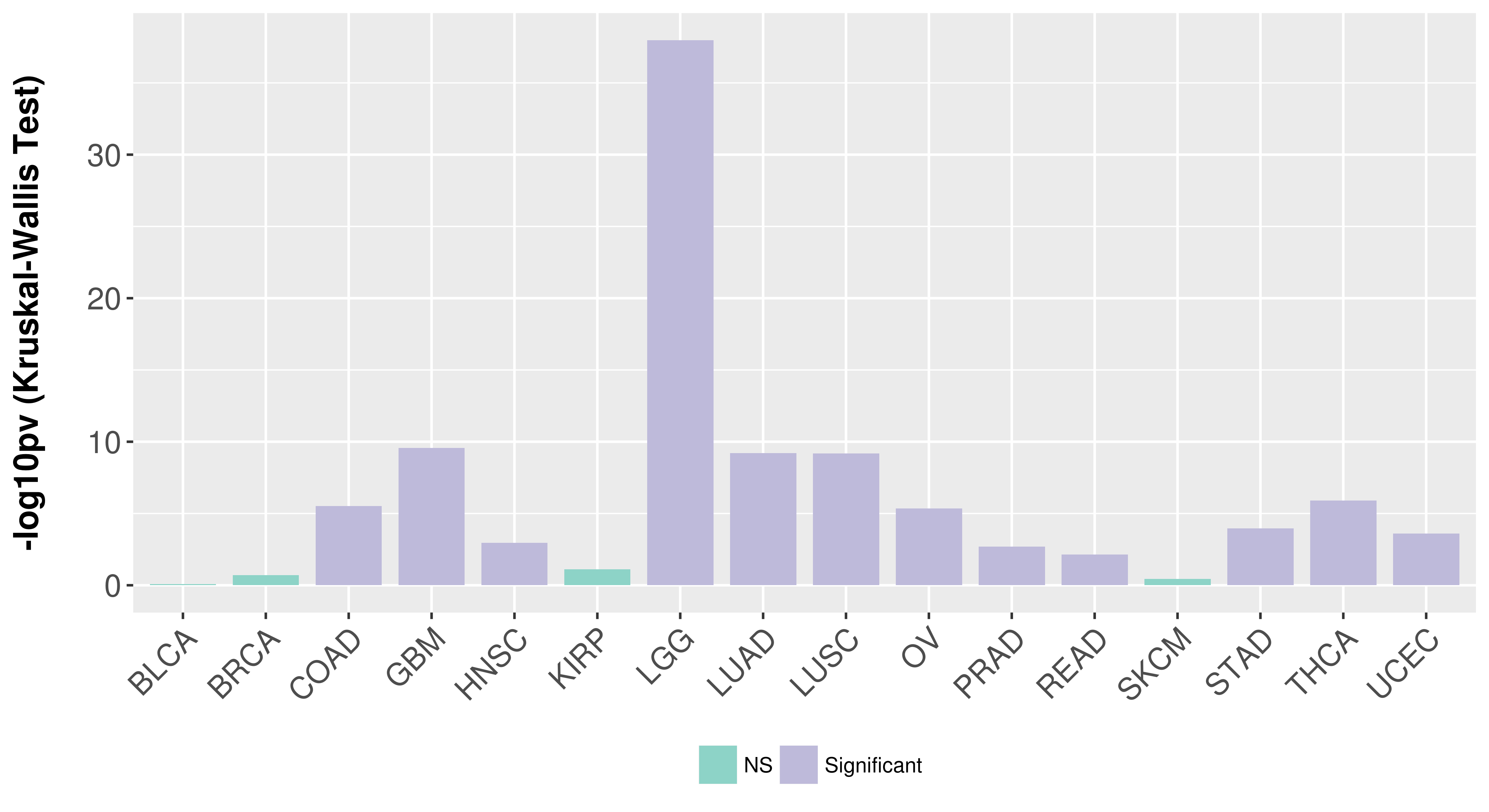
| COSM757217 | c.1706G>T | p.S569I | Substitution - Missense | Lung
|
| COSM1714907 | c.229G>A | p.E77K | Substitution - Missense | Skin
|
| COSM488388 | c.1966A>C | p.K656Q | Substitution - Missense | Kidney
|
| COSM163441 | c.862A>G | p.I288V | Substitution - Missense | Breast
|
| COSM1121697 | c.1343C>A | p.P448H | Substitution - Missense | Endometrium
|
| COSM4946472 | c.1600T>C | p.Y534H | Substitution - Missense | Liver
|
| COSM757221 | c.783A>T | p.K261N | Substitution - Missense | Lung
|
| COSM3844755 | c.1715C>G | p.S572C | Substitution - Missense | Breast
|
| COSM1121700 | c.2104G>A | p.G702S | Substitution - Missense | Endometrium
|
| COSM1121698 | c.1811G>A | p.R604H | Substitution - Missense | Large_intestine
|
| COSM1121693 | c.956C>T | p.T319M | Substitution - Missense | Stomach
|
| COSM219574 | c.2094G>T | p.L698F | Substitution - Missense | Breast
|
| COSM1756581 | c.1255G>A | p.E419K | Substitution - Missense | Urinary_tract
|
| COSM488386 | c.579A>C | p.L193L | Substitution - coding silent | Kidney
|
| COSM1682573 | c.1435C>A | p.L479M | Substitution - Missense | Large_intestine
|
| COSM4408295 | c.2092T>G | p.L698V | Substitution - Missense | Pancreas
|
| COSM3765012 | c.1474C>T | p.R492* | Substitution - Nonsense | Central_nervous_system
|
| COSM5757546 | c.650T>C | p.M217T | Substitution - Missense | Large_intestine
|
| COSM673747 | c.47-9C>G | p.? | Unknown | Endometrium
|
| COSM4840336 | c.2317C>T | p.L773L | Substitution - coding silent | Cervix
|
| COSM3561787 | c.2370G>A | p.R790R | Substitution - coding silent | Skin
|
| COSM1756582 | c.2408C>G | p.S803C | Substitution - Missense | Urinary_tract
|
| COSM1663472 | c.16C>T | p.P6S | Substitution - Missense | Kidney
|
| COSM5761337 | c.432C>T | p.D144D | Substitution - coding silent | Pancreas
|
| COSM1121691 | c.657C>A | p.F219L | Substitution - Missense | Endometrium
|
| COSM5347588 | c.1290_1293delCTAT | p.Y431fs*5 | Deletion - Frameshift | Liver
|
| COSM5094956 | c.1653A>T | p.E551D | Substitution - Missense | Thymus
|
| COSM1625908 | c.1126G>A | p.E376K | Substitution - Missense | Liver
|
| COSM1636552 | c.2348A>C | p.N783T | Substitution - Missense | Liver
|
| COSM1121698 | c.1811G>A | p.R604H | Substitution - Missense | Endometrium
|
| COSM3973672 | c.361A>G | p.T121A | Substitution - Missense | Central_nervous_system
|
| COSM5064698 | c.1890C>T | p.C630C | Substitution - coding silent | Stomach
|
| COSM1315528 | c.403G>C | p.V135L | Substitution - Missense | Urinary_tract
|
| COSM3561785 | c.1549G>A | p.E517K | Substitution - Missense | Skin
|
| COSM1121686 | c.43G>A | p.E15K | Substitution - Missense | Stomach
|
| COSM5412704 | c.849C>T | p.I283I | Substitution - coding silent | Skin
|
| COSM3914010 | c.1226C>T | p.S409F | Substitution - Missense | Skin
|
| COSM1121694 | c.1063G>A | p.E355K | Substitution - Missense | Endometrium
|
| COSM1121689 | c.223G>A | p.E75K | Substitution - Missense | Endometrium
|
| COSM3844753 | c.912A>C | p.P304P | Substitution - coding silent | Breast
|
| COSM4629020 | c.1146T>C | p.A382A | Substitution - coding silent | Large_intestine
|
| COSM3406387 | c.310G>A | p.V104I | Substitution - Missense | Central_nervous_system
|
| COSM5159319 | c.973-10C>A | p.? | Unknown | Large_intestine
|
| COSM757219 | c.997G>T | p.A333S | Substitution - Missense | Lung
|
| COSM1262040 | c.656T>C | p.F219S | Substitution - Missense | Stomach
|
| COSM5366825 | c.1652A>C | p.E551A | Substitution - Missense | Large_intestine
|
| COSM4149056 | c.1137C>A | p.H379Q | Substitution - Missense | Ovary
|
| COSM1262039 | c.1267C>T | p.P423S | Substitution - Missense | Oesophagus
|
| COSM5086493 | c.92_93insA | p.I33fs*4 | Insertion - Frameshift | Large_intestine
|
| COSM4795767 | c.1579G>A | p.A527T | Substitution - Missense | Liver
|
| COSM4713950 | c.1022A>C | p.E341A | Substitution - Missense | Large_intestine
|
| COSM4589429 | c.2076T>C | p.P692P | Substitution - coding silent | Bone
|
| COSM3844751 | c.313C>G | p.L105V | Substitution - Missense | Breast
|
| COSM5423274 | c.2375A>G | p.D792G | Substitution - Missense | Prostate
|
| COSM1682573 | c.1435C>A | p.L479M | Substitution - Missense | Large_intestine
|
| COSM4406115 | c.94A>C | p.K32Q | Substitution - Missense | Skin
|
| COSM4649415 | c.2450C>T | p.S817F | Substitution - Missense | Large_intestine
|
| COSM4919167 | c.6A>G | p.K2K | Substitution - coding silent | Liver
|
| COSM1293014 | c.1559C>T | p.A520V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM356543 | c.1809C>A | p.N603K | Substitution - Missense | Lung
|
| COSM1194357 | c.1780C>G | p.P594A | Substitution - Missense | Lung
|
| COSM3800613 | c.1360G>A | p.E454K | Substitution - Missense | Urinary_tract
|
| COSM4473603 | c.1861C>T | p.P621S | Substitution - Missense | Skin
|
| COSM1315530 | c.1384G>T | p.E462* | Substitution - Nonsense | Urinary_tract
|
| COSM457539 | c.660T>C | p.C220C | Substitution - coding silent | Breast
|
| COSM3940002 | c.1836T>G | p.S612S | Substitution - coding silent | Oesophagus
|
| COSM5718270 | c.869G>A | p.C290Y | Substitution - Missense | Skin
|
| COSM1121687 | c.121G>T | p.A41S | Substitution - Missense | Endometrium
|
| COSM4713951 | c.1561G>A | p.G521R | Substitution - Missense | Large_intestine
|
| COSM3561783 | c.1194G>A | p.Q398Q | Substitution - coding silent | Oesophagus
|
| COSM5519866 | c.529C>T | p.R177C | Substitution - Missense | Biliary_tract
|
| COSM3561783 | c.1194G>A | p.Q398Q | Substitution - coding silent | Skin
|
| COSM4002303 | c.246A>C | p.V82V | Substitution - coding silent | Haematopoietic_and_lymphoid_tissue
|
| COSM757218 | c.1574C>A | p.T525K | Substitution - Missense | Lung
|
| COSM4109602 | c.2468G>T | p.R823M | Substitution - Missense | Stomach
|
| COSM4156853 | c.472A>C | p.M158L | Substitution - Missense | Thyroid
|
| COSM3561782 | c.552C>T | p.N184N | Substitution - coding silent | Skin
|
| COSM1121692 | c.925G>T | p.A309S | Substitution - Missense | Endometrium
|
| COSM1121696 | c.1234C>T | p.R412* | Substitution - Nonsense | Endometrium
|
| COSM4002303 | c.246A>C | p.V82V | Substitution - coding silent | Stomach
|
| COSM1121692 | c.925G>T | p.A309S | Substitution - Missense | Endometrium
|
| COSM4951388 | c.2068C>T | p.Q690* | Substitution - Nonsense | Liver
|
| COSM4149057 | c.1138C>A | p.H380N | Substitution - Missense | Ovary
|
| COSM1497267 | c.1233A>G | p.V411V | Substitution - coding silent | Kidney
|
| COSM1644036 | c.1294_1295insA | p.N432fs*10 | Insertion - Frameshift | Stomach
|
| COSM5757546 | c.650T>C | p.M217T | Substitution - Missense | Large_intestine
|
| COSM5898682 | c.285G>A | p.R95R | Substitution - coding silent | Skin
|
| COSM1121701 | c.2228A>C | p.K743T | Substitution - Missense | Endometrium
|
| COSM3561781 | c.139G>A | p.D47N | Substitution - Missense | Skin
|
| COSM4713949 | c.323G>A | p.R108Q | Substitution - Missense | Large_intestine
|
| COSM757216 | c.2452C>A | p.H818N | Substitution - Missense | Lung
|
| COSM1121688 | c.158A>C | p.K53T | Substitution - Missense | Endometrium
|
| COSM5892501 | c.1342C>T | p.P448S | Substitution - Missense | Skin
|
| COSM1682573 | c.1435C>A | p.L479M | Substitution - Missense | Large_intestine
|
| COSM3561784 | c.1353G>A | p.E451E | Substitution - coding silent | Skin
|
| COSM294698 | c.2106C>A | p.G702G | Substitution - coding silent | Large_intestine
|
| COSM5617558 | c.1011C>A | p.T337T | Substitution - coding silent | Lung
|
| COSM3844752 | c.488T>G | p.V163G | Substitution - Missense | Breast
|
| COSM488387 | c.1612A>T | p.R538* | Substitution - Nonsense | Kidney
|
| COSM1682573 | c.1435C>A | p.L479M | Substitution - Missense | Large_intestine
|
| COSM3844754 | c.1479G>T | p.E493D | Substitution - Missense | Breast
|
| COSM4951388 | c.2068C>T | p.Q690* | Substitution - Nonsense | Liver
|
| COSM1121699 | c.2030T>C | p.V677A | Substitution - Missense | Endometrium
|
| COSM1756581 | c.1255G>A | p.E419K | Substitution - Missense | Urinary_tract
|
| COSM125335 | c.2021C>G | p.S674C | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM4109600 | c.1375C>T | p.Q459* | Substitution - Nonsense | Stomach
|
| COSM3414290 | c.133C>T | p.R45W | Substitution - Missense | Oesophagus
|
| COSM5742253 | c.2061C>A | p.F687L | Substitution - Missense | Small_intestine
|
| COSM257718 | c.1613G>T | p.R538I | Substitution - Missense | Large_intestine
|
| COSM4109601 | c.1402C>T | p.R468* | Substitution - Nonsense | Stomach
|
| COSM4713952 | c.1718T>C | p.V573A | Substitution - Missense | Large_intestine
|
| COSM4569266 | c.1634T>A | p.M545K | Substitution - Missense | Skin
|
| COSM4109599 | c.322C>T | p.R108* | Substitution - Nonsense | Stomach
|
| COSM125334 | c.1775C>T | p.S592L | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM1121695 | c.1158C>T | p.S386S | Substitution - coding silent | Endometrium
|
| COSM125334 | c.1775C>T | p.S592L | Substitution - Missense | Endometrium
|
| COSM5412705 | c.1515G>A | p.Q505Q | Substitution - coding silent | Skin
|
| COSM1121686 | c.43G>A | p.E15K | Substitution - Missense | Endometrium
|
| COSM1468088 | c.919C>T | p.R307W | Substitution - Missense | Large_intestine
|
| COSM1728167 | c.2131C>T | p.H711Y | Substitution - Missense | Liver
|
| COSM4754375 | c.2203A>C | p.K735Q | Substitution - Missense | Stomach
|
| COSM1315529 | c.1372G>C | p.V458L | Substitution - Missense | Urinary_tract
|
| COSM1756582 | c.2408C>G | p.S803C | Substitution - Missense | Urinary_tract
|
| COSM5963978 | c.2389G>A | p.E797K | Substitution - Missense | Breast
|
| COSM1190533 | c.1066G>A | p.V356M | Substitution - Missense | Lung
|
| COSM1121693 | c.956C>T | p.T319M | Substitution - Missense | Cervix
|
| COSM1121694 | c.1063G>A | p.E355K | Substitution - Missense | Pancreas
|
| COSM1121686 | c.43G>A | p.E15K | Substitution - Missense | Thyroid
|
| COSM3783607 | c.673G>A | p.V225I | Substitution - Missense | Stomach
|
| COSM4850710 | c.2296G>A | p.G766S | Substitution - Missense | Cervix
|
| COSM1121690 | c.535T>C | p.C179R | Substitution - Missense | Endometrium
|
| COSM1490974 | c.974G>A | p.C325Y | Substitution - Missense | Breast
|
| COSM757220 | c.822G>T | p.W274C | Substitution - Missense | Lung
|
| COSM1220350 | c.1271C>T | p.T424M | Substitution - Missense | Large_intestine
|
| COSM1121693 | c.956C>T | p.T319M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5796587 | c.394G>T | p.A132S | Substitution - Missense | Breast
|
| COSM1468089 | c.1100delA | p.N368fs*58 | Deletion - Frameshift | Large_intestine
|
| COSM163441 | c.862A>G | p.I288V | Substitution - Missense | Breast
|
| COSM1625908 | c.1126G>A | p.E376K | Substitution - Missense | Liver
|
| COSM3783607 | c.673G>A | p.V225I | Substitution - Missense | Prostate
|
| COSM1220349 | c.620G>A | p.R207Q | Substitution - Missense | Large_intestine
|
| COSM4713948 | c.49C>A | p.P17T | Substitution - Missense | Large_intestine
|
| COSM2965600 | c.951G>A | p.L317L | Substitution - coding silent | Thyroid
|
| COSM4379352 | c.747G>T | p.L249L | Substitution - coding silent | Large_intestine
|
| COSM1121693 | c.956C>T | p.T319M | Substitution - Missense | Endometrium
|
| COSM4471601 | c.172C>G | p.H58D | Substitution - Missense | Skin
|
| COSM4908749 | c.601G>A | p.V201M | Substitution - Missense | Central_nervous_system
|
| COSM1138153 | c.619C>T | p.R207W | Substitution - Missense | Kidney
|
| COSM3406388 | c.2286G>C | p.Q762H | Substitution - Missense | Central_nervous_system
|
| COSM3561786 | c.1998C>T | p.S666S | Substitution - coding silent | Skin
|
| COSM219574 | c.2094G>T | p.L698F | Substitution - Missense | Breast
|
| COSM5898683 | c.1178C>T | p.T393I | Substitution - Missense | Skin
|
| COSM4795767 | c.1579G>A | p.A527T | Substitution - Missense | Liver
|
| COSM3414291 | c.2370G>T | p.R790S | Substitution - Missense | Oesophagus
|