Browse KAT2A in pancancer
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00583 Acetyltransferase (GNAT) family PF00439 Bromodomain PF06466 PCAF (P300/CBP-associated factor) N-terminal domain |
||||||||||
| Function |
Functions as a histone acetyltransferase (HAT) to promote transcriptional activation. Acetylation of histones gives a specific tag for epigenetic transcription activation. Has significant histone acetyltransferase activity with core histones, but not with nucleosome core particles. Also acetylates non-histone proteins, such as CEBPB (PubMed:17301242). Component of the ATAC complex, a complex with histone acetyltransferase activity on histones H3 and H4. In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat's transactivating activity and may help inducing chromatin remodeling of proviral genes. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000422 mitophagy GO:0001701 in utero embryonic development GO:0001756 somitogenesis GO:0001838 embryonic epithelial tube formation GO:0001841 neural tube formation GO:0001843 neural tube closure GO:0003002 regionalization GO:0005996 monosaccharide metabolic process GO:0006006 glucose metabolic process GO:0006094 gluconeogenesis GO:0006109 regulation of carbohydrate metabolic process GO:0006111 regulation of gluconeogenesis GO:0006338 chromatin remodeling GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006626 protein targeting to mitochondrion GO:0006839 mitochondrial transport GO:0006914 autophagy GO:0007389 pattern specification process GO:0007507 heart development GO:0009952 anterior/posterior pattern specification GO:0009991 response to extracellular stimulus GO:0010506 regulation of autophagy GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010676 positive regulation of cellular carbohydrate metabolic process GO:0010821 regulation of mitochondrion organization GO:0010822 positive regulation of mitochondrion organization GO:0010906 regulation of glucose metabolic process GO:0010907 positive regulation of glucose metabolic process GO:0014020 primary neural tube formation GO:0014706 striated muscle tissue development GO:0016051 carbohydrate biosynthetic process GO:0016331 morphogenesis of embryonic epithelium GO:0016570 histone modification GO:0016573 histone acetylation GO:0016578 histone deubiquitination GO:0016579 protein deubiquitination GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019318 hexose metabolic process GO:0019319 hexose biosynthetic process GO:0021537 telencephalon development GO:0021915 neural tube development GO:0022037 metencephalon development GO:0030900 forebrain development GO:0030901 midbrain development GO:0030902 hindbrain development GO:0031056 regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031346 positive regulation of cell projection organization GO:0031647 regulation of protein stability GO:0031667 response to nutrient levels GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0034612 response to tumor necrosis factor GO:0035051 cardiocyte differentiation GO:0035065 regulation of histone acetylation GO:0035066 positive regulation of histone acetylation GO:0035148 tube formation GO:0035239 tube morphogenesis GO:0035264 multicellular organism growth GO:0035282 segmentation GO:0035947 regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter GO:0035948 positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter GO:0040029 regulation of gene expression, epigenetic GO:0042692 muscle cell differentiation GO:0043255 regulation of carbohydrate biosynthetic process GO:0043388 positive regulation of DNA binding GO:0043543 protein acylation GO:0043966 histone H3 acetylation GO:0043967 histone H4 acetylation GO:0043983 histone H4-K12 acetylation GO:0044154 histone H3-K14 acetylation GO:0044262 cellular carbohydrate metabolic process GO:0044283 small molecule biosynthetic process GO:0044723 single-organism carbohydrate metabolic process GO:0045722 positive regulation of gluconeogenesis GO:0045815 positive regulation of gene expression, epigenetic GO:0045913 positive regulation of carbohydrate metabolic process GO:0046364 monosaccharide biosynthetic process GO:0048311 mitochondrion distribution GO:0048312 intracellular distribution of mitochondria GO:0048738 cardiac muscle tissue development GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051101 regulation of DNA binding GO:0051146 striated muscle cell differentiation GO:0051222 positive regulation of protein transport GO:0051640 organelle localization GO:0051646 mitochondrion localization GO:0055007 cardiac muscle cell differentiation GO:0060537 muscle tissue development GO:0060562 epithelial tube morphogenesis GO:0060606 tube closure GO:0061053 somite development GO:0061726 mitochondrion disassembly GO:0070585 protein localization to mitochondrion GO:0070646 protein modification by small protein removal GO:0071356 cellular response to tumor necrosis factor GO:0071929 alpha-tubulin acetylation GO:0072175 epithelial tube formation GO:0072655 establishment of protein localization to mitochondrion GO:0090316 positive regulation of intracellular protein transport GO:1901983 regulation of protein acetylation GO:1901985 positive regulation of protein acetylation GO:1902275 regulation of chromatin organization GO:1903008 organelle disassembly GO:1903146 regulation of mitophagy GO:1903214 regulation of protein targeting to mitochondrion GO:1903533 regulation of protein targeting GO:1903747 regulation of establishment of protein localization to mitochondrion GO:1903749 positive regulation of establishment of protein localization to mitochondrion GO:1903829 positive regulation of cellular protein localization GO:1903955 positive regulation of protein targeting to mitochondrion GO:1904951 positive regulation of establishment of protein localization GO:1905269 positive regulation of chromatin organization GO:1990089 response to nerve growth factor GO:1990090 cellular response to nerve growth factor stimulus GO:2000677 regulation of transcription regulatory region DNA binding GO:2000679 positive regulation of transcription regulatory region DNA binding GO:2000756 regulation of peptidyl-lysine acetylation GO:2000758 positive regulation of peptidyl-lysine acetylation |
| Molecular Function |
GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0010484 H3 histone acetyltransferase activity GO:0010485 H4 histone acetyltransferase activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0019902 phosphatase binding GO:0019903 protein phosphatase binding GO:0034212 peptide N-acetyltransferase activity GO:0042826 histone deacetylase binding GO:0043997 histone acetyltransferase activity (H4-K12 specific) GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000428 DNA-directed RNA polymerase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0005667 transcription factor complex GO:0005671 Ada2/Gcn5/Ada3 transcription activator complex GO:0005819 spindle GO:0016591 DNA-directed RNA polymerase II, holoenzyme GO:0030880 RNA polymerase complex GO:0030914 STAGA complex GO:0031248 protein acetyltransferase complex GO:0033276 transcription factor TFTC complex GO:0044454 nuclear chromosome part GO:0044798 nuclear transcription factor complex GO:0055029 nuclear DNA-directed RNA polymerase complex GO:0061695 transferase complex, transferring phosphorus-containing groups GO:0070461 SAGA-type complex GO:0072686 mitotic spindle GO:0090575 RNA polymerase II transcription factor complex GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG |
hsa04330 Notch signaling pathway hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-5250924: B-WICH complex positively regulates rRNA expression R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-5688426: Deubiquitination R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-392499: Metabolism of proteins R-HSA-157052: NICD traffics to nucleus R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-350054: Notch-HLH transcription pathway R-HSA-5250913: Positive epigenetic regulation of rRNA expression R-HSA-597592: Post-translational protein modification R-HSA-1912422: Pre-NOTCH Expression and Processing R-HSA-1912408: Pre-NOTCH Transcription and Translation R-HSA-73854: RNA Polymerase I Promoter Clearance R-HSA-73864: RNA Polymerase I Transcription R-HSA-73762: RNA Polymerase I Transcription Initiation R-HSA-504046: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer R-HSA-5689880: Ub-specific processing proteases |
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for KAT2A. |
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|
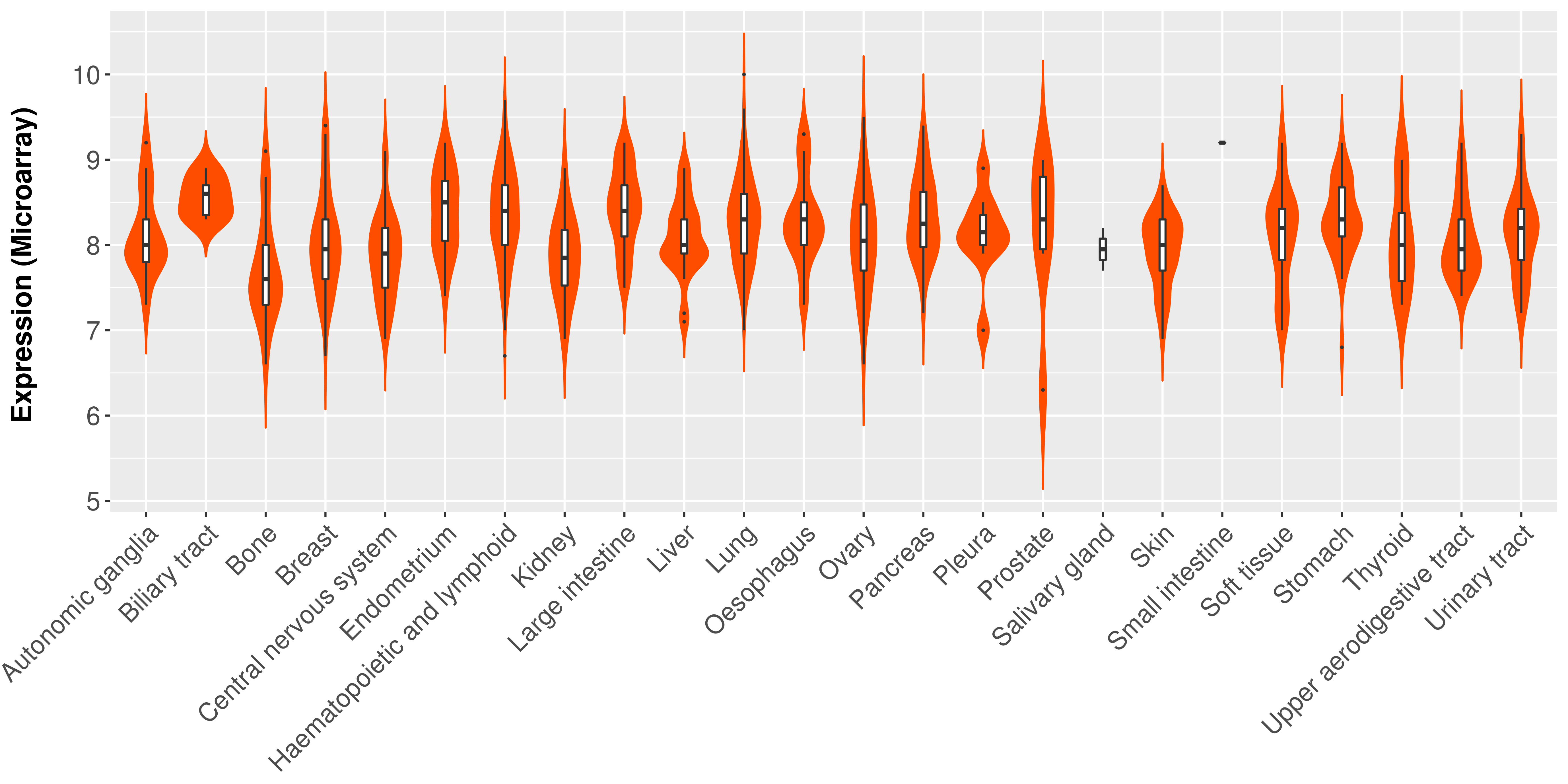
|
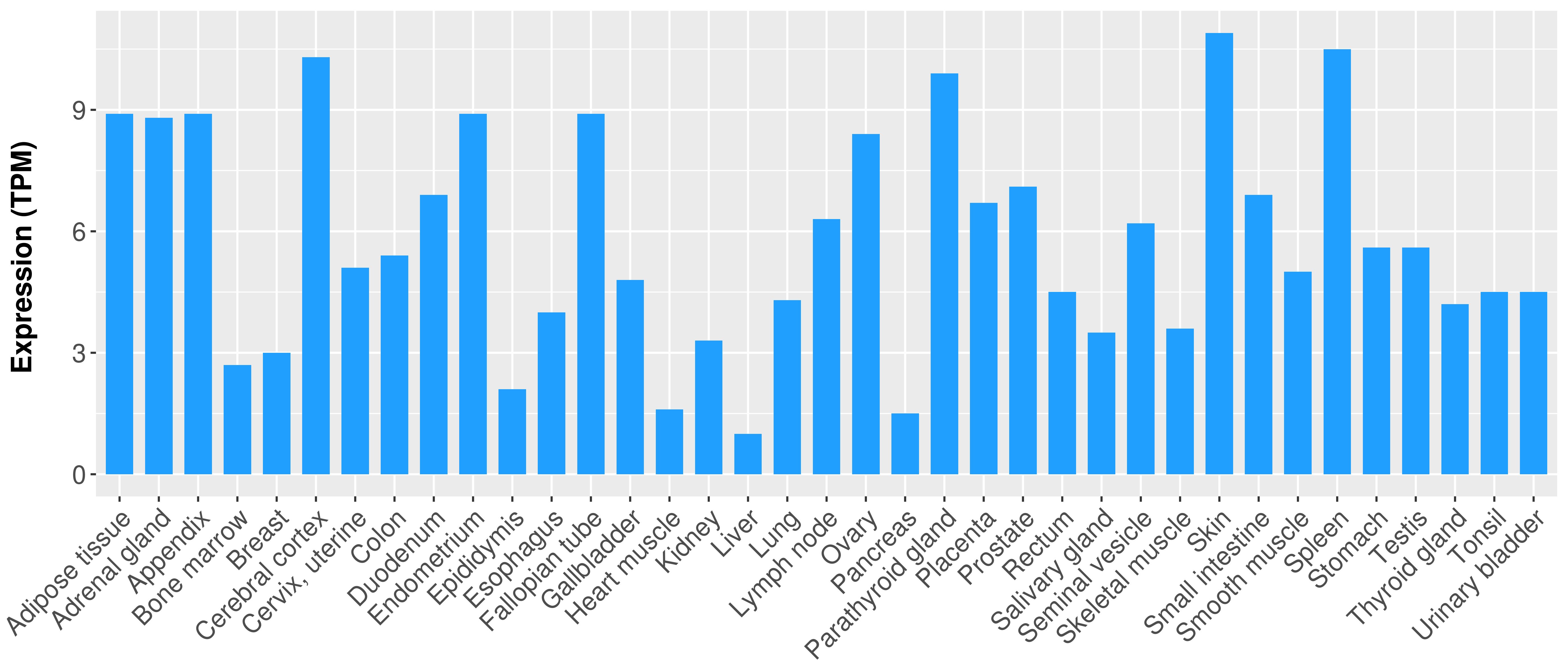
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
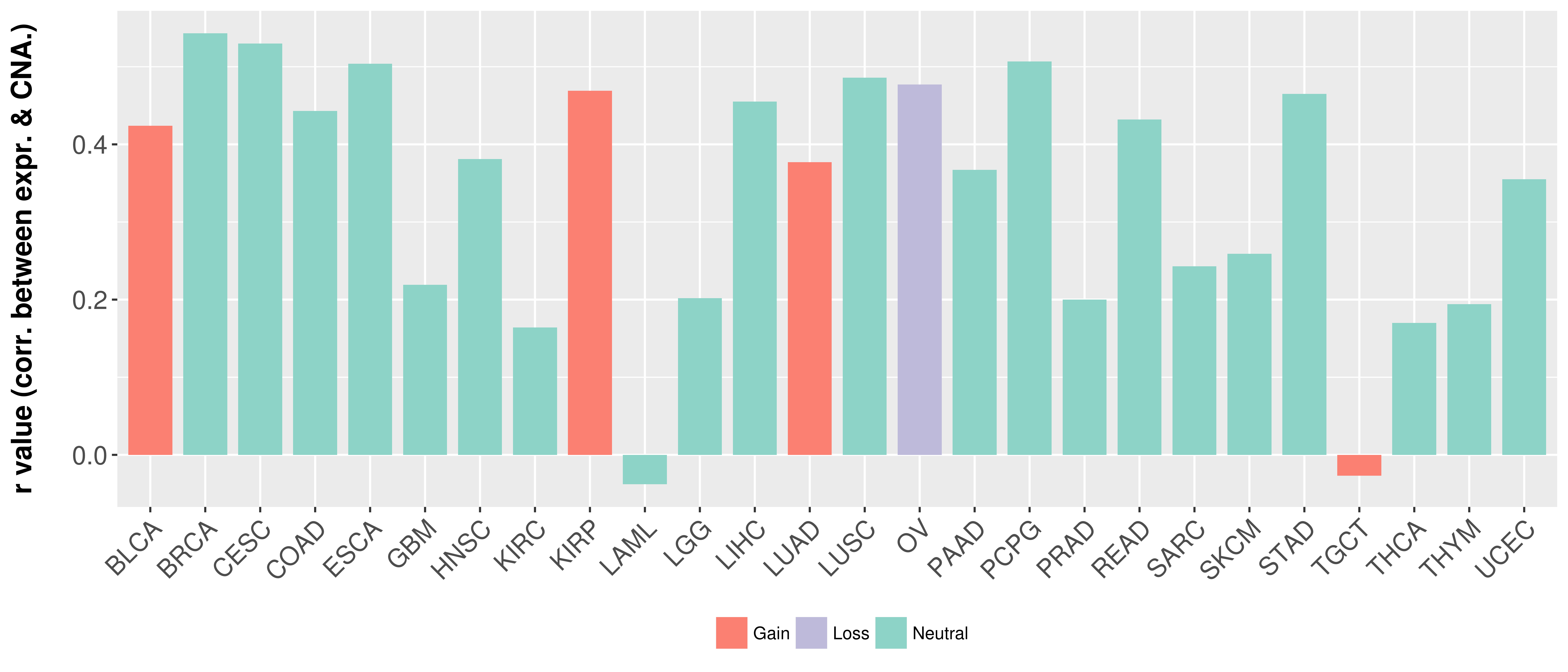
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
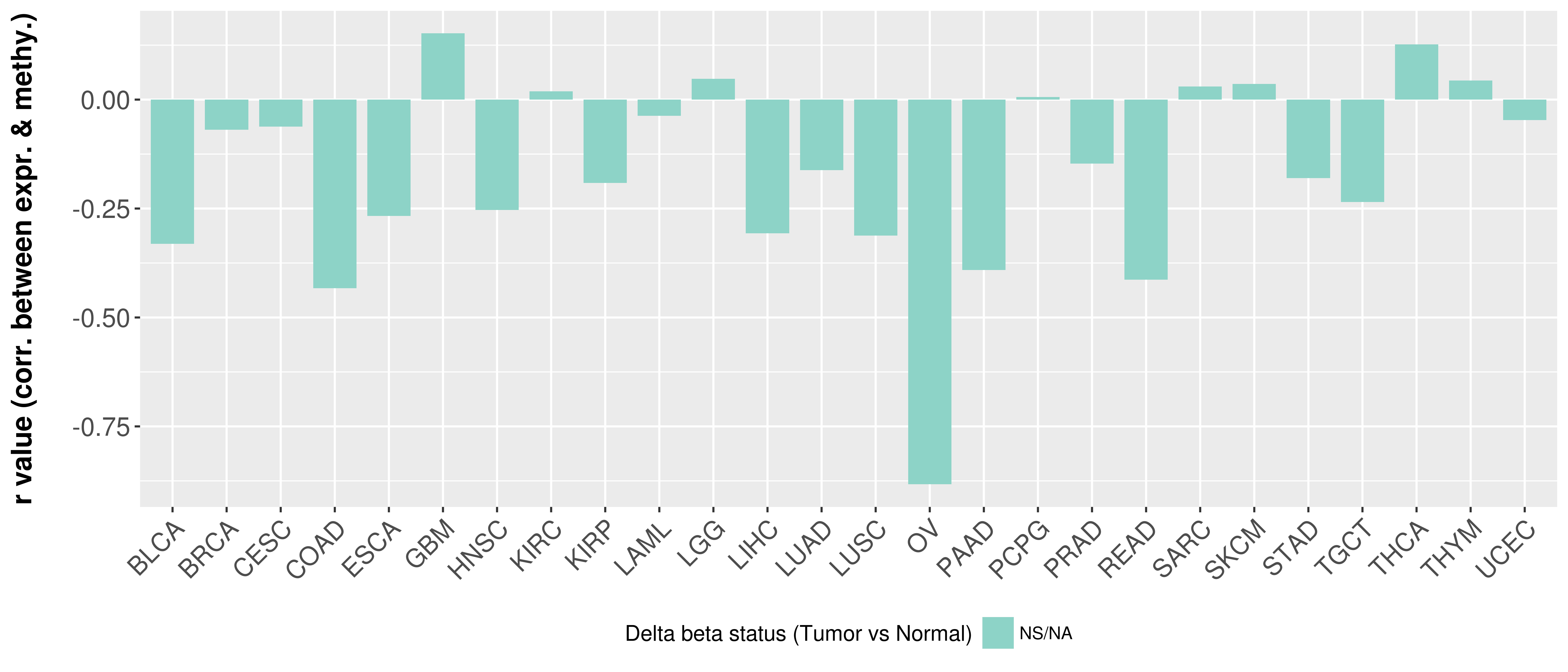
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
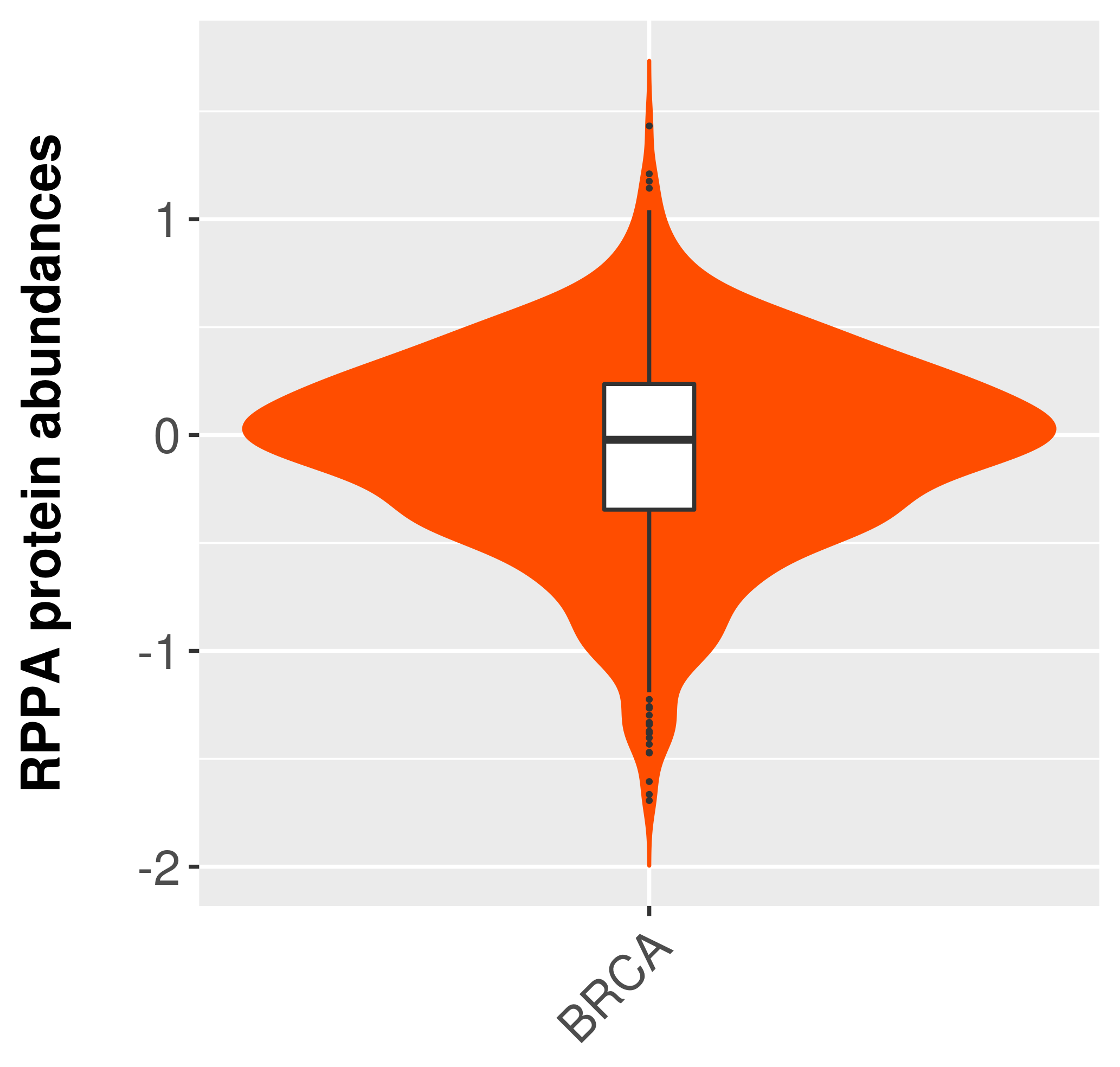
|
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
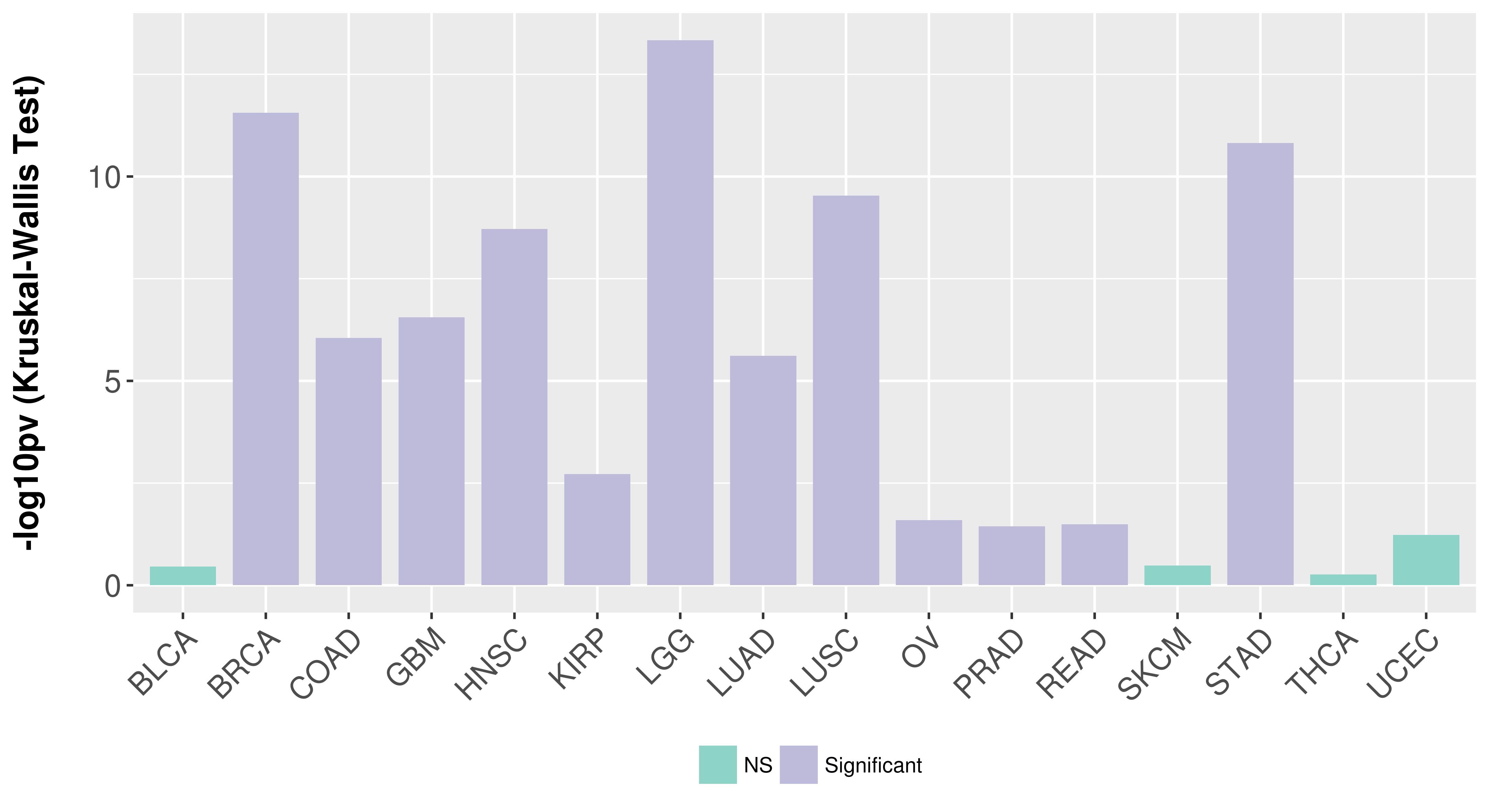
Association between expresson and subtype.
|
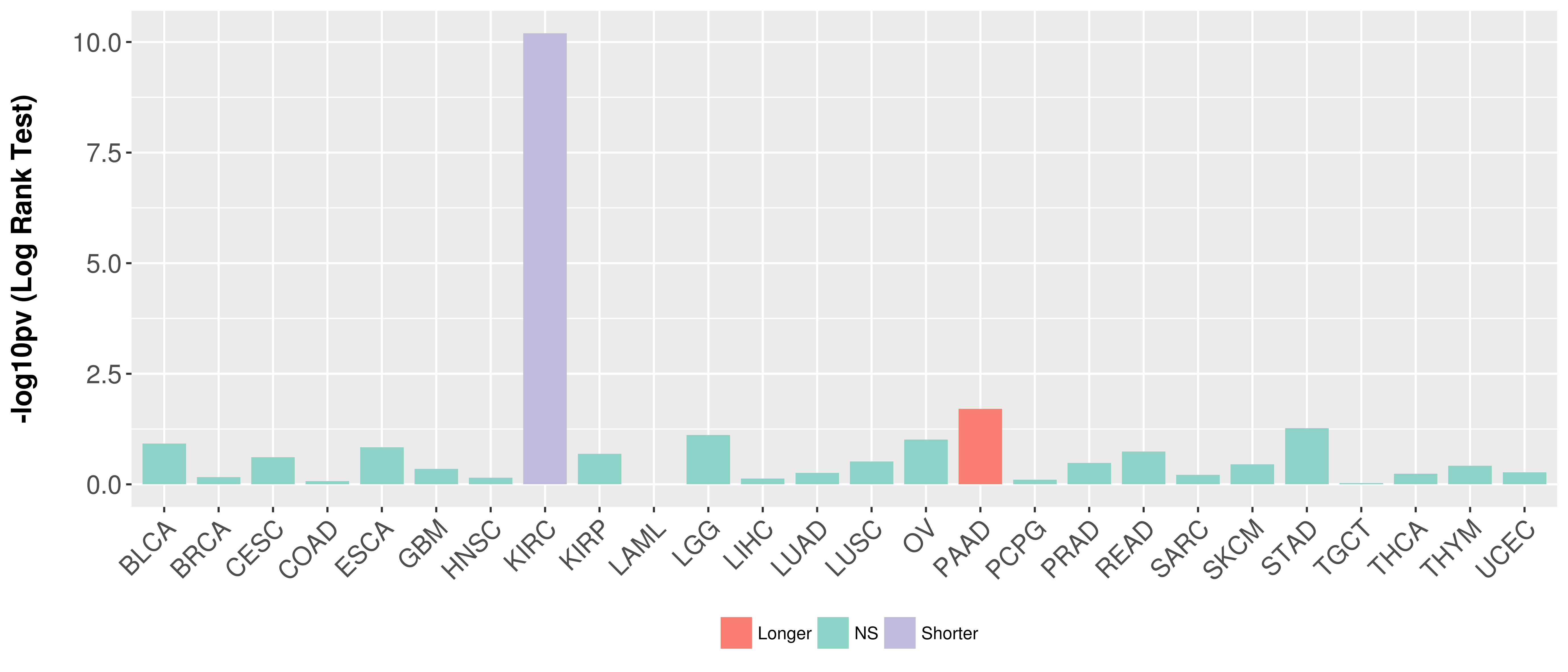
Overall survival analysis based on expression.
|

Association between expresson and stage.
|

Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
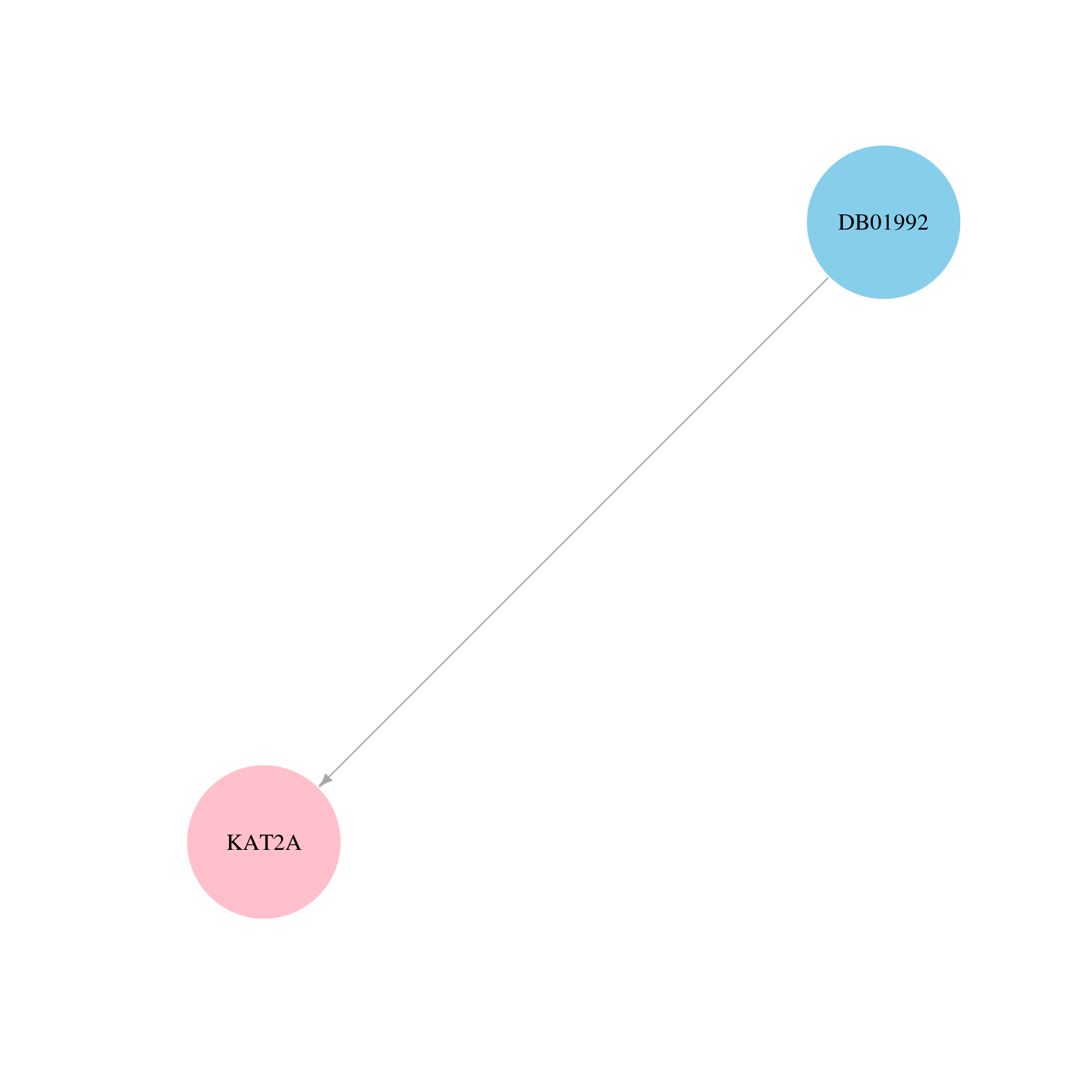
|
| Summary | |
|---|---|
| Symbol | KAT2A |
| Name | lysine acetyltransferase 2A |
| Aliases | PCAF-b; GCN5L2; GCN5 general control of amino-acid synthesis 5-like 2 (yeast); hGCN5; GCN5 (general control ...... |
| Location | 17q21.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for KAT2A. |
