Browse KDM1A in pancancer
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01593 Flavin containing amine oxidoreductase PF04433 SWIRM domain |
||||||||||
| Function |
Histone demethylase that demethylates both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me. May play a role in the repression of neuronal genes. Alone, it is unable to demethylate H3K4me on nucleosomes and requires the presence of RCOR1/CoREST to achieve such activity. Also acts as a coactivator of androgen receptor (ANDR)-dependent transcription, by being recruited to ANDR target genes and mediating demethylation of H3K9me, a specific tag for epigenetic transcriptional repression. The presence of PRKCB in ANDR-containing complexes, which mediates phosphorylation of 'Thr-6' of histone H3 (H3T6ph), a specific tag that prevents demethylation H3K4me, prevents H3K4me demethylase activity of KDM1A. Demethylates di-methylated 'Lys-370' of p53/TP53 which prevents interaction of p53/TP53 with TP53BP1 and represses p53/TP53-mediated transcriptional activation. Demethylates and stabilizes the DNA methylase DNMT1. Required for gastrulation during embryogenesis. Component of a RCOR/GFI/KDM1A/HDAC complex that suppresses, via histone deacetylase (HDAC) recruitment, a number of genes implicated in multilineage blood cell development. Effector of SNAI1-mediated transcription repression of E-cadherin/CDH1, CDN7 and KRT8. Required for the maintenance of the silenced state of the SNAI1 target genes E-cadherin/CDH1 and CDN7. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000018 regulation of DNA recombination GO:0000375 RNA splicing, via transesterification reactions GO:0000377 RNA splicing, via transesterification reactions with bulged adenosine as nucleophile GO:0000380 alternative mRNA splicing, via spliceosome GO:0000398 mRNA splicing, via spliceosome GO:0000724 double-strand break repair via homologous recombination GO:0000725 recombinational repair GO:0001701 in utero embryonic development GO:0002262 myeloid cell homeostasis GO:0002521 leukocyte differentiation GO:0002573 myeloid leukocyte differentiation GO:0006144 purine nucleobase metabolic process GO:0006282 regulation of DNA repair GO:0006302 double-strand break repair GO:0006310 DNA recombination GO:0006397 mRNA processing GO:0006476 protein deacetylation GO:0006479 protein methylation GO:0006482 protein demethylation GO:0007596 blood coagulation GO:0007599 hemostasis GO:0008213 protein alkylation GO:0008214 protein dealkylation GO:0008361 regulation of cell size GO:0008380 RNA splicing GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0009112 nucleobase metabolic process GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0009636 response to toxic substance GO:0010212 response to ionizing radiation GO:0010332 response to gamma radiation GO:0010569 regulation of double-strand break repair via homologous recombination GO:0010720 positive regulation of cell development GO:0010725 regulation of primitive erythrocyte differentiation GO:0010817 regulation of hormone levels GO:0010975 regulation of neuron projection development GO:0010976 positive regulation of neuron projection development GO:0014074 response to purine-containing compound GO:0016570 histone modification GO:0016571 histone methylation GO:0016574 histone ubiquitination GO:0016575 histone deacetylation GO:0016577 histone demethylation GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0021536 diencephalon development GO:0021537 telencephalon development GO:0021543 pallium development GO:0021700 developmental maturation GO:0021983 pituitary gland development GO:0021987 cerebral cortex development GO:0030099 myeloid cell differentiation GO:0030218 erythrocyte differentiation GO:0030219 megakaryocyte differentiation GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0030851 granulocyte differentiation GO:0030900 forebrain development GO:0031056 regulation of histone modification GO:0031057 negative regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031060 regulation of histone methylation GO:0031061 negative regulation of histone methylation GO:0031346 positive regulation of cell projection organization GO:0031396 regulation of protein ubiquitination GO:0031398 positive regulation of protein ubiquitination GO:0032091 negative regulation of protein binding GO:0032259 methylation GO:0032350 regulation of hormone metabolic process GO:0032352 positive regulation of hormone metabolic process GO:0032535 regulation of cellular component size GO:0032844 regulation of homeostatic process GO:0032846 positive regulation of homeostatic process GO:0033169 histone H3-K9 demethylation GO:0033182 regulation of histone ubiquitination GO:0033184 positive regulation of histone ubiquitination GO:0034101 erythrocyte homeostasis GO:0034644 cellular response to UV GO:0034720 histone H3-K4 demethylation GO:0034968 histone lysine methylation GO:0035162 embryonic hemopoiesis GO:0035270 endocrine system development GO:0035561 regulation of chromatin binding GO:0035563 positive regulation of chromatin binding GO:0035601 protein deacylation GO:0042445 hormone metabolic process GO:0042446 hormone biosynthetic process GO:0042551 neuron maturation GO:0042692 muscle cell differentiation GO:0042770 signal transduction in response to DNA damage GO:0042771 intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:0043392 negative regulation of DNA binding GO:0043393 regulation of protein binding GO:0043414 macromolecule methylation GO:0043433 negative regulation of sequence-specific DNA binding transcription factor activity GO:0043516 regulation of DNA damage response, signal transduction by p53 class mediator GO:0043518 negative regulation of DNA damage response, signal transduction by p53 class mediator GO:0045637 regulation of myeloid cell differentiation GO:0045639 positive regulation of myeloid cell differentiation GO:0045646 regulation of erythrocyte differentiation GO:0045648 positive regulation of erythrocyte differentiation GO:0045652 regulation of megakaryocyte differentiation GO:0045654 positive regulation of megakaryocyte differentiation GO:0045666 positive regulation of neuron differentiation GO:0045793 positive regulation of cell size GO:0045995 regulation of embryonic development GO:0046098 guanine metabolic process GO:0046683 response to organophosphorus GO:0046885 regulation of hormone biosynthetic process GO:0046886 positive regulation of hormone biosynthetic process GO:0048469 cell maturation GO:0048568 embryonic organ development GO:0048732 gland development GO:0048872 homeostasis of number of cells GO:0050769 positive regulation of neurogenesis GO:0050817 coagulation GO:0050878 regulation of body fluid levels GO:0051052 regulation of DNA metabolic process GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051100 negative regulation of binding GO:0051101 regulation of DNA binding GO:0051567 histone H3-K9 methylation GO:0051568 histone H3-K4 methylation GO:0051569 regulation of histone H3-K4 methylation GO:0051570 regulation of histone H3-K9 methylation GO:0051572 negative regulation of histone H3-K4 methylation GO:0051573 negative regulation of histone H3-K9 methylation GO:0051591 response to cAMP GO:0051962 positive regulation of nervous system development GO:0055001 muscle cell development GO:0060215 primitive hemopoiesis GO:0060319 primitive erythrocyte differentiation GO:0060992 response to fungicide GO:0061351 neural precursor cell proliferation GO:0061647 histone H3-K9 modification GO:0070076 histone lysine demethylation GO:0070988 demethylation GO:0071214 cellular response to abiotic stimulus GO:0071320 cellular response to cAMP GO:0071407 cellular response to organic cyclic compound GO:0071417 cellular response to organonitrogen compound GO:0071478 cellular response to radiation GO:0071479 cellular response to ionizing radiation GO:0071480 cellular response to gamma radiation GO:0071482 cellular response to light stimulus GO:0072089 stem cell proliferation GO:0072091 regulation of stem cell proliferation GO:0072331 signal transduction by p53 class mediator GO:0072332 intrinsic apoptotic signaling pathway by p53 class mediator GO:0090066 regulation of anatomical structure size GO:0097193 intrinsic apoptotic signaling pathway GO:0098732 macromolecule deacylation GO:1901796 regulation of signal transduction by p53 class mediator GO:1901797 negative regulation of signal transduction by p53 class mediator GO:1902165 regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902166 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902229 regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902230 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902253 regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902254 negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902275 regulation of chromatin organization GO:1902532 negative regulation of intracellular signal transduction GO:1903320 regulation of protein modification by small protein conjugation or removal GO:1903322 positive regulation of protein modification by small protein conjugation or removal GO:1903706 regulation of hemopoiesis GO:1903708 positive regulation of hemopoiesis GO:1905268 negative regulation of chromatin organization GO:1905269 positive regulation of chromatin organization GO:2000177 regulation of neural precursor cell proliferation GO:2000179 positive regulation of neural precursor cell proliferation GO:2000648 positive regulation of stem cell proliferation GO:2000779 regulation of double-strand break repair GO:2001020 regulation of response to DNA damage stimulus GO:2001021 negative regulation of response to DNA damage stimulus GO:2001233 regulation of apoptotic signaling pathway GO:2001234 negative regulation of apoptotic signaling pathway GO:2001242 regulation of intrinsic apoptotic signaling pathway GO:2001243 negative regulation of intrinsic apoptotic signaling pathway |
| Molecular Function |
GO:0001085 RNA polymerase II transcription factor binding GO:0002039 p53 binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0019213 deacetylase activity GO:0030374 ligand-dependent nuclear receptor transcription coactivator activity GO:0032451 demethylase activity GO:0032452 histone demethylase activity GO:0032453 histone demethylase activity (H3-K4 specific) GO:0032454 histone demethylase activity (H3-K9 specific) GO:0033558 protein deacetylase activity GO:0034648 histone demethylase activity (H3-dimethyl-K4 specific) GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0042162 telomeric DNA binding GO:0043425 bHLH transcription factor binding GO:0043426 MRF binding GO:0048037 cofactor binding GO:0050660 flavin adenine dinucleotide binding GO:0050662 coenzyme binding GO:0050681 androgen receptor binding GO:0051427 hormone receptor binding GO:0061752 telomeric repeat-containing RNA binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0005667 transcription factor complex GO:0044454 nuclear chromosome part GO:0098687 chromosomal region GO:1990391 DNA repair complex |
| KEGG | - |
| Reactome |
R-HSA-5625886: Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-983231: Factors involved in megakaryocyte development and platelet production R-HSA-3214815: HDACs deacetylate histones R-HSA-3214842: HDMs demethylate histones R-HSA-109582: Hemostasis R-HSA-195258: RHO GTPase Effectors R-HSA-5625740: RHO GTPases activate PKNs R-HSA-162582: Signal Transduction R-HSA-194315: Signaling by Rho GTPases |
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for KDM1A. |
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
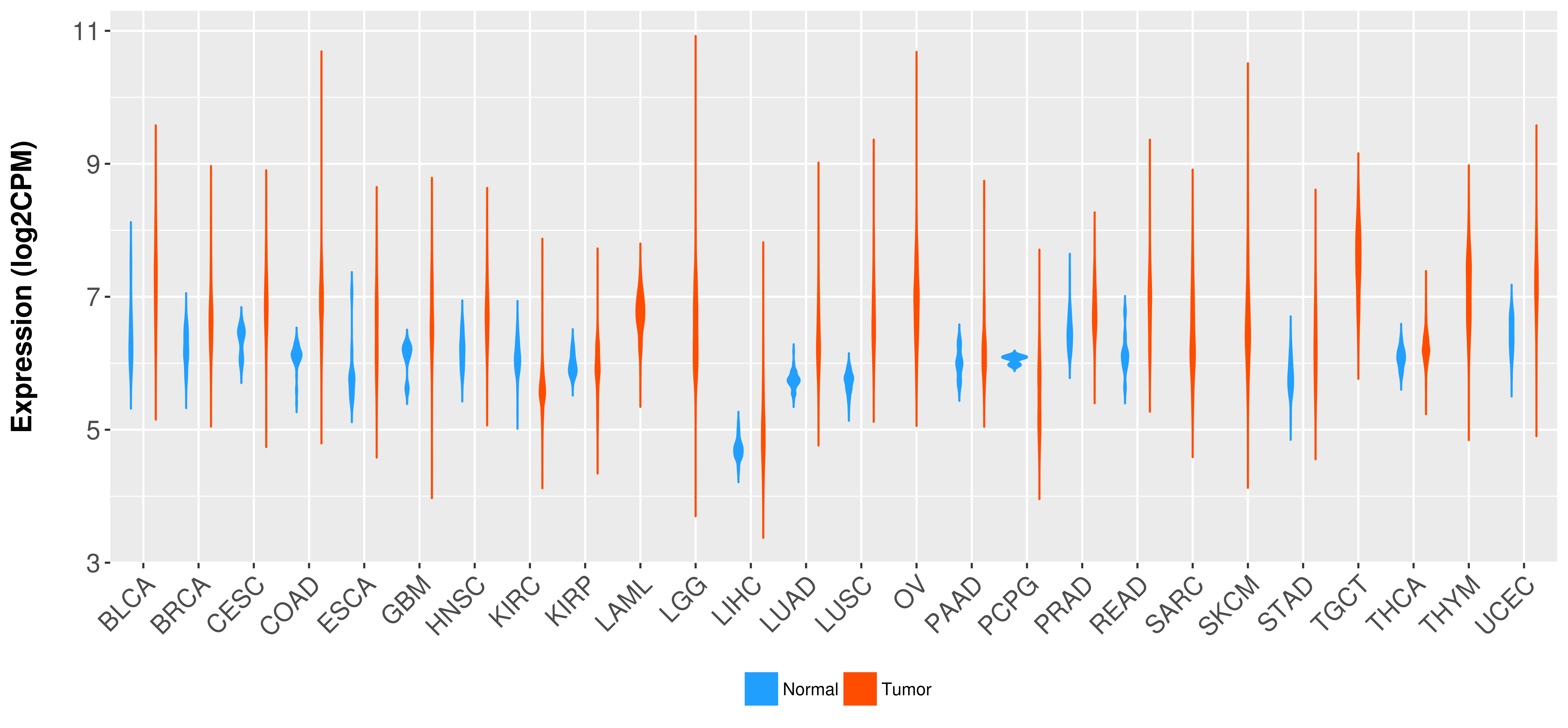
Differential expression analysis for cancers with more than 10 normal samples
|
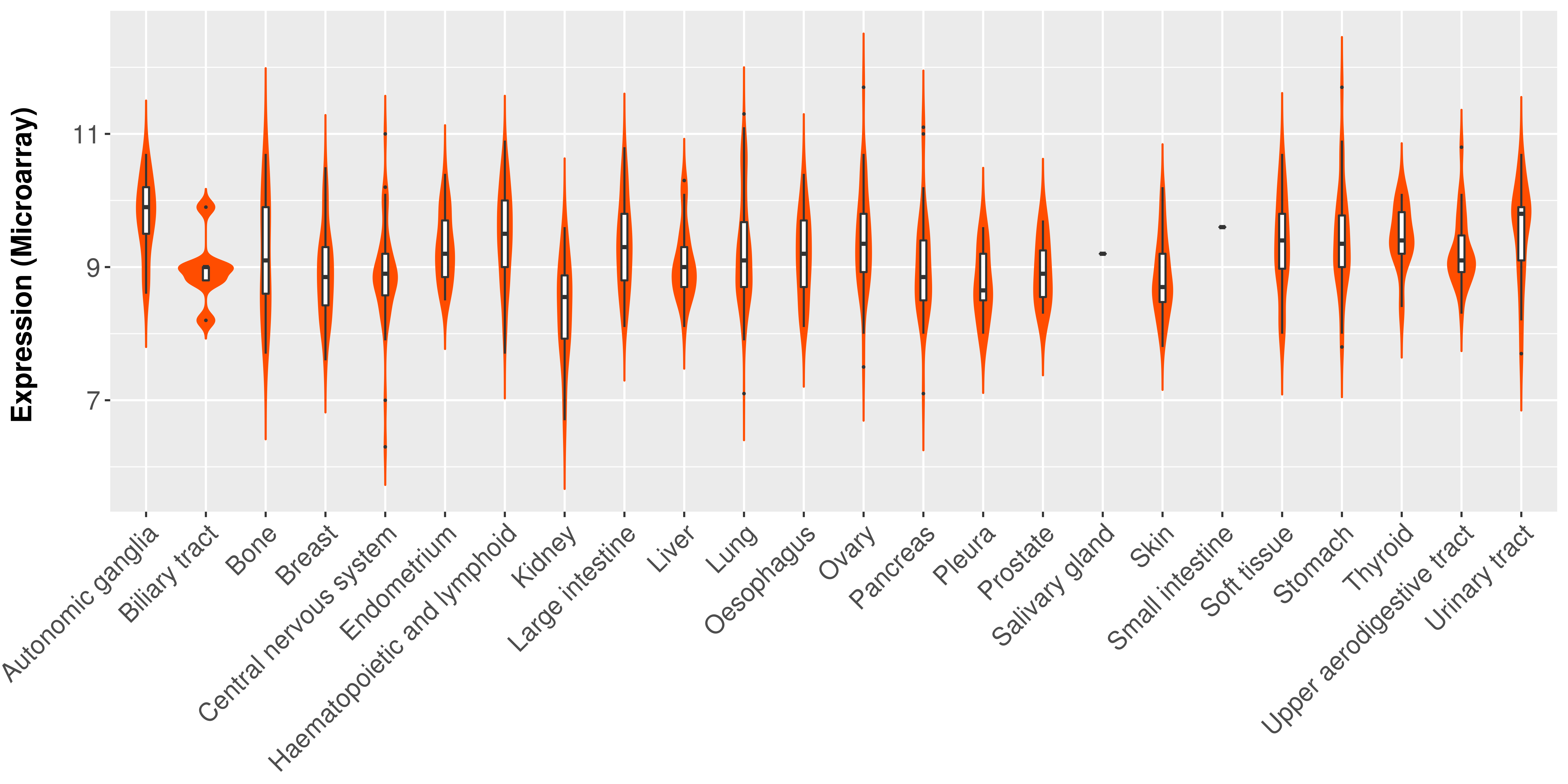
|
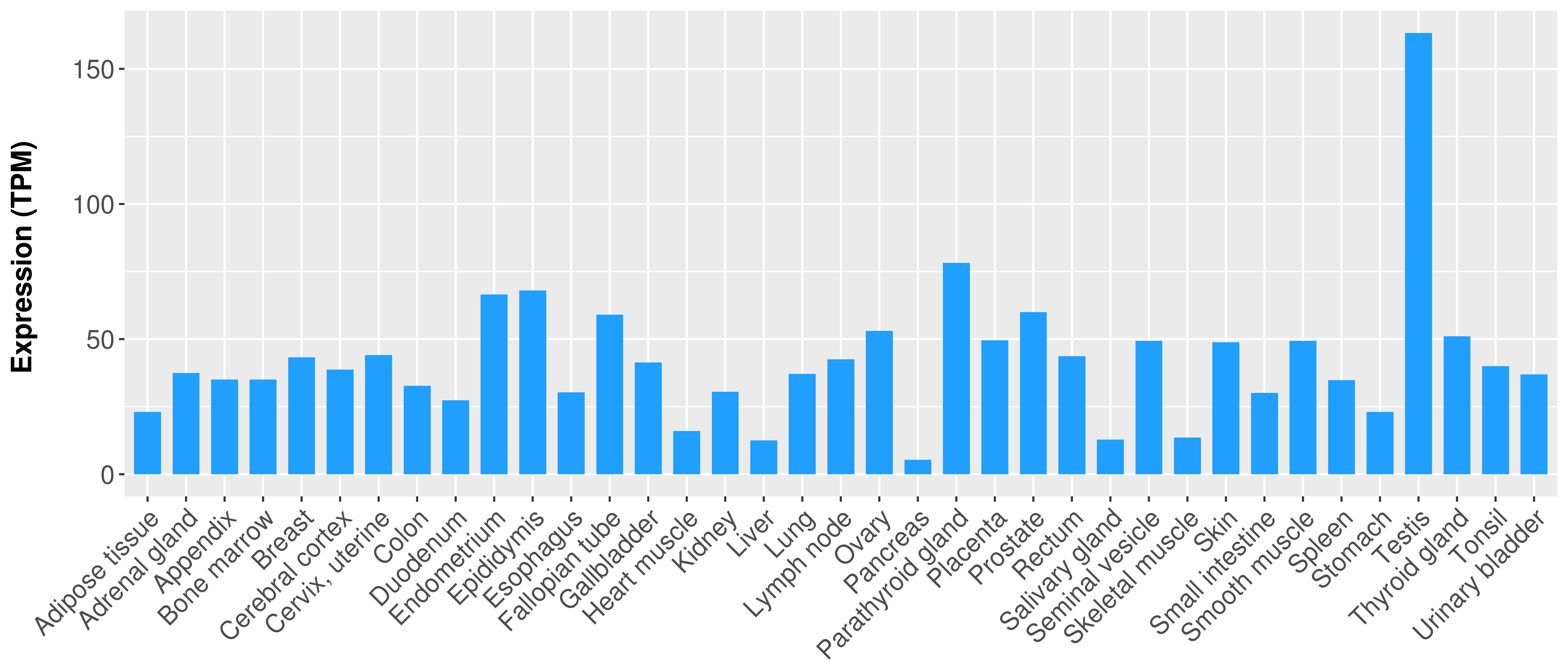
|
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
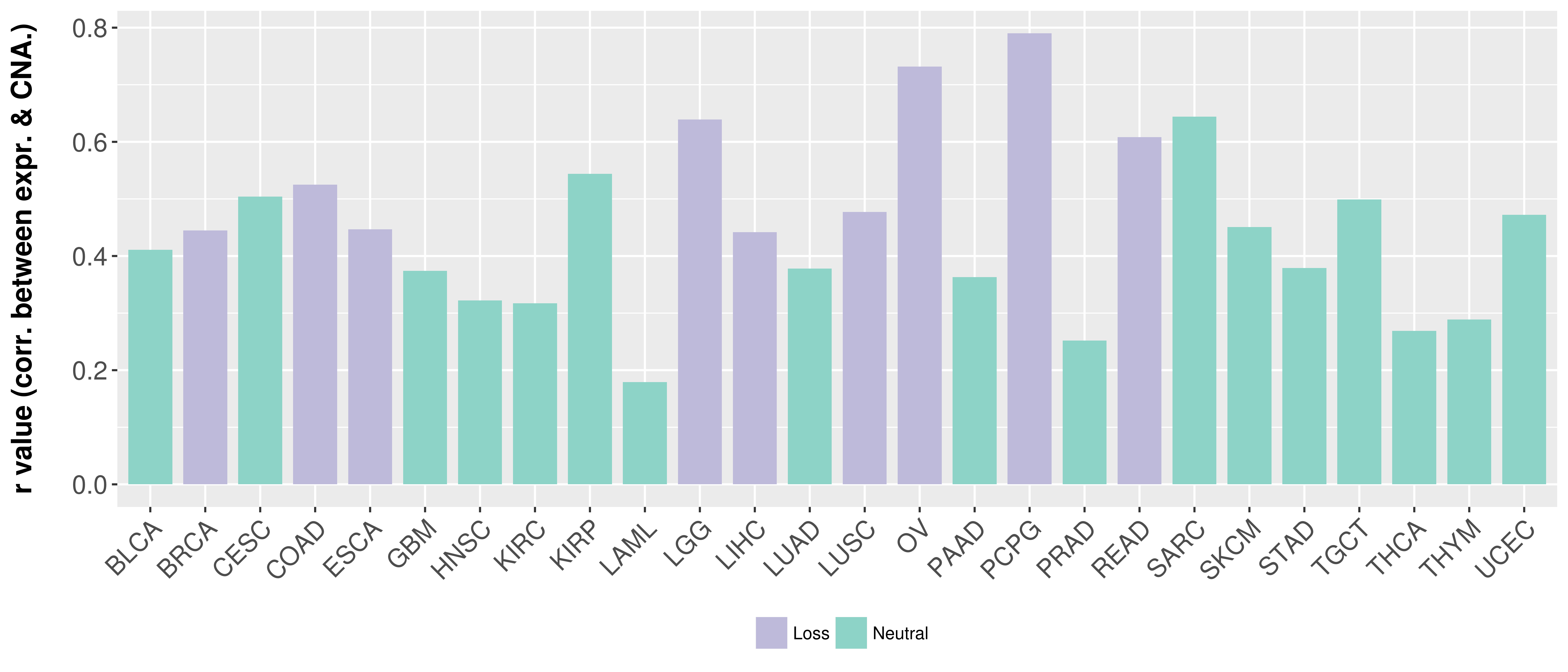
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
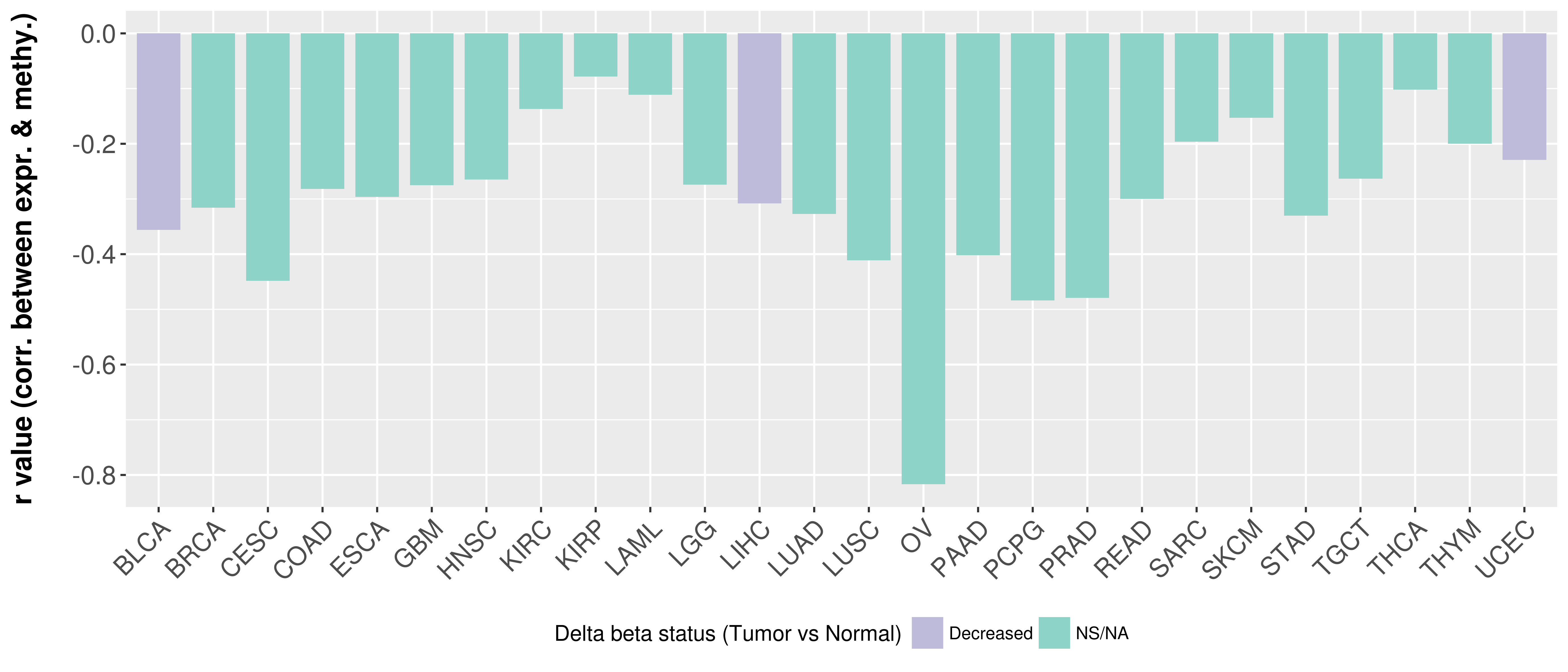
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
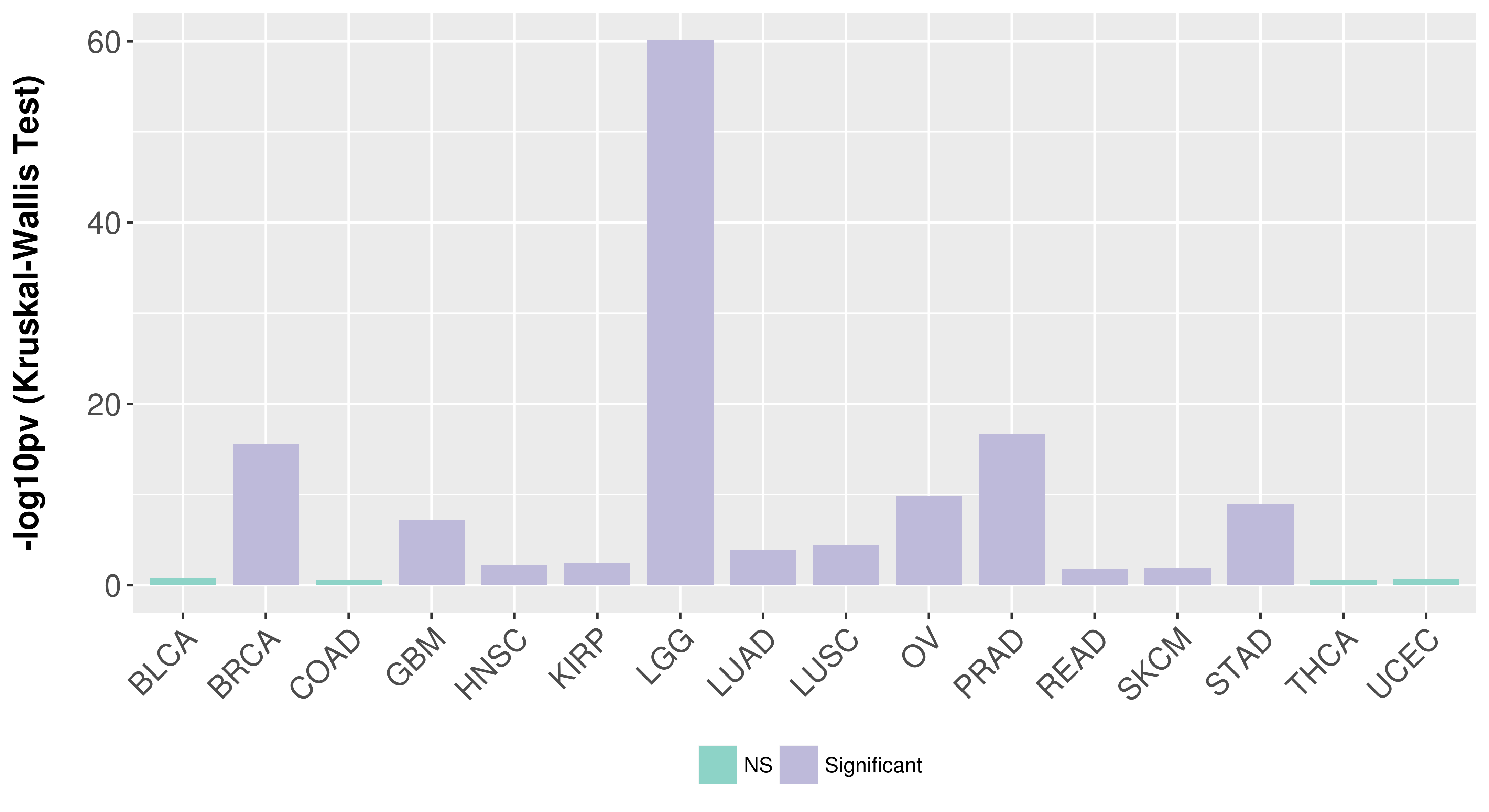
Association between expresson and subtype.
|
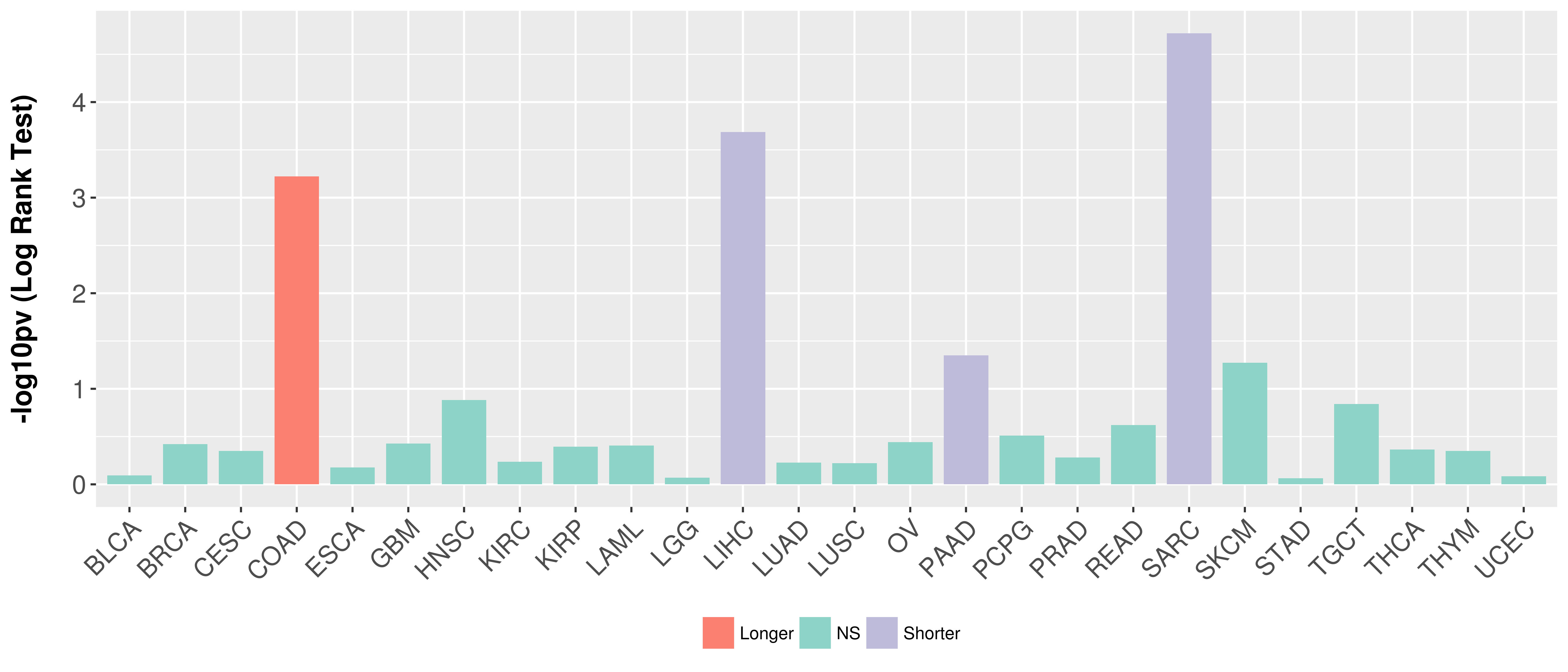
Overall survival analysis based on expression.
|
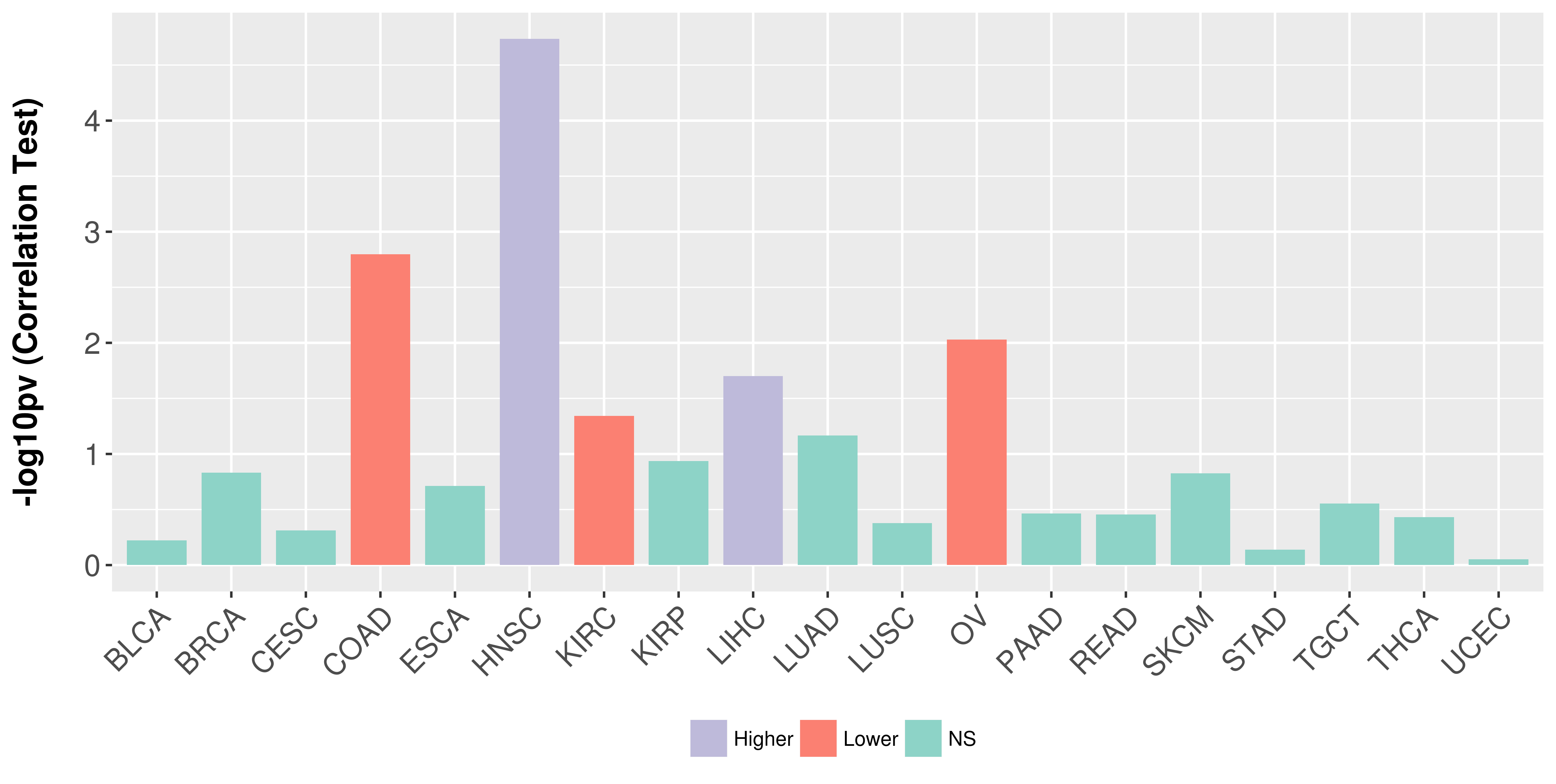
Association between expresson and stage.
|
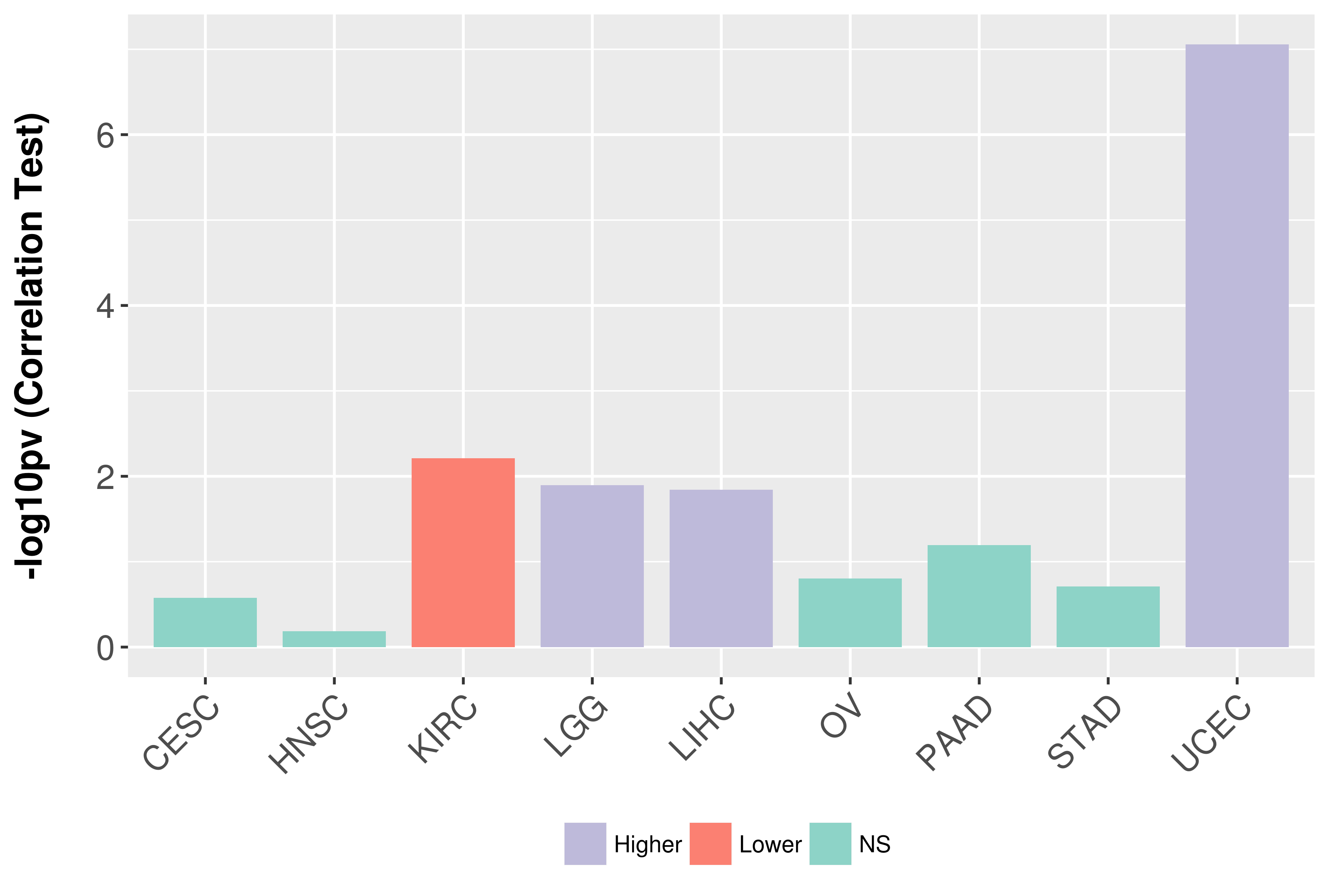
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for KDM1A. |
| Summary | |
|---|---|
| Symbol | KDM1A |
| Name | lysine demethylase 1A |
| Aliases | KIAA0601; BHC110; LSD1; AOF2; KDM1; amine oxidase (flavin containing) domain 2; lysine (K)-specific demethyl ...... |
| Location | 1p36.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|