Browse KDM5D in pancancer
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01388 ARID/BRIGHT DNA binding domain PF02373 JmjC domain PF02375 jmjN domain PF00628 PHD-finger PF08429 PLU-1-like protein PF02928 C5HC2 zinc finger |
||||||||||
| Function |
Histone demethylase that specifically demethylates 'Lys-4' of histone H3, thereby playing a central role in histone code. Does not demethylate histone H3 'Lys-9', H3 'Lys-27', H3 'Lys-36', H3 'Lys-79' or H4 'Lys-20'. Demethylates trimethylated and dimethylated but not monomethylated H3 'Lys-4'. May play a role in spermatogenesis. Involved in transcriptional repression of diverse metastasis-associated genes; in this function seems to cooperate with ZMYND8. Suppresses prostate cancer cell invasion. Regulates androgen receptor (AR) transcriptional activity by demethylating H3K4me3 active transcription marks. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0002250 adaptive immune response GO:0002443 leukocyte mediated immunity GO:0002449 lymphocyte mediated immunity GO:0002456 T cell mediated immunity GO:0002457 T cell antigen processing and presentation GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0006482 protein demethylation GO:0008214 protein dealkylation GO:0016570 histone modification GO:0016577 histone demethylation GO:0019882 antigen processing and presentation GO:0034720 histone H3-K4 demethylation GO:0070076 histone lysine demethylation GO:0070988 demethylation |
| Molecular Function |
GO:0016705 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen GO:0016706 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors GO:0032451 demethylase activity GO:0032452 histone demethylase activity GO:0032453 histone demethylase activity (H3-K4 specific) GO:0051213 dioxygenase activity |
| Cellular Component | - |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214842: HDMs demethylate histones |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for KDM5D. |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
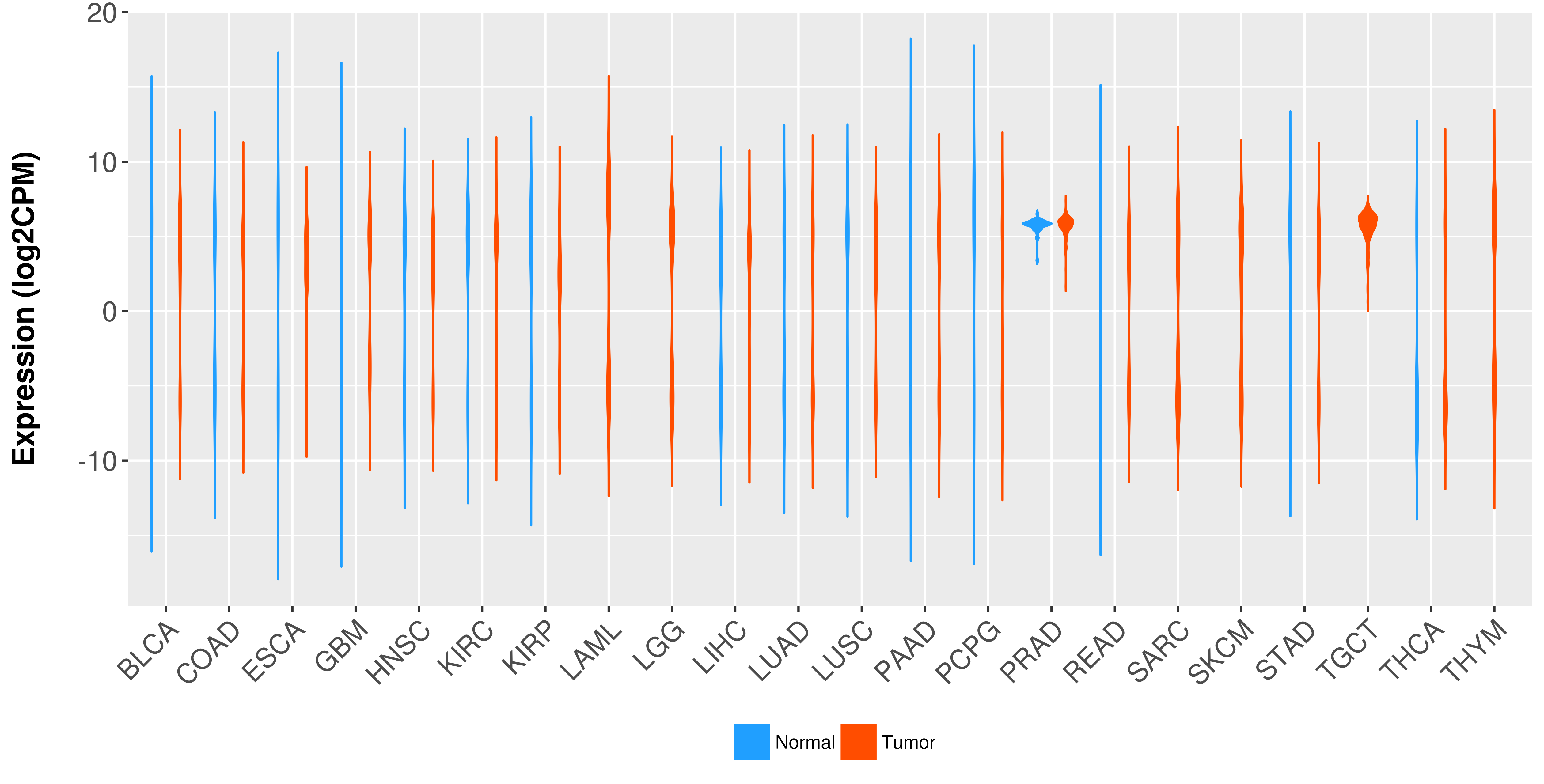
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
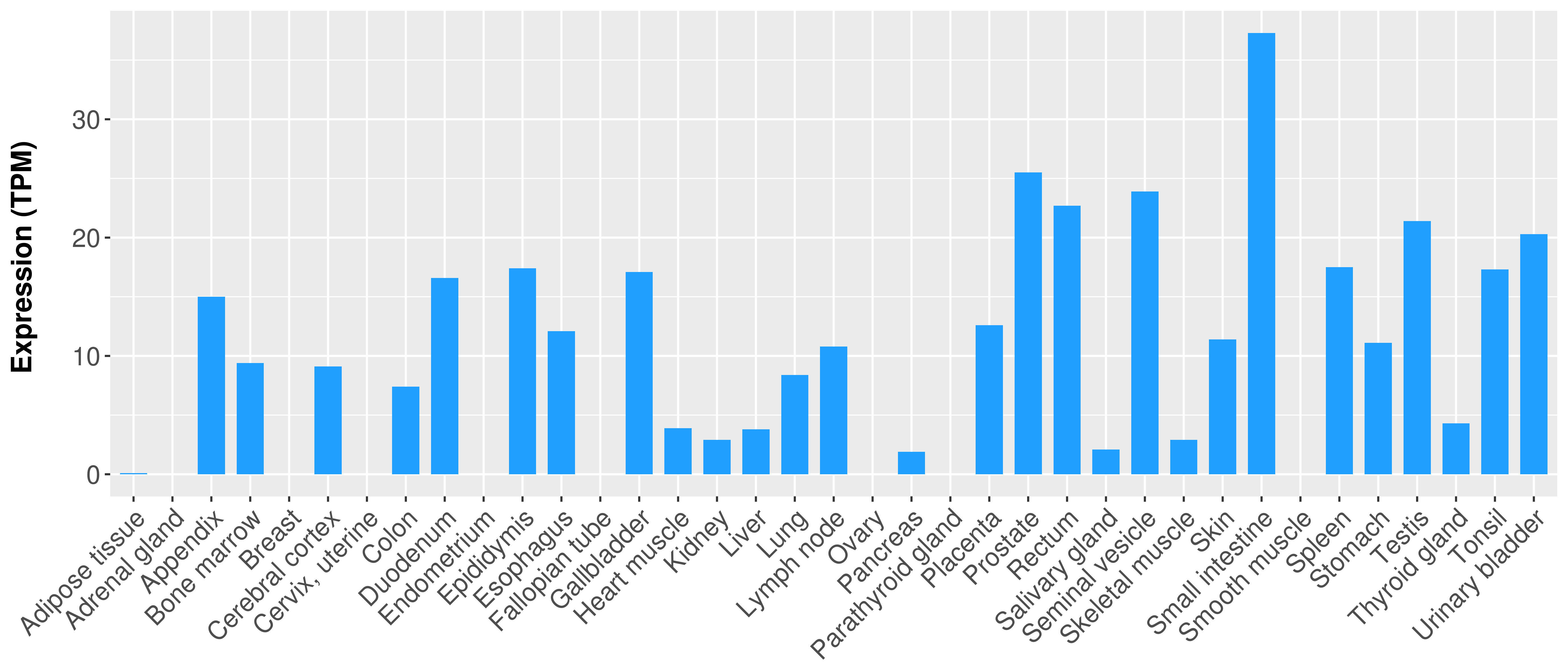
|
|
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
| There is no record. |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
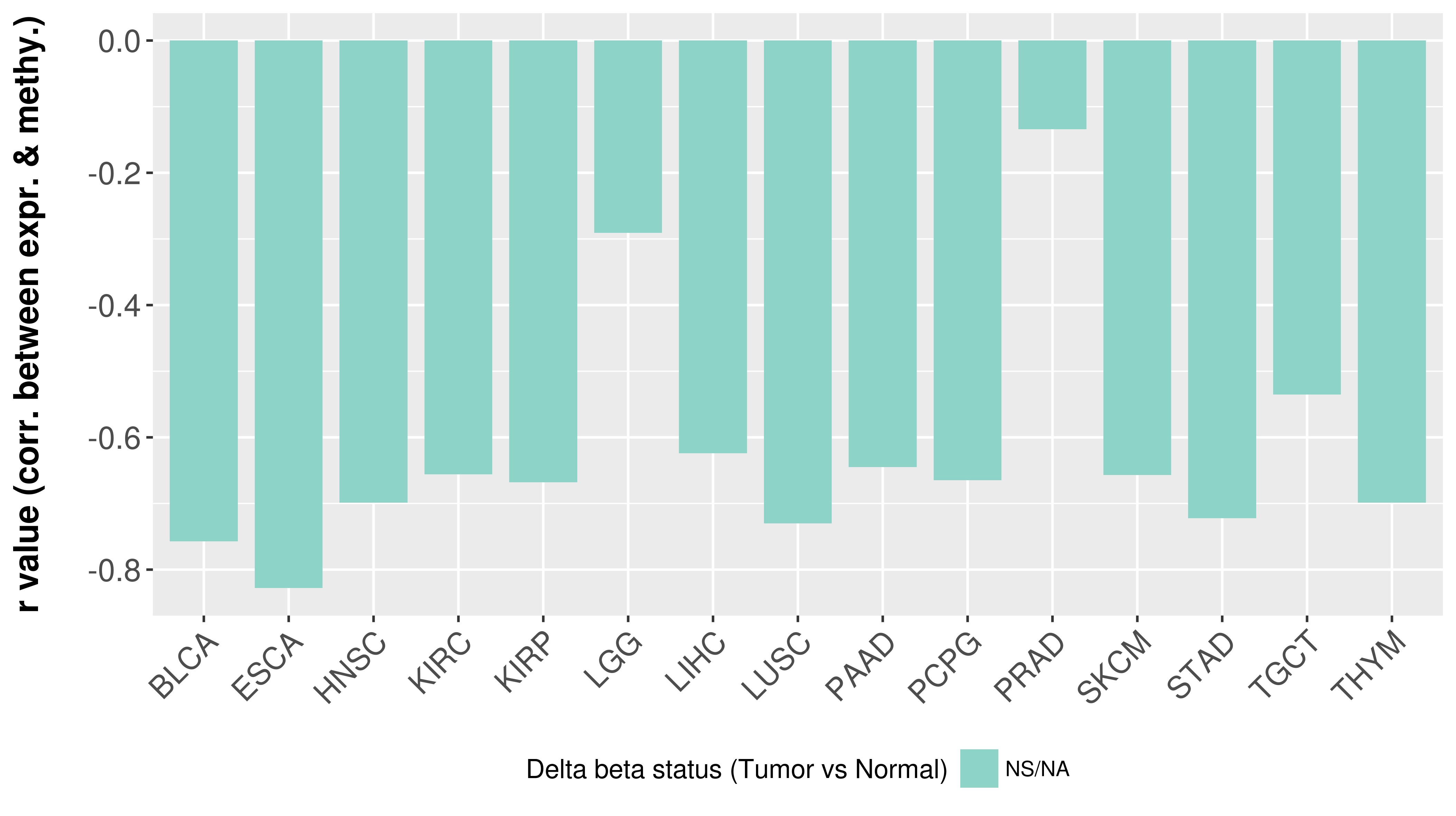
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
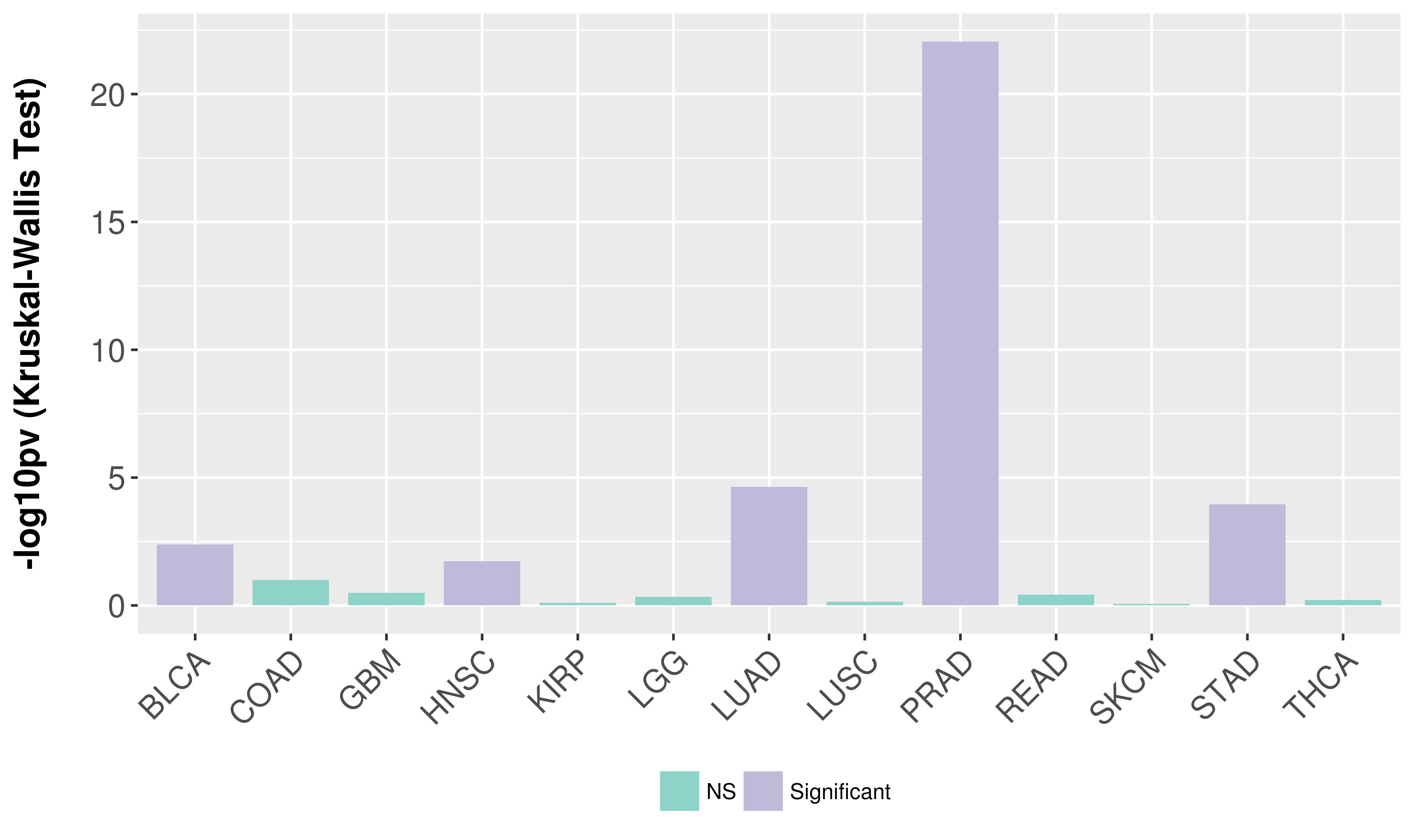
Association between expresson and subtype.
|
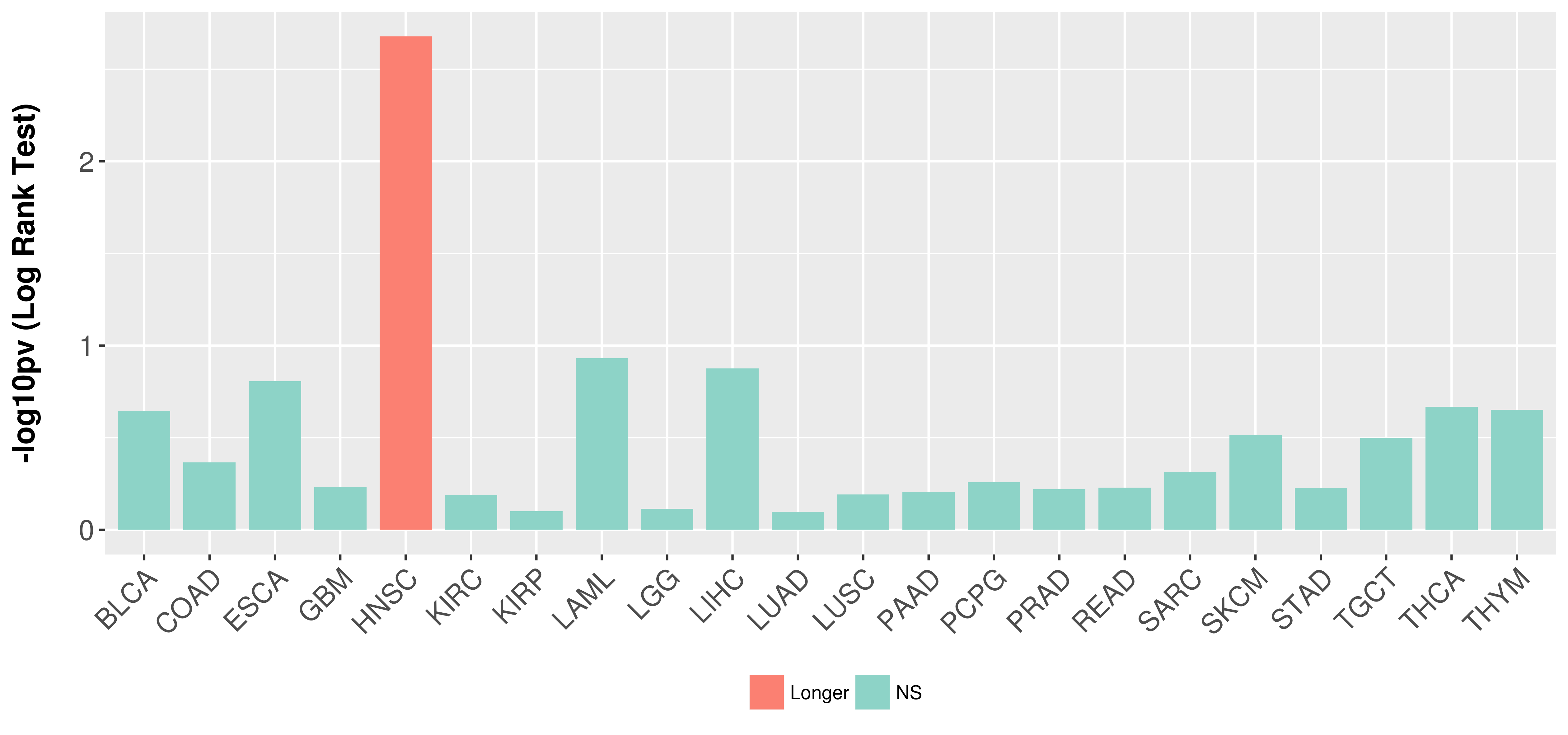
Overall survival analysis based on expression.
|
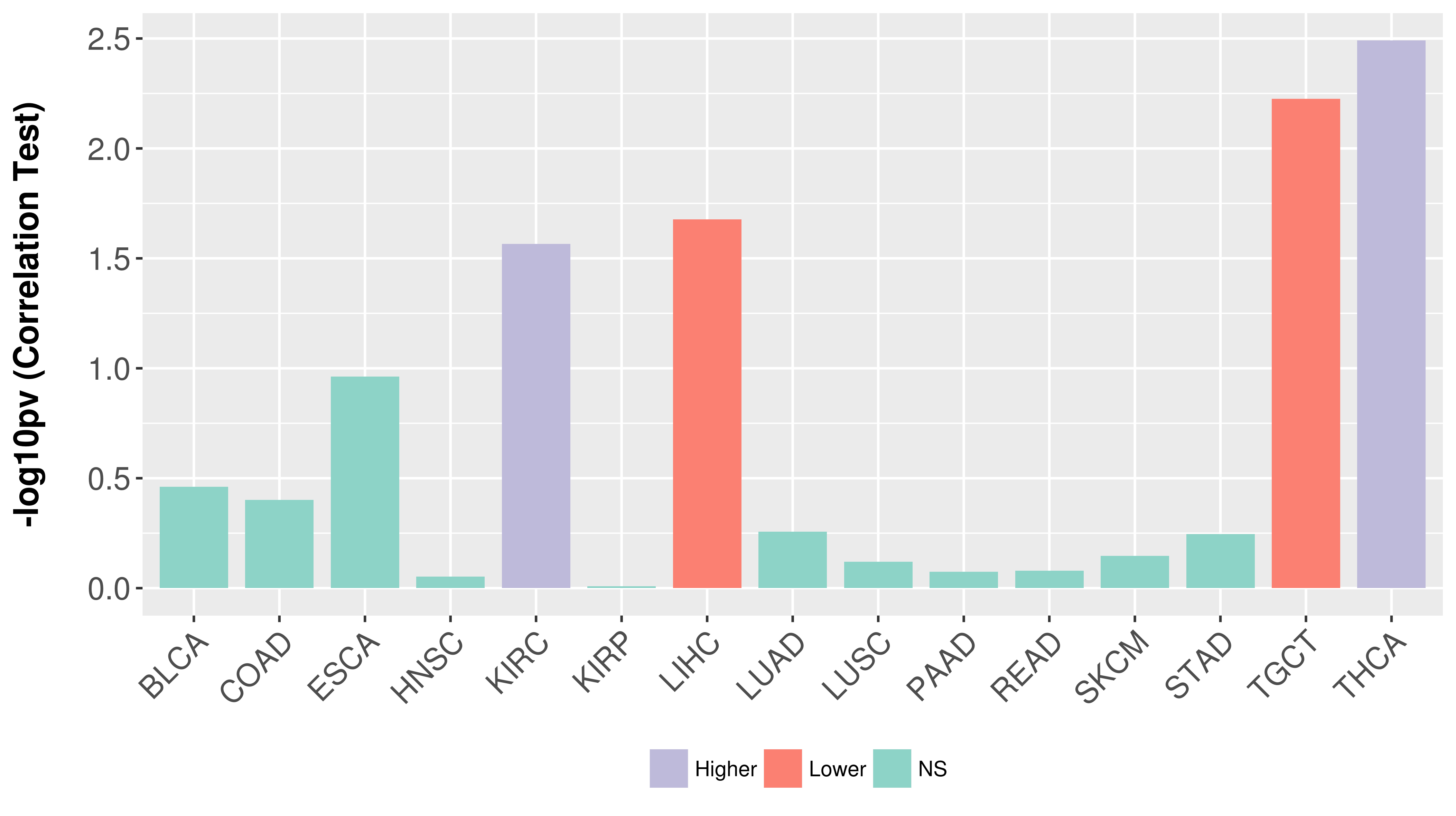
Association between expresson and stage.
|
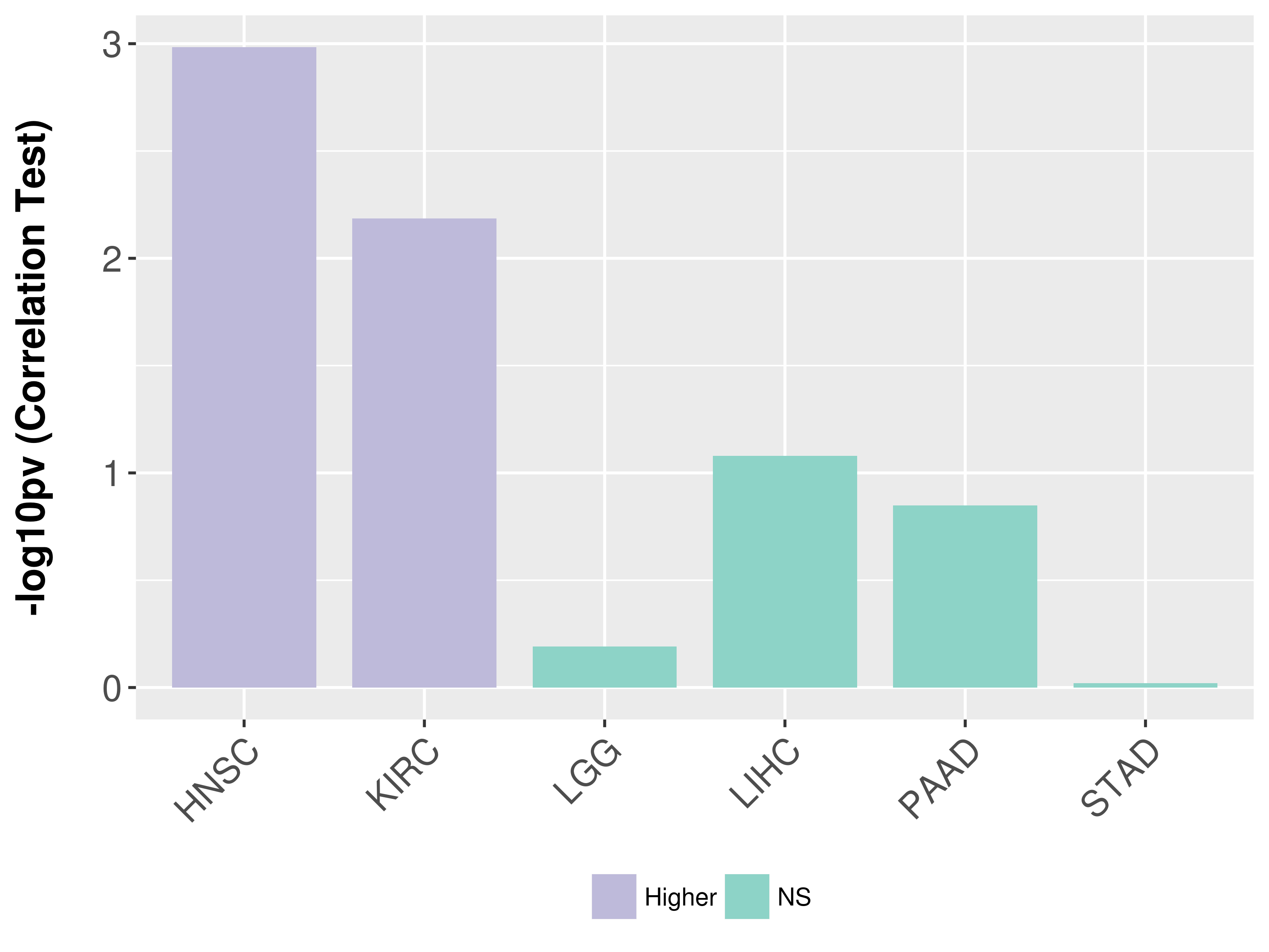
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
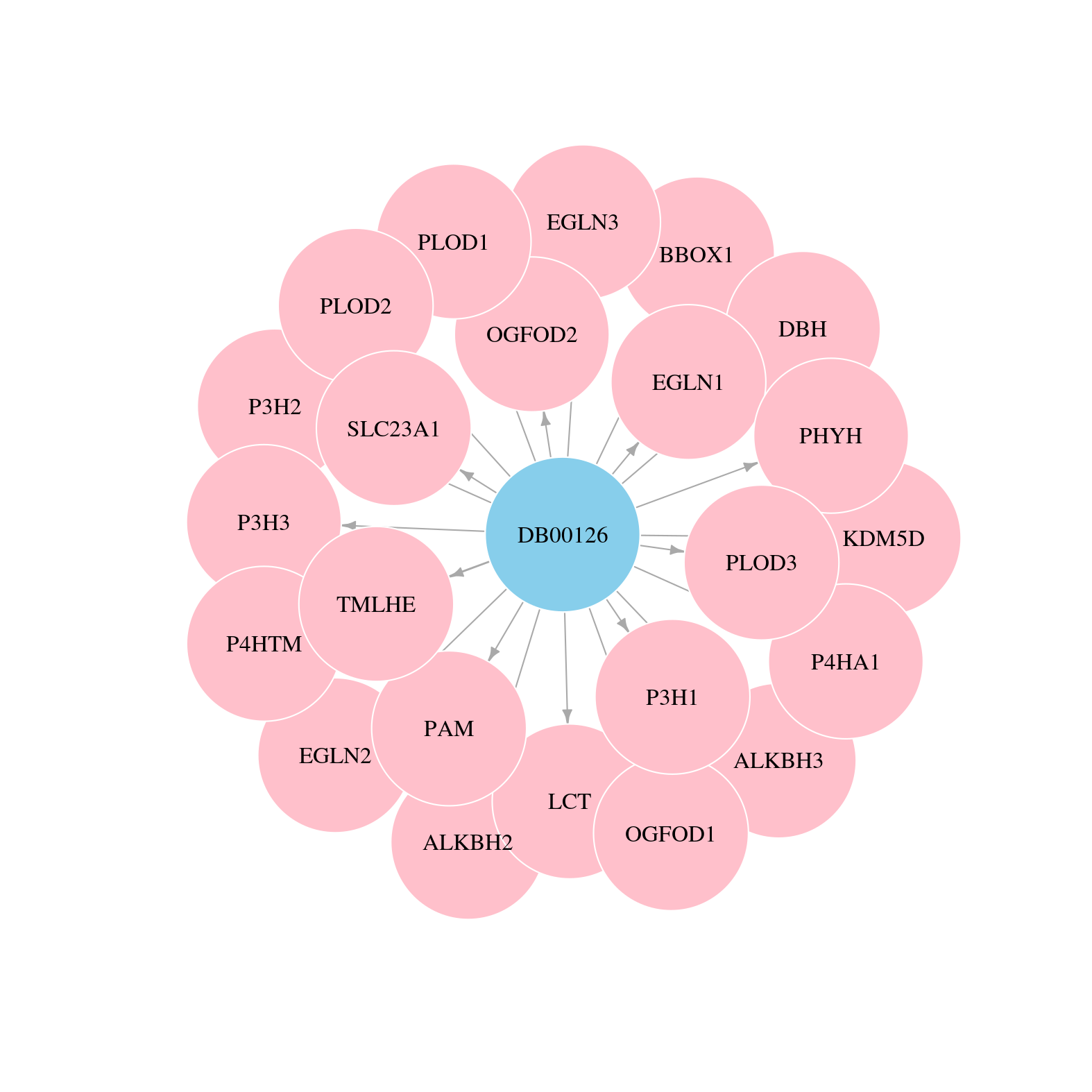
> Drugs from DrugBank database |
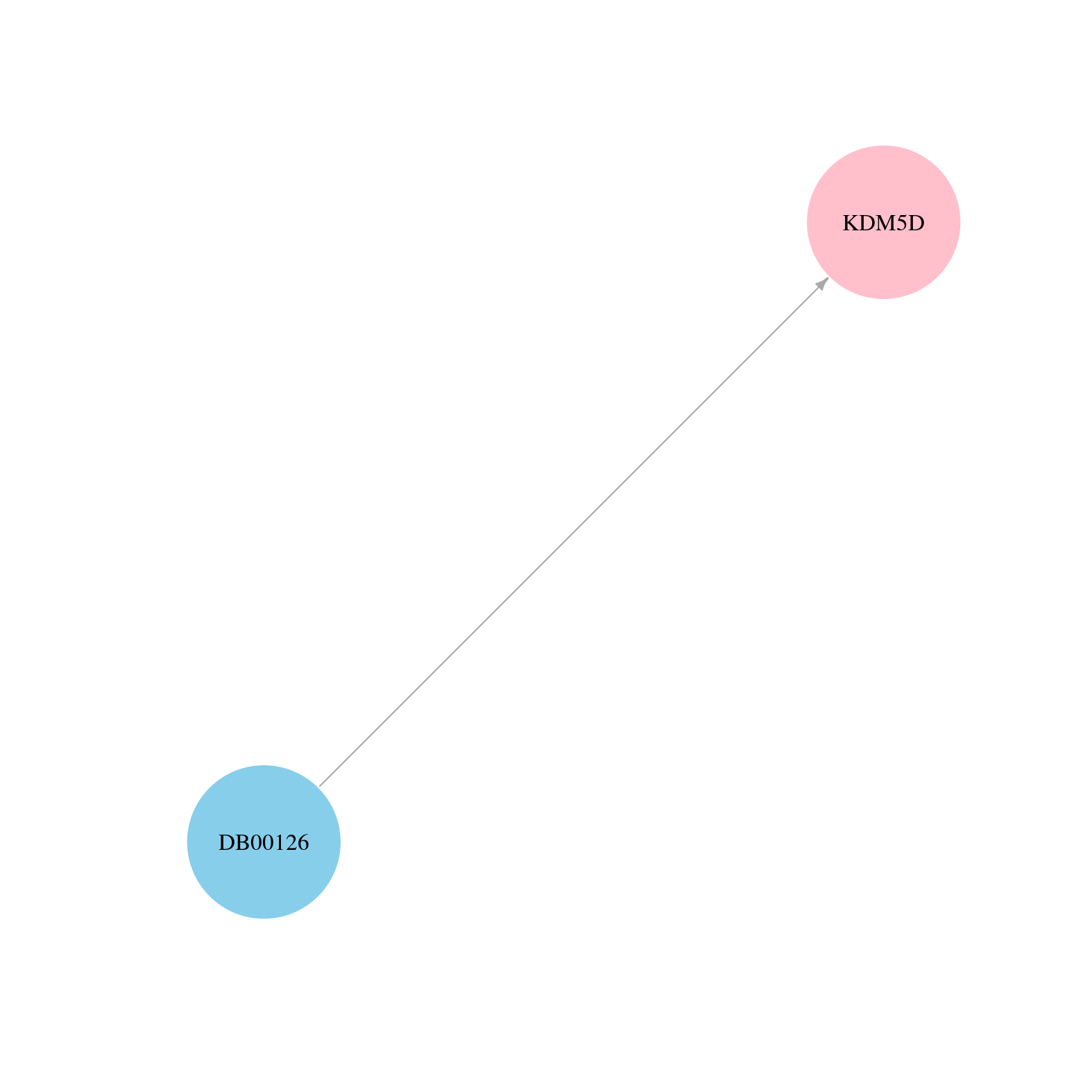
|
| Summary | |
|---|---|
| Symbol | KDM5D |
| Name | lysine demethylase 5D |
| Aliases | KIAA0234; HYA; HY; SMCY; JARID1D; Jumonji, AT rich interactive domain 1D (RBP2-like); Smcy homolog, Y-linked ...... |
| Location | Yq11.223 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|