Browse NCOA2 in pancancer
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF07469 Domain of unknown function (DUF1518) PF16279 Domain of unknown function (DUF4927) PF08815 Nuclear receptor coactivator PF00989 PAS fold PF08832 Steroid receptor coactivator |
||||||||||
| Function |
Transcriptional coactivator for steroid receptors and nuclear receptors. Coactivator of the steroid binding domain (AF-2) but not of the modulating N-terminal domain (AF-1). Required with NCOA1 to control energy balance between white and brown adipose tissues. Critical regulator of glucose metabolism regulation, acts as RORA coactivator to specifically modulate G6PC expression. Involved in the positive regulation of the transcriptional activity of the glucocorticoid receptor NR3C1 by sumoylation enhancer RWDD3. Positively regulates the circadian clock by acting as a transcriptional coactivator for the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0005996 monosaccharide metabolic process GO:0006006 glucose metabolic process GO:0006109 regulation of carbohydrate metabolic process GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006820 anion transport GO:0006869 lipid transport GO:0007622 rhythmic behavior GO:0007623 circadian rhythm GO:0007626 locomotory behavior GO:0010469 regulation of receptor activity GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010876 lipid localization GO:0010906 regulation of glucose metabolic process GO:0015711 organic anion transport GO:0015718 monocarboxylic acid transport GO:0015721 bile acid and bile salt transport GO:0015849 organic acid transport GO:0015850 organic hydroxy compound transport GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019318 hexose metabolic process GO:0030522 intracellular receptor signaling pathway GO:0032922 circadian regulation of gene expression GO:0043543 protein acylation GO:0044262 cellular carbohydrate metabolic process GO:0044723 single-organism carbohydrate metabolic process GO:0045475 locomotor rhythm GO:0046942 carboxylic acid transport GO:0048511 rhythmic process GO:0048512 circadian behavior GO:1904016 response to Thyroglobulin triiodothyronine GO:1904017 cellular response to Thyroglobulin triiodothyronine GO:2000273 positive regulation of receptor activity |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0001161 intronic transcription regulatory region sequence-specific DNA binding GO:0001162 RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0010861 thyroid hormone receptor activator activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0016922 ligand-dependent nuclear receptor binding GO:0030374 ligand-dependent nuclear receptor transcription coactivator activity GO:0030375 thyroid hormone receptor coactivator activity GO:0030545 receptor regulator activity GO:0030546 receptor activator activity GO:0034212 peptide N-acetyltransferase activity GO:0035257 nuclear hormone receptor binding GO:0044213 intronic transcription regulatory region DNA binding GO:0051427 hormone receptor binding GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component | - |
| KEGG |
hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-5625886: Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 R-HSA-2426168: Activation of gene expression by SREBF (SREBP) R-HSA-1368108: BMAL1 R-HSA-194068: Bile acid and bile salt metabolism R-HSA-211859: Biological oxidations R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-211897: Cytochrome P450 - arranged by substrate type R-HSA-1266738: Developmental Biology R-HSA-211976: Endogenous sterols R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-1592230: Mitochondrial biogenesis R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-1989781: PPARA activates gene expression R-HSA-211945: Phase 1 - Functionalization of compounds R-HSA-195258: RHO GTPase Effectors R-HSA-5625740: RHO GTPases activate PKNs R-HSA-1368082: RORA activates gene expression R-HSA-159418: Recycling of bile acids and salts R-HSA-1655829: Regulation of cholesterol biosynthesis by SREBP (SREBF) R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) R-HSA-162582: Signal Transduction R-HSA-194315: Signaling by Rho GTPases R-HSA-192105: Synthesis of bile acids and bile salts R-HSA-193807: Synthesis of bile acids and bile salts via 27-hydroxycholesterol R-HSA-193368: Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis R-HSA-381340: Transcriptional regulation of white adipocyte differentiation R-HSA-2032785: YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
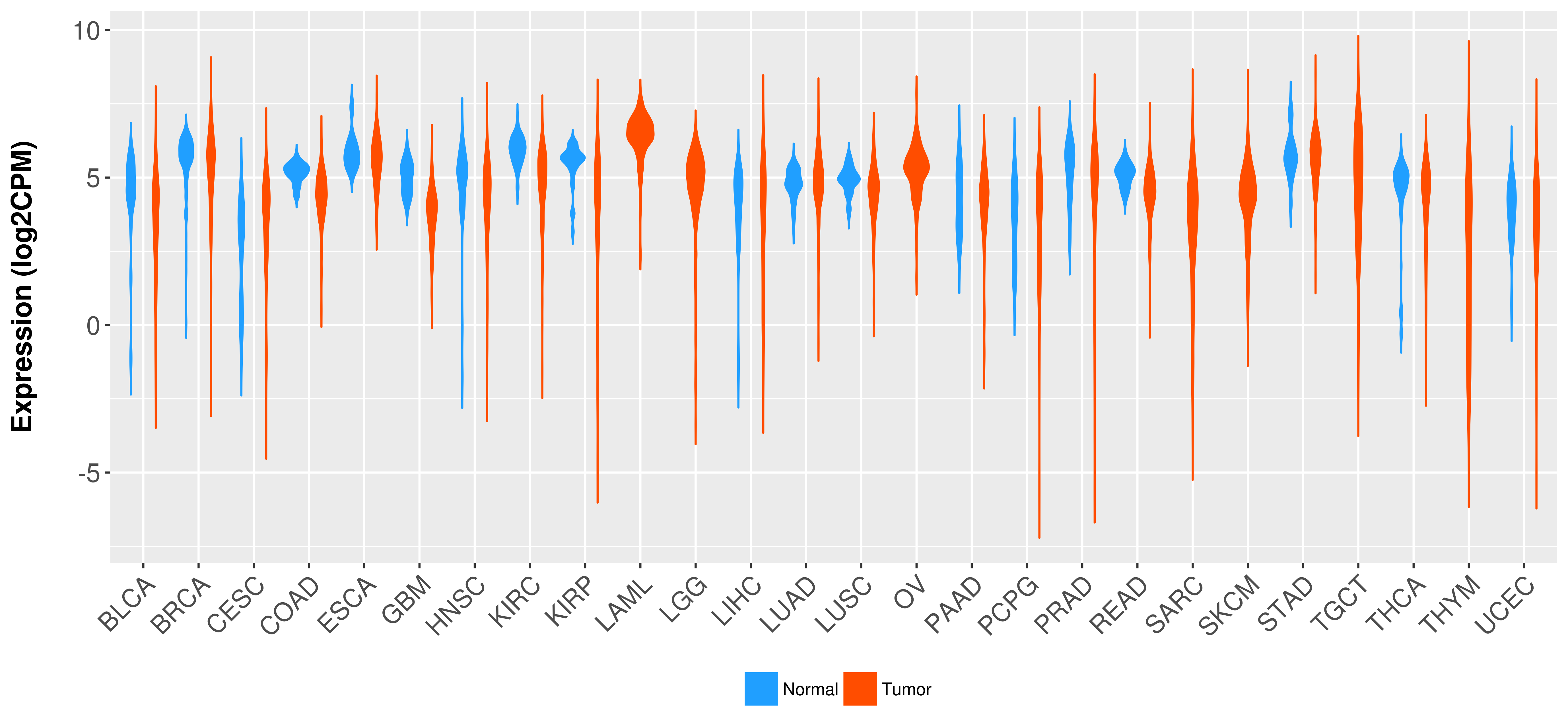
Differential expression analysis for cancers with more than 10 normal samples
|
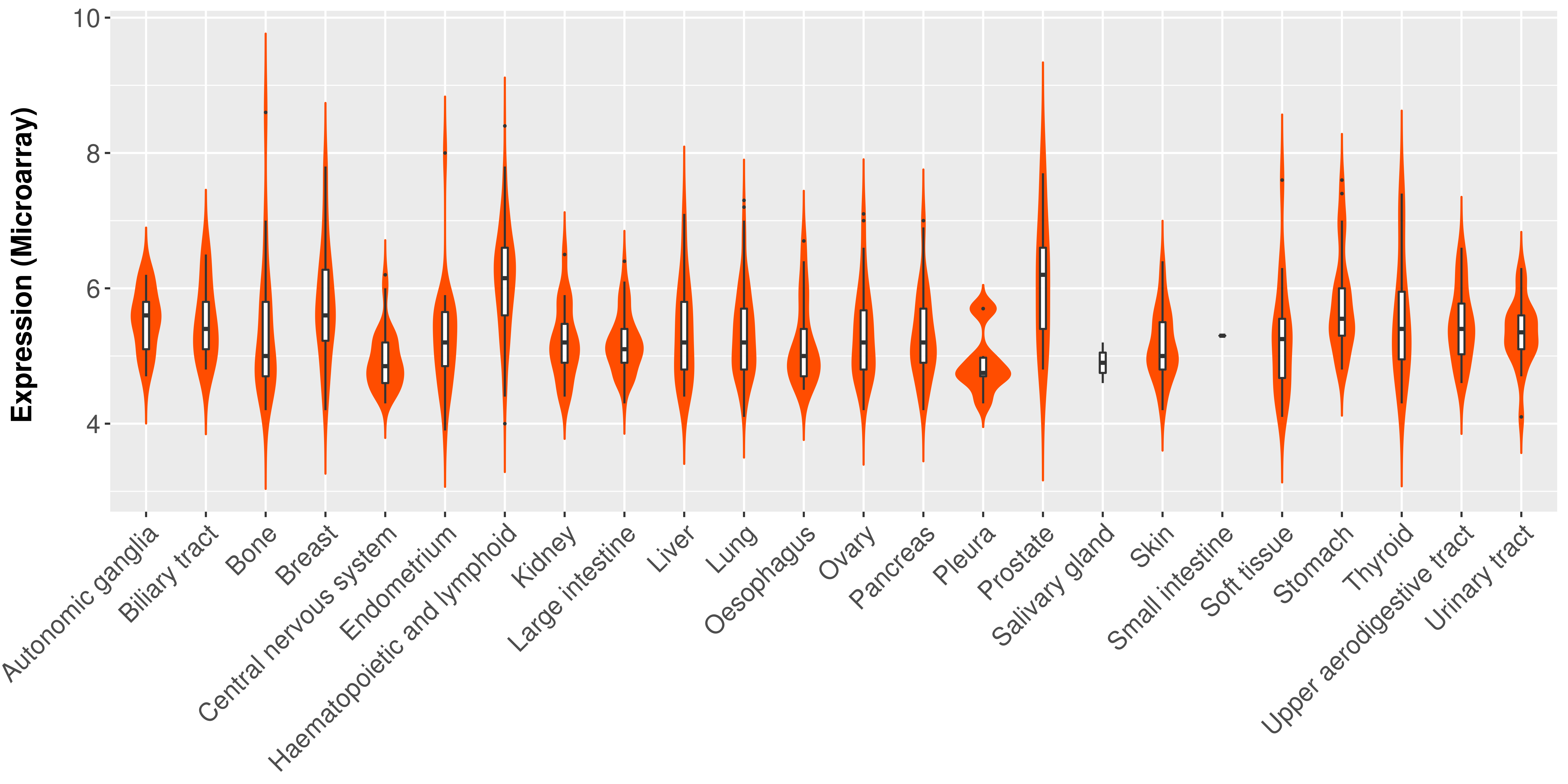
|
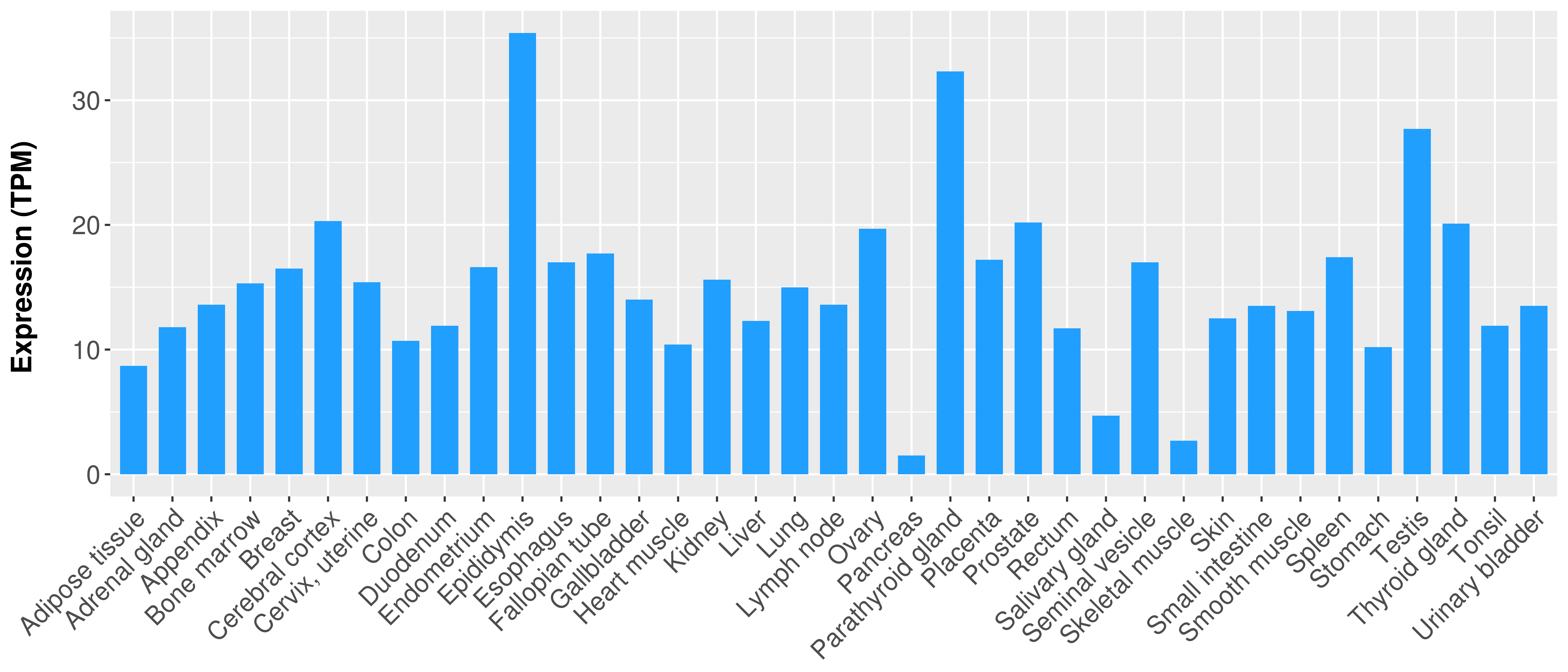
|
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
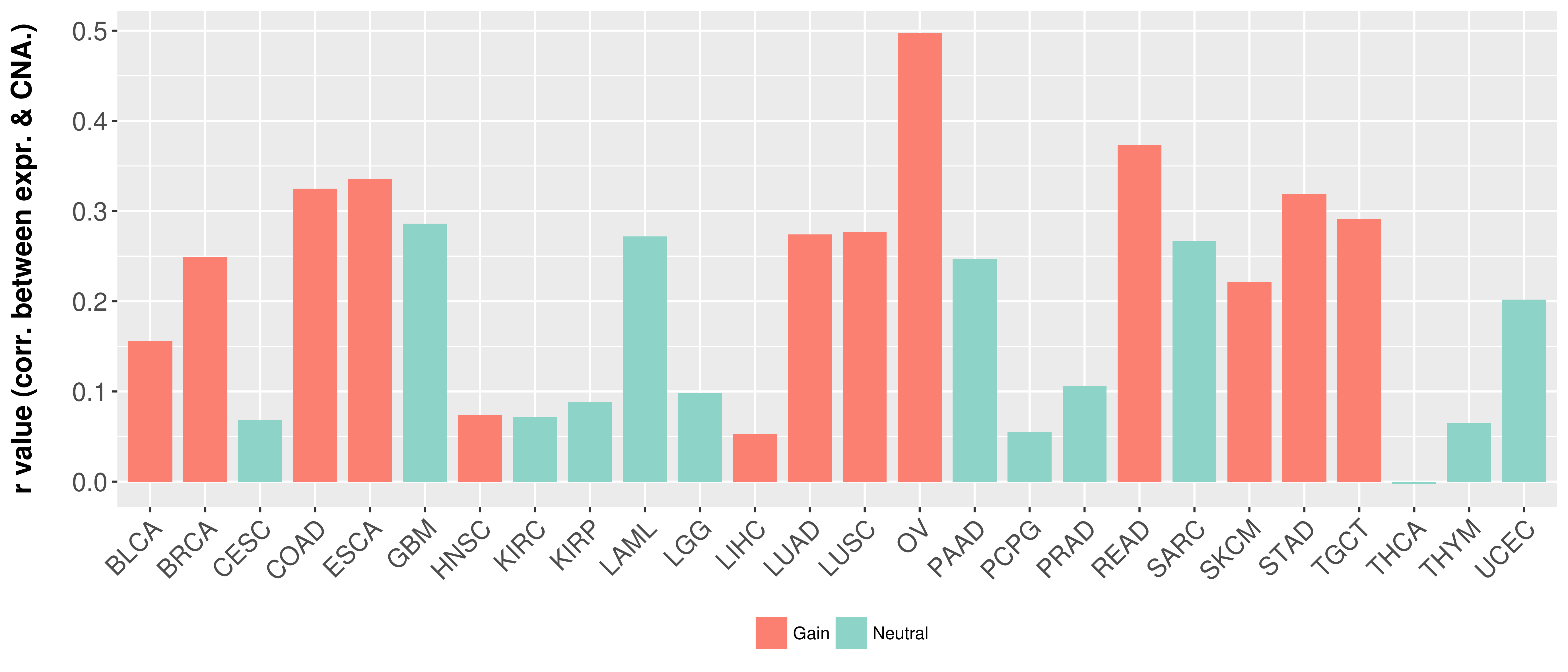
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
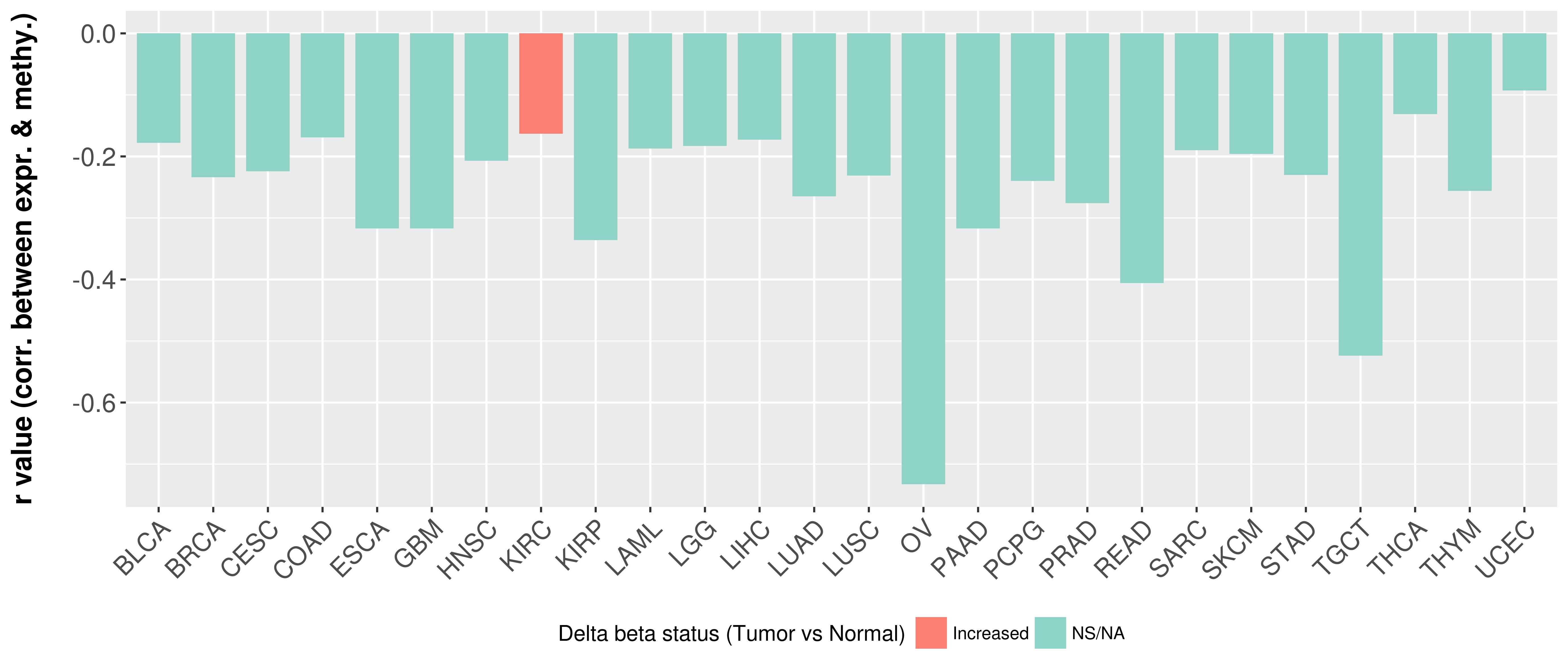
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
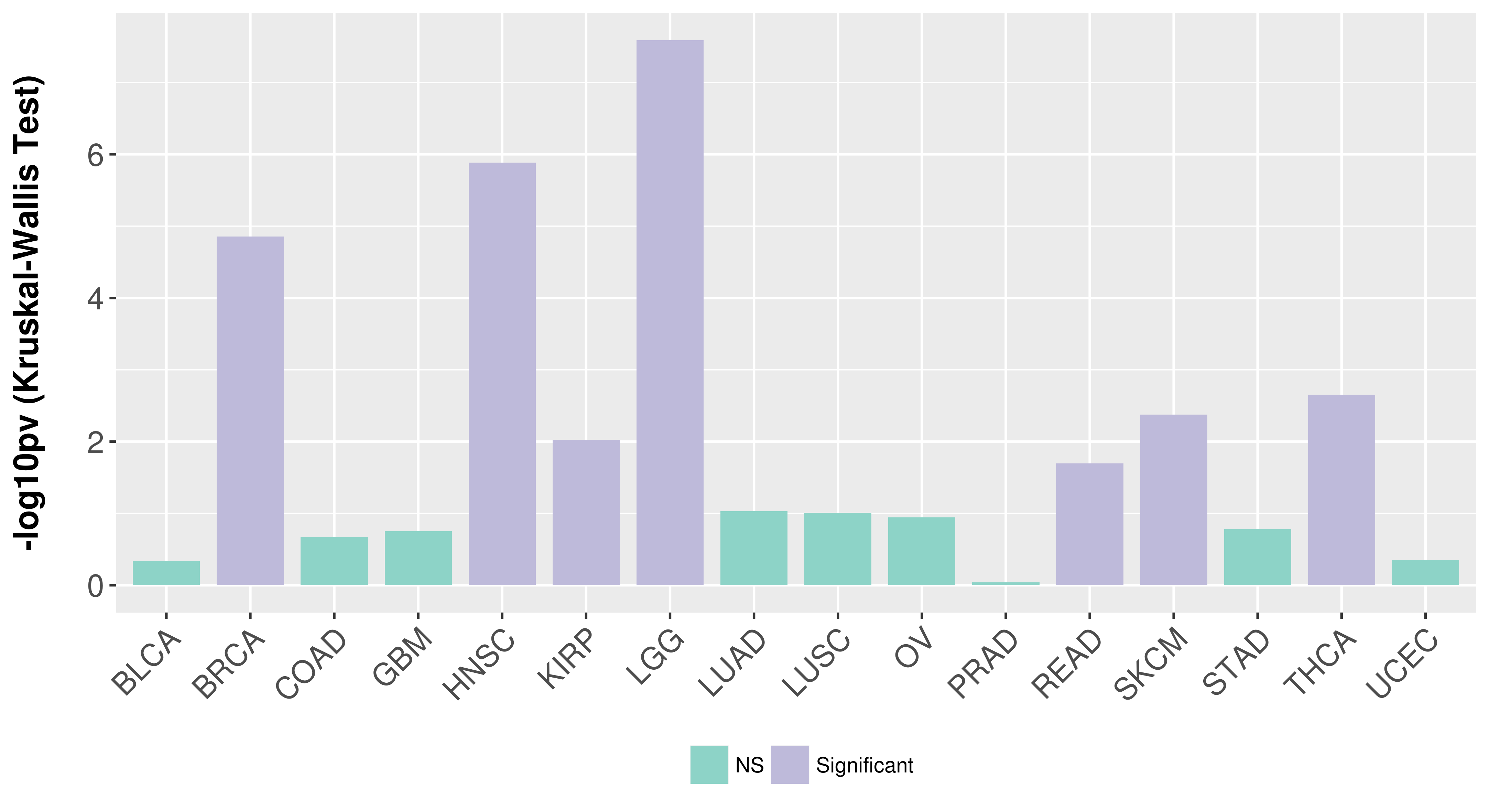
Association between expresson and subtype.
|
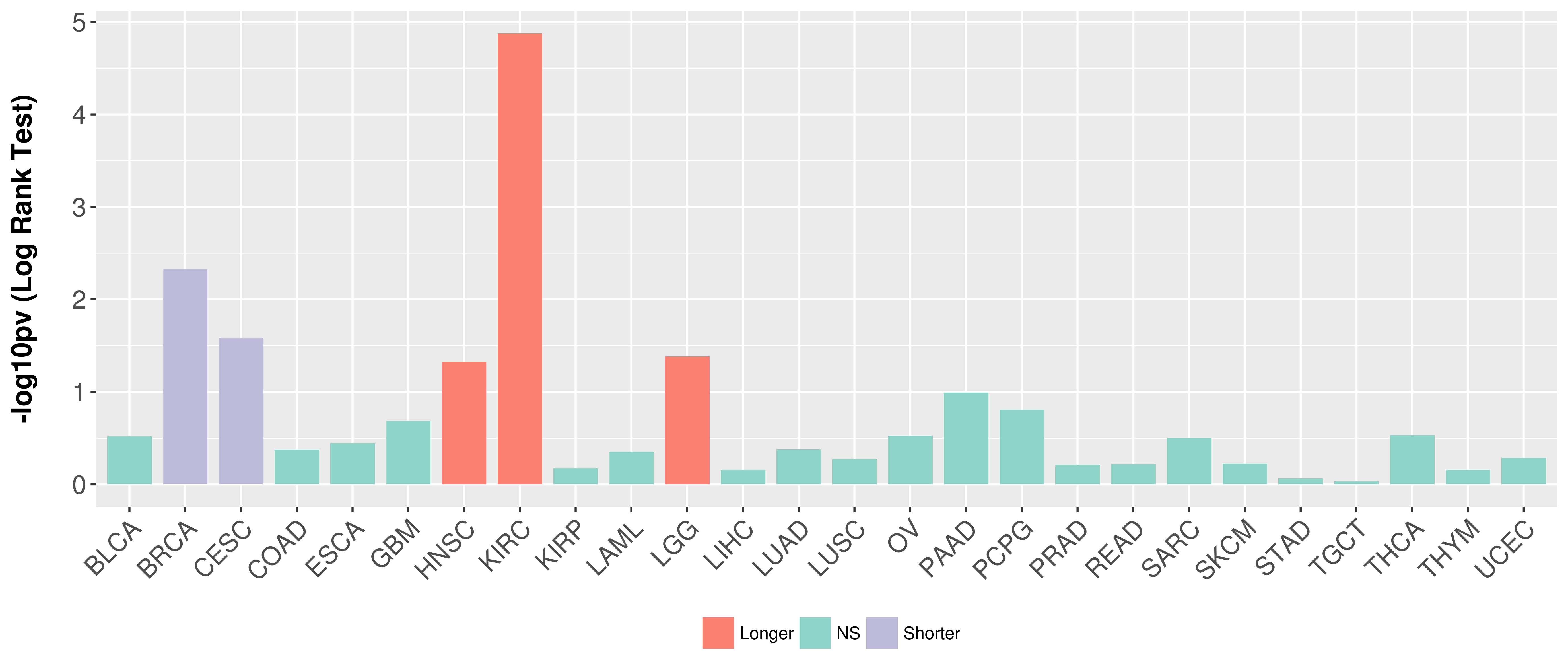
Overall survival analysis based on expression.
|
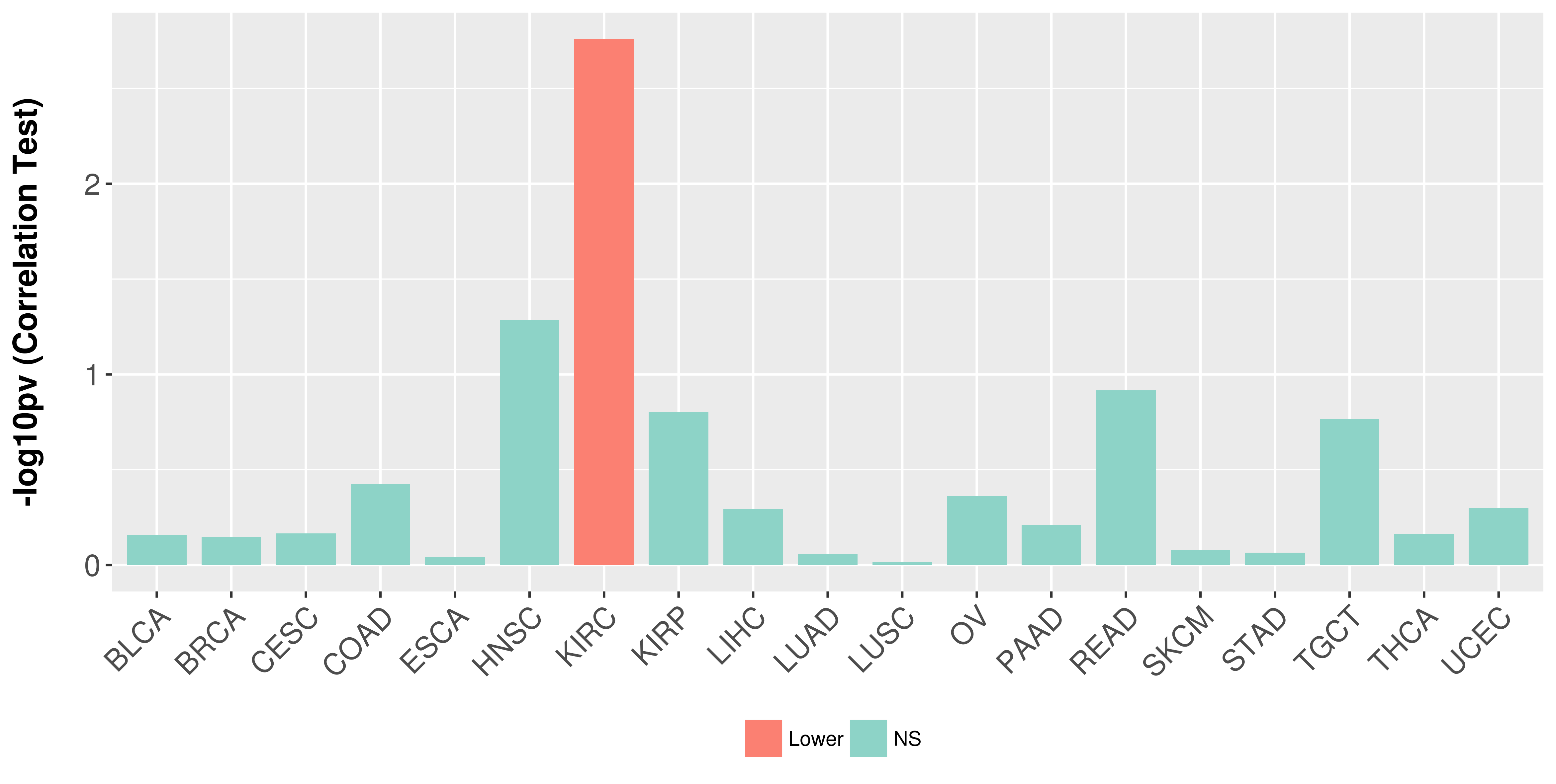
Association between expresson and stage.
|
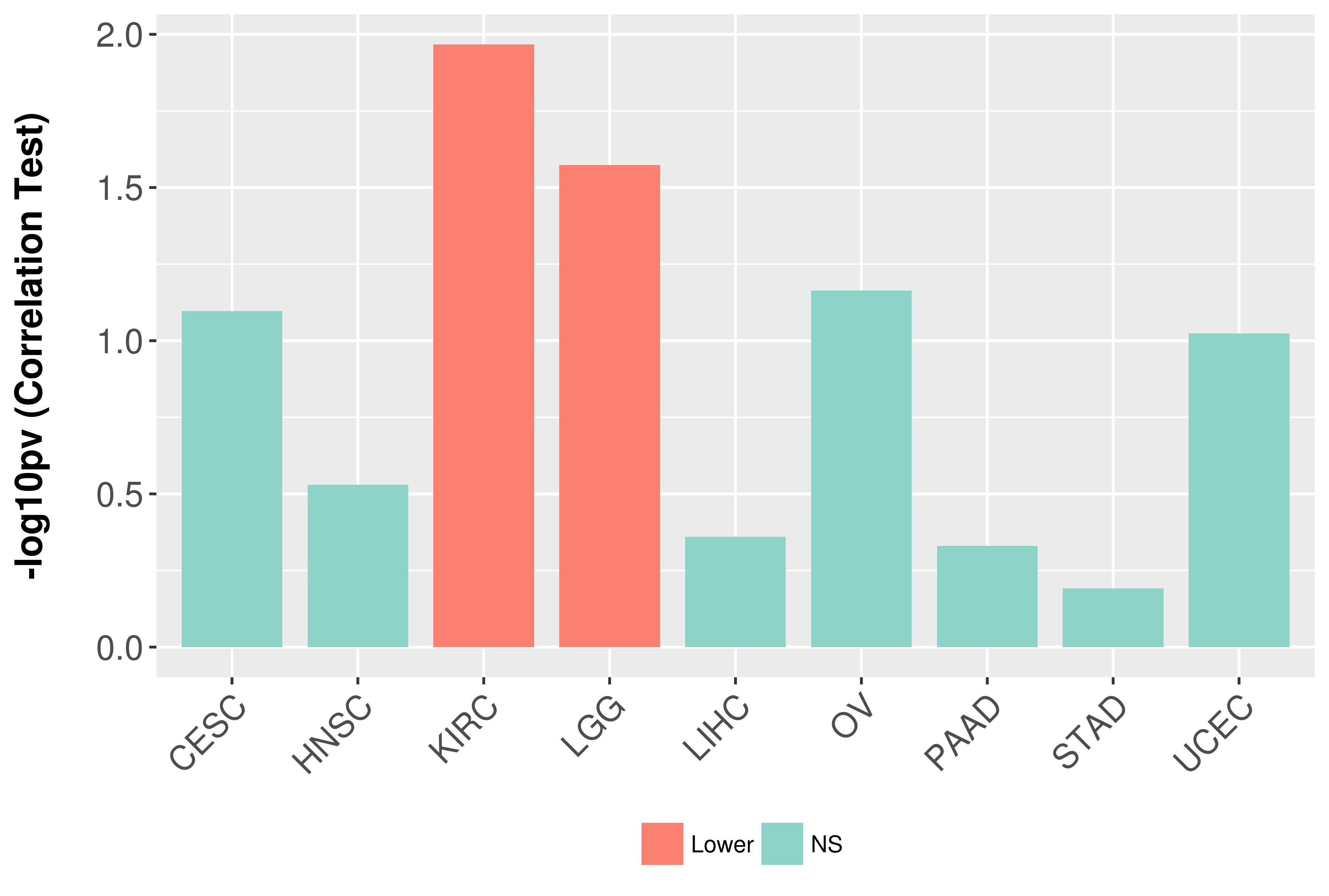
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
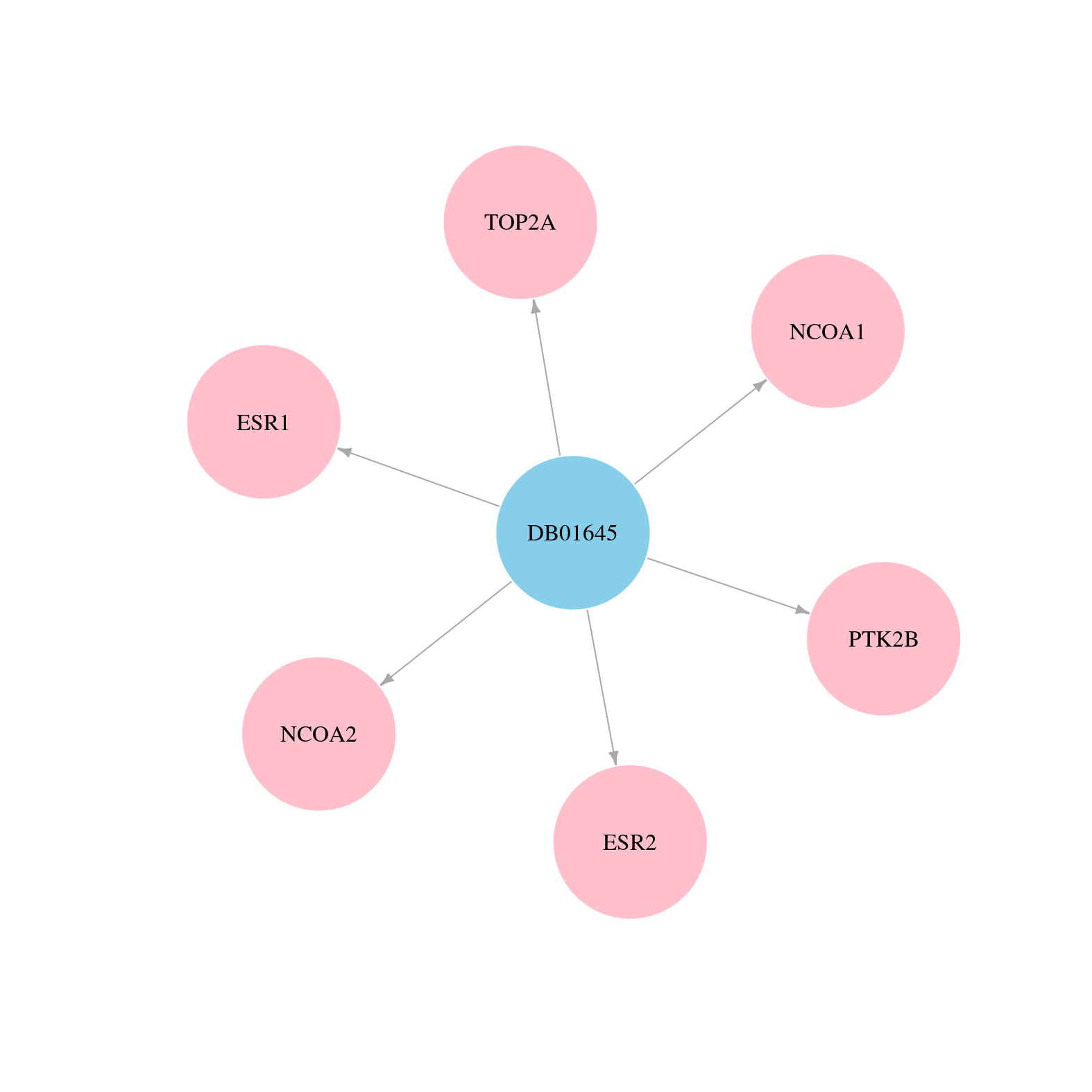
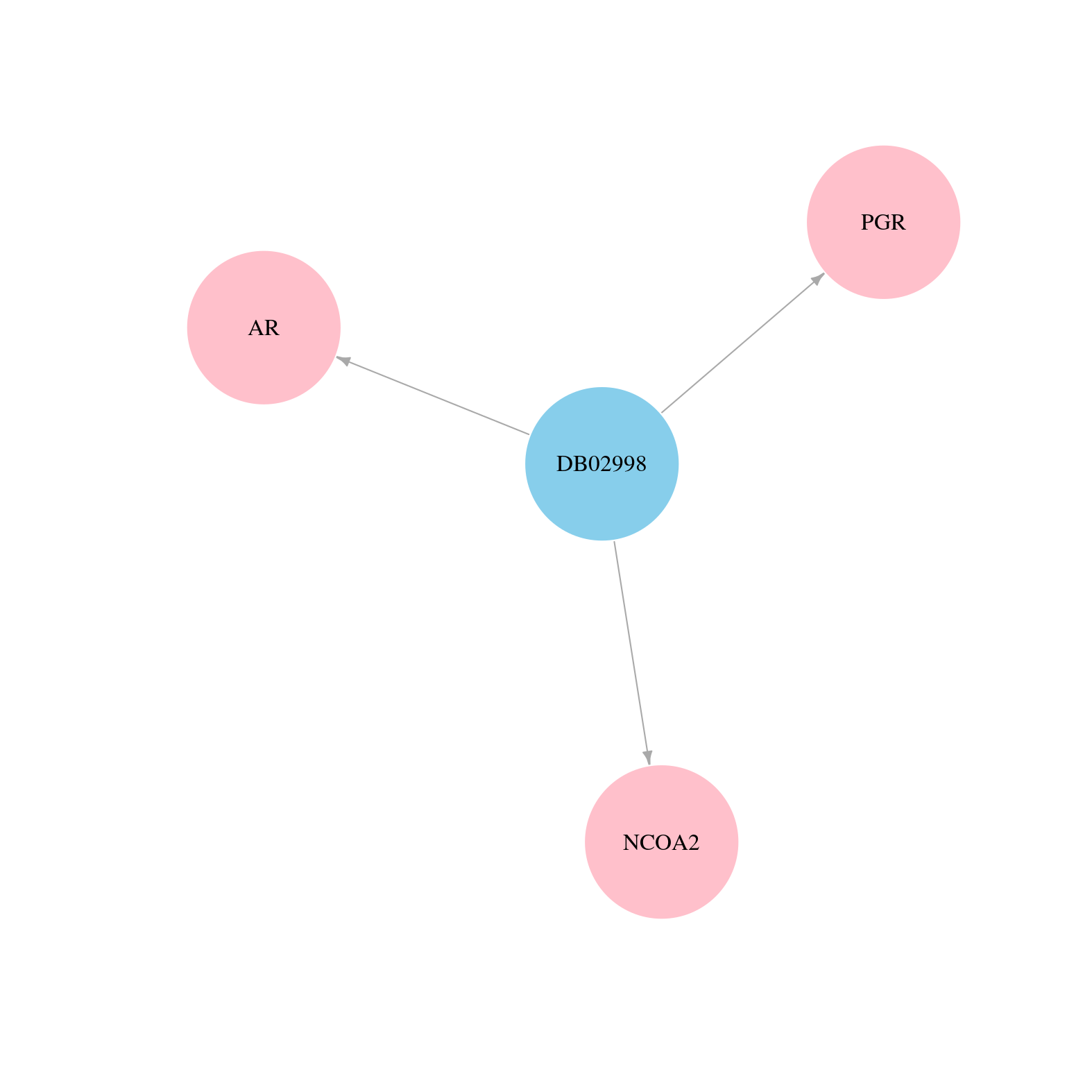
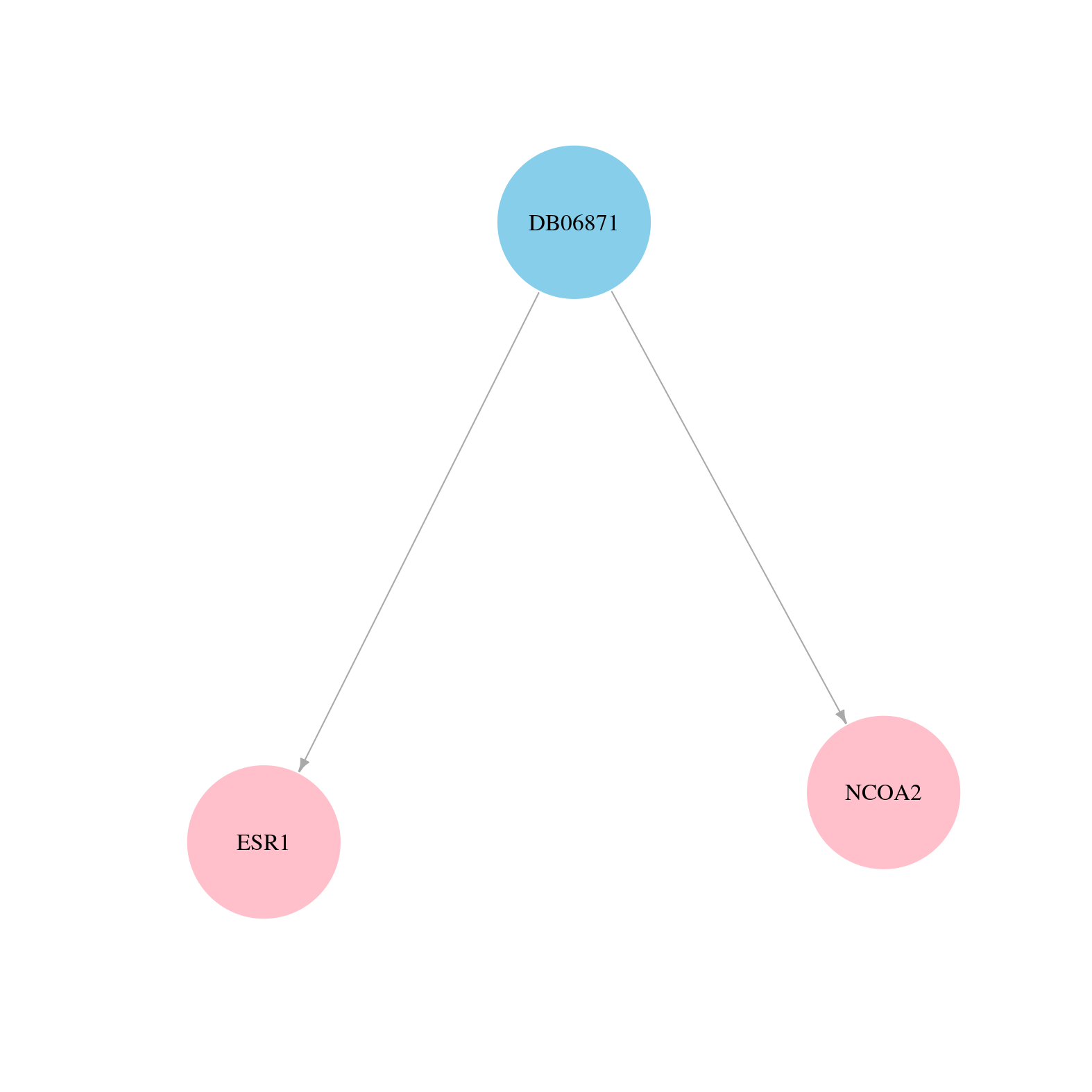
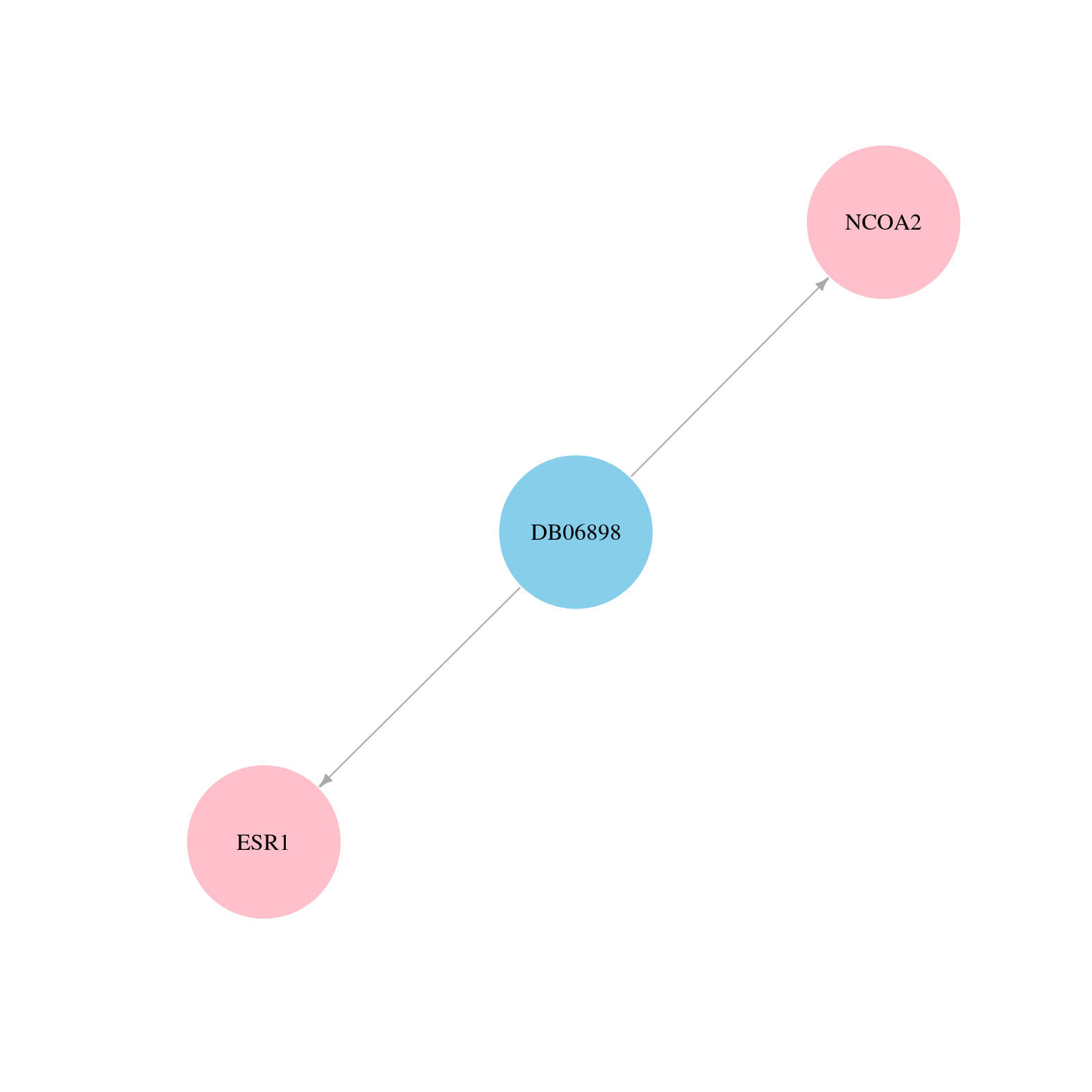
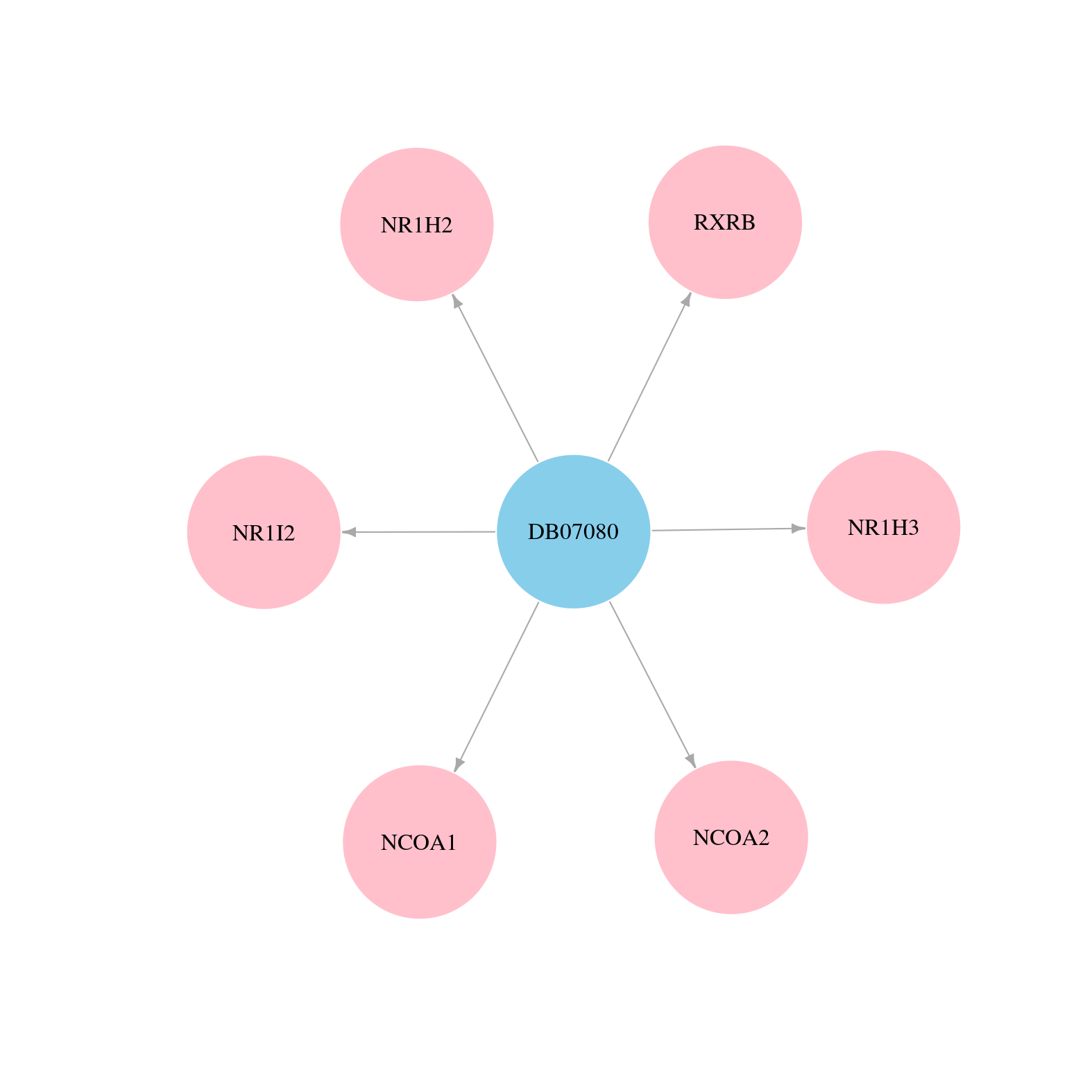
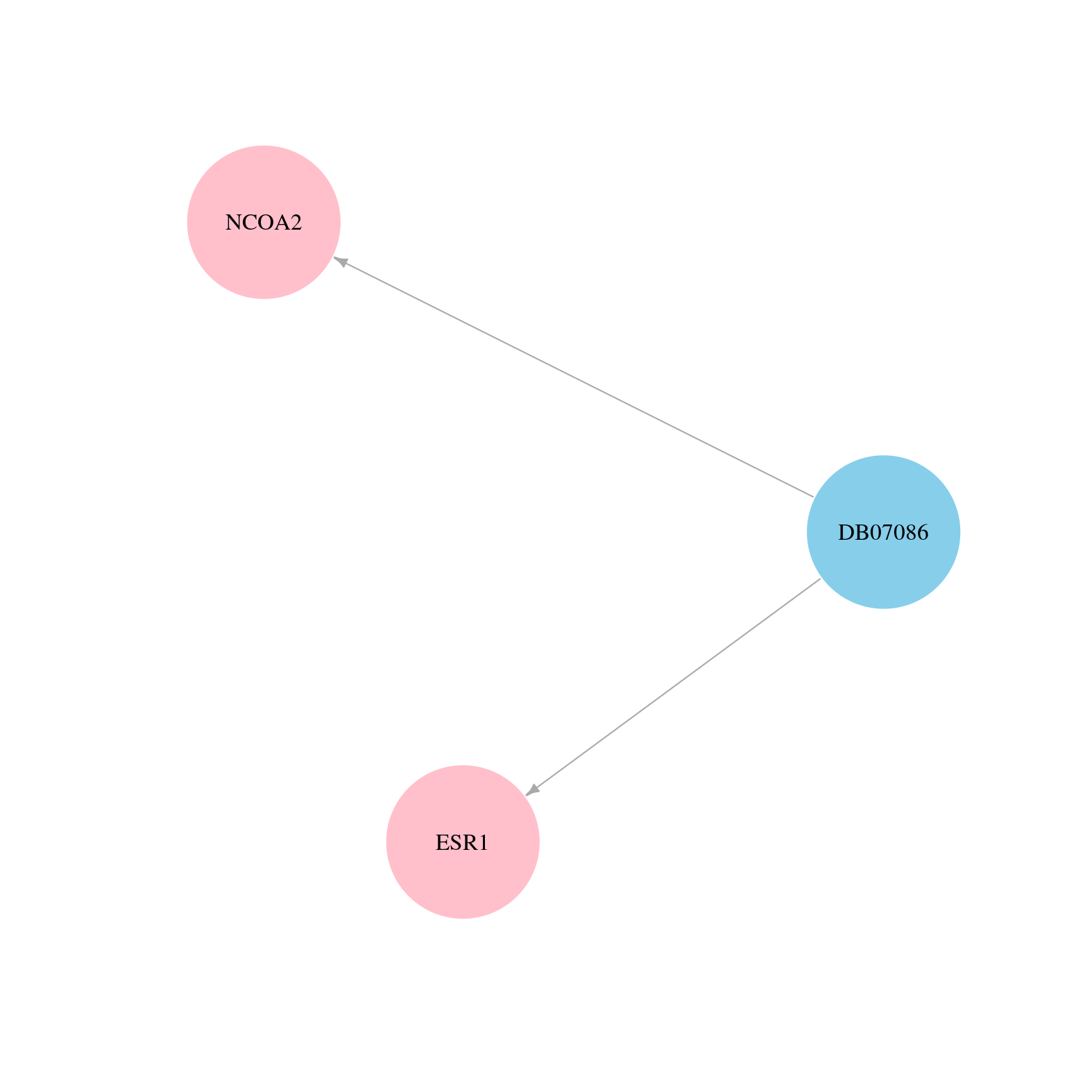
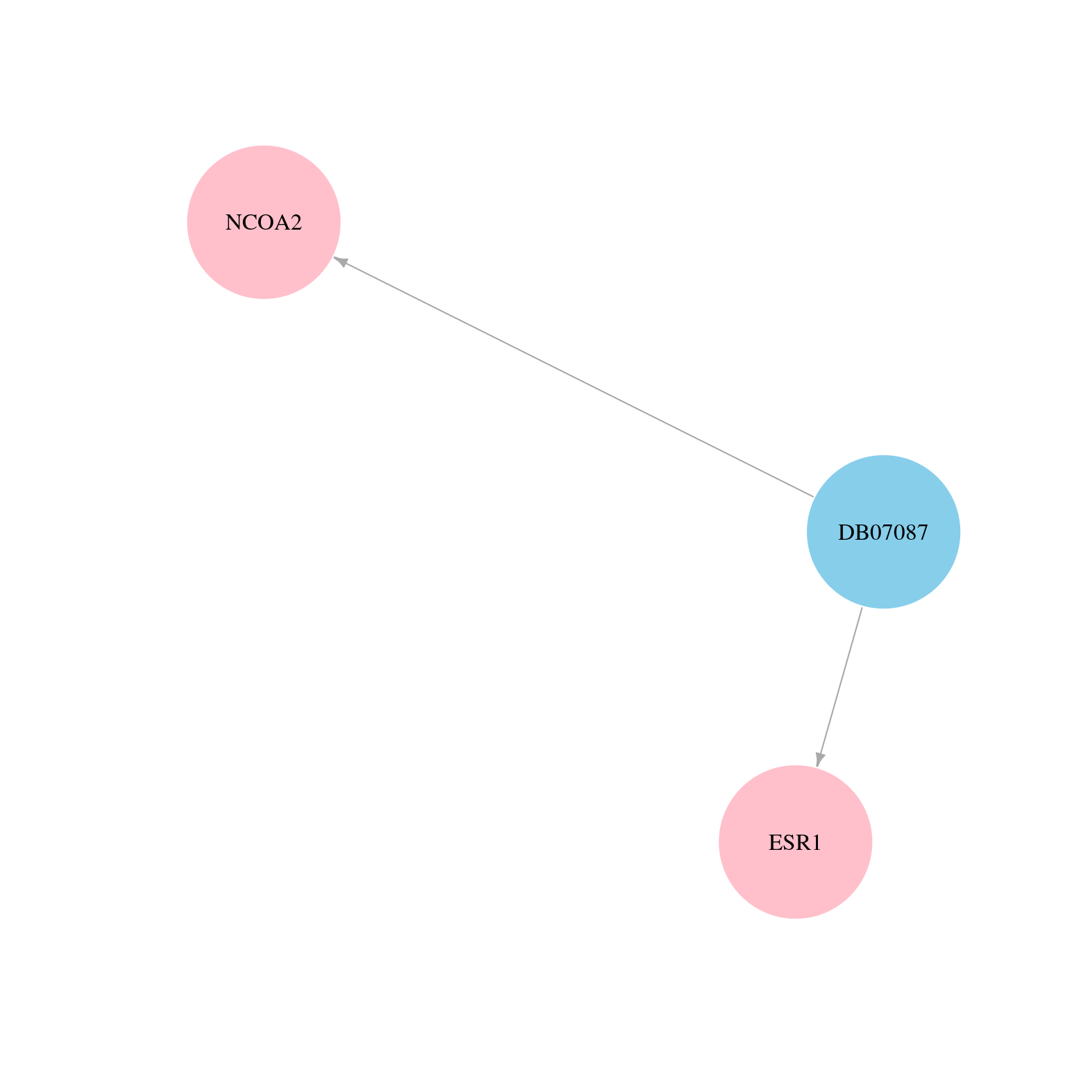
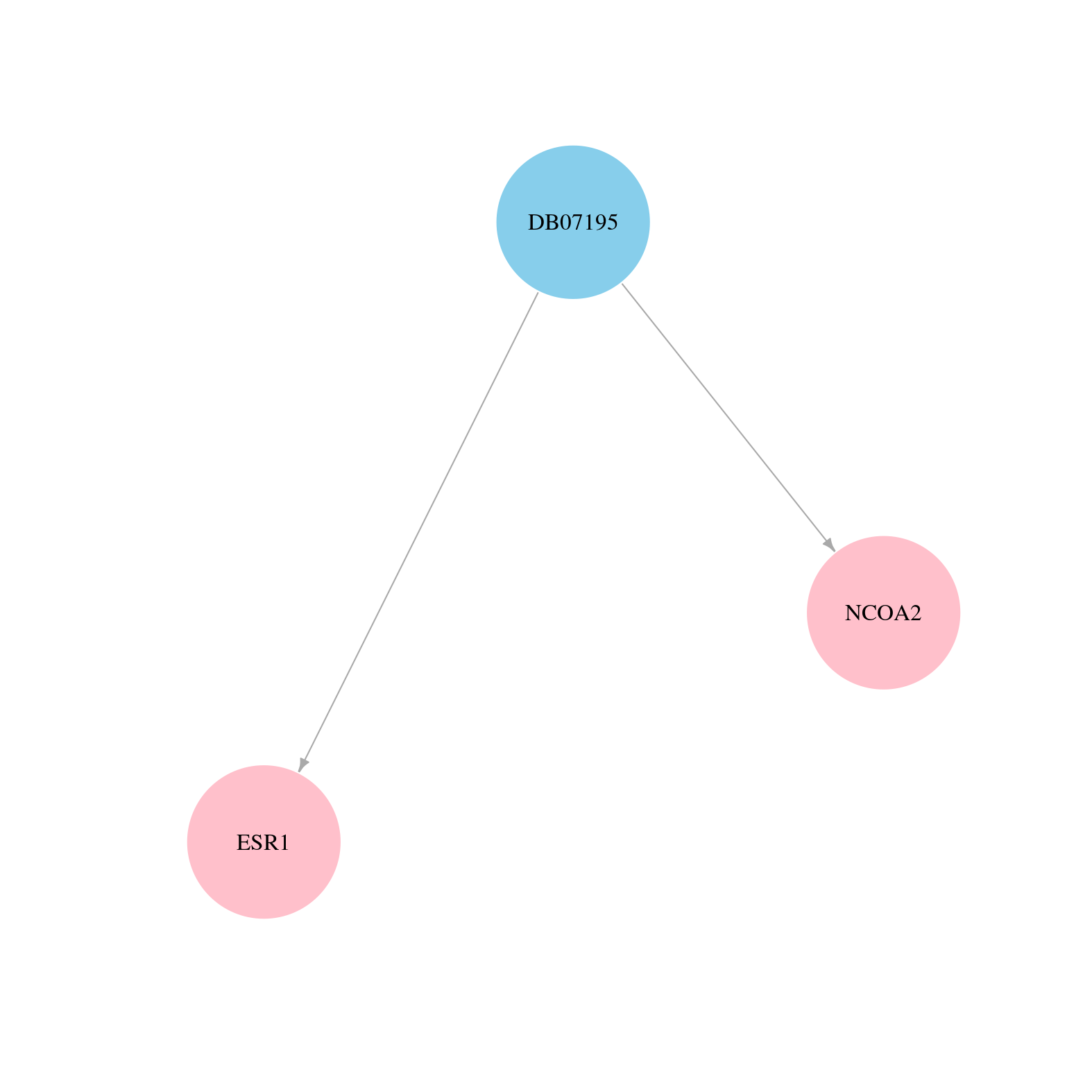
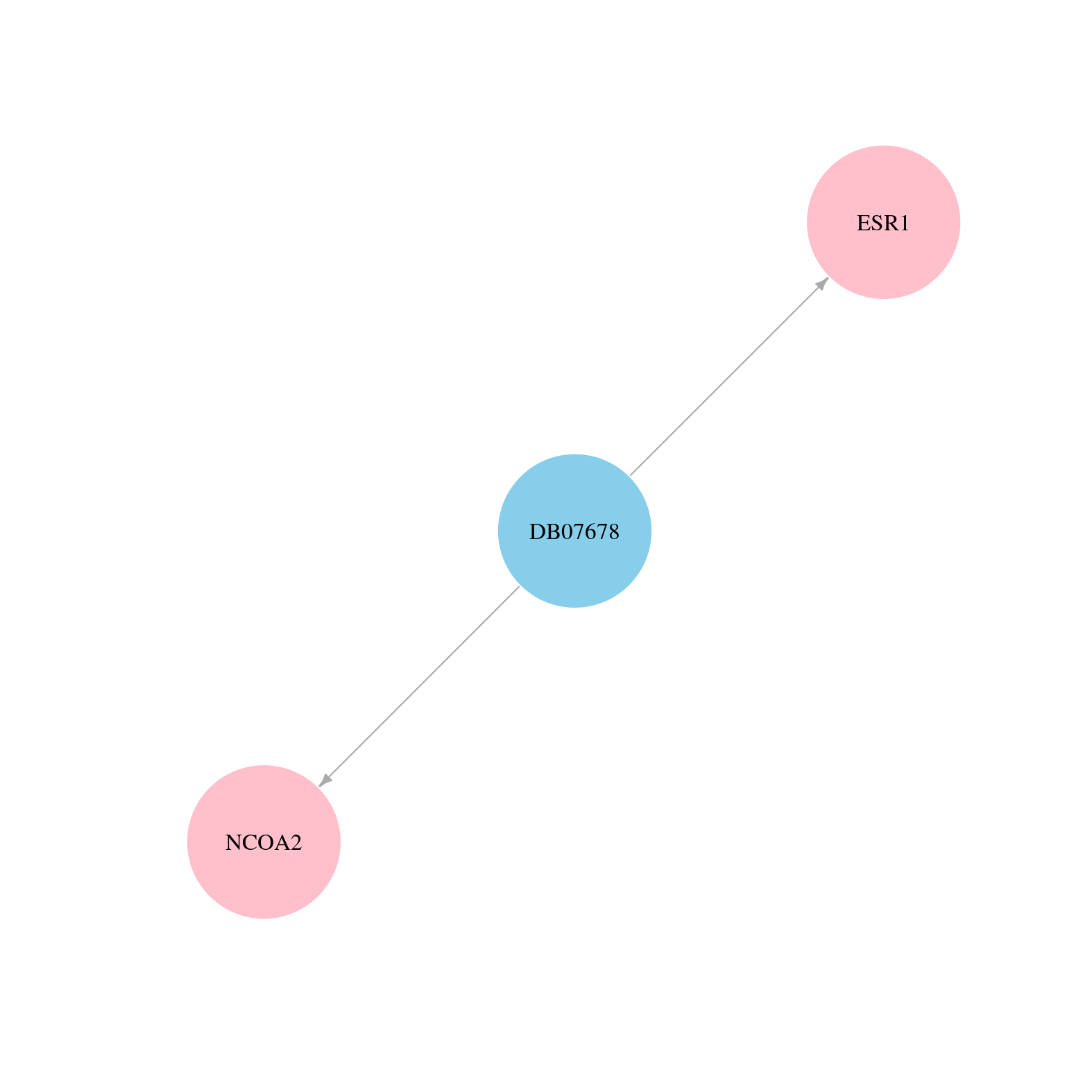
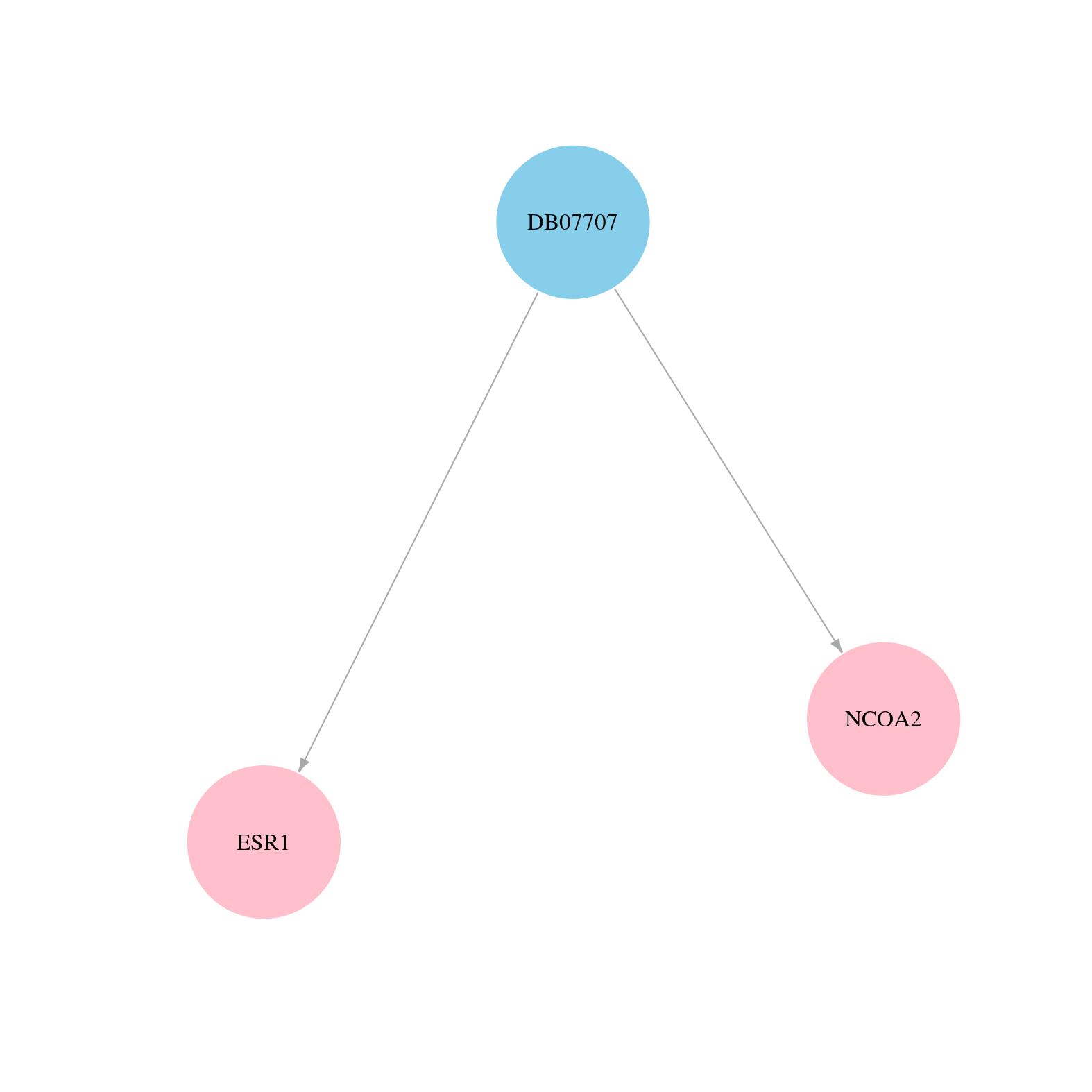
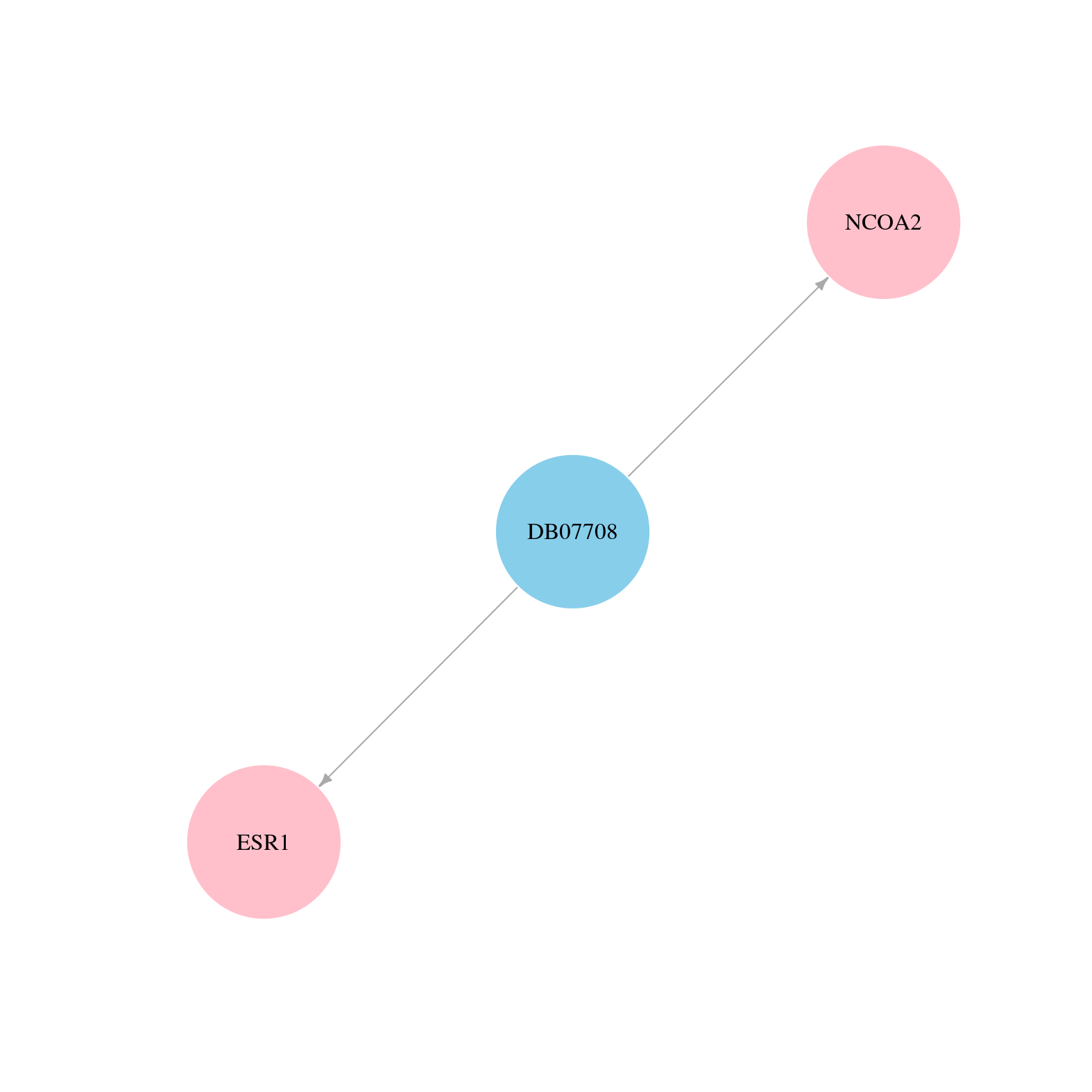
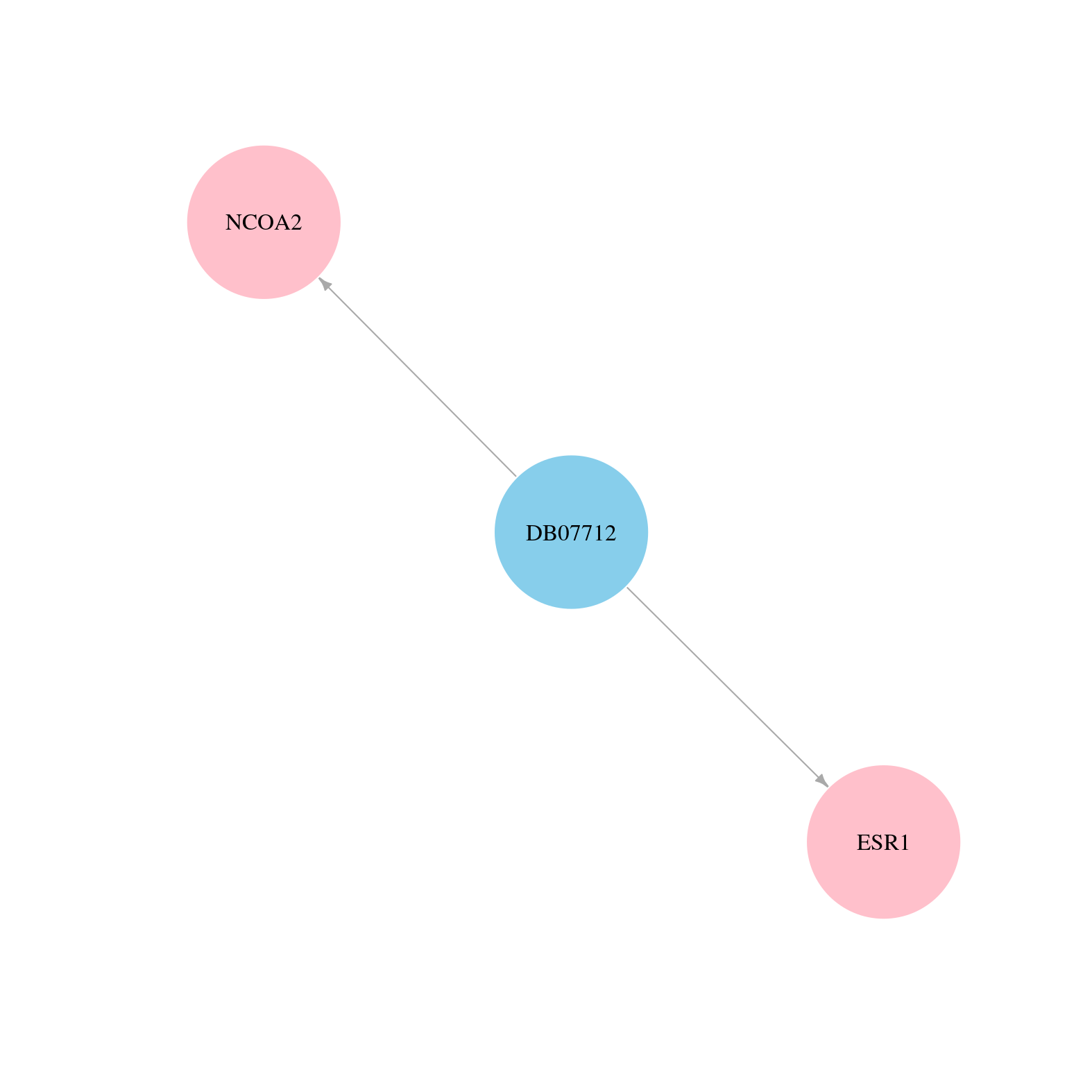
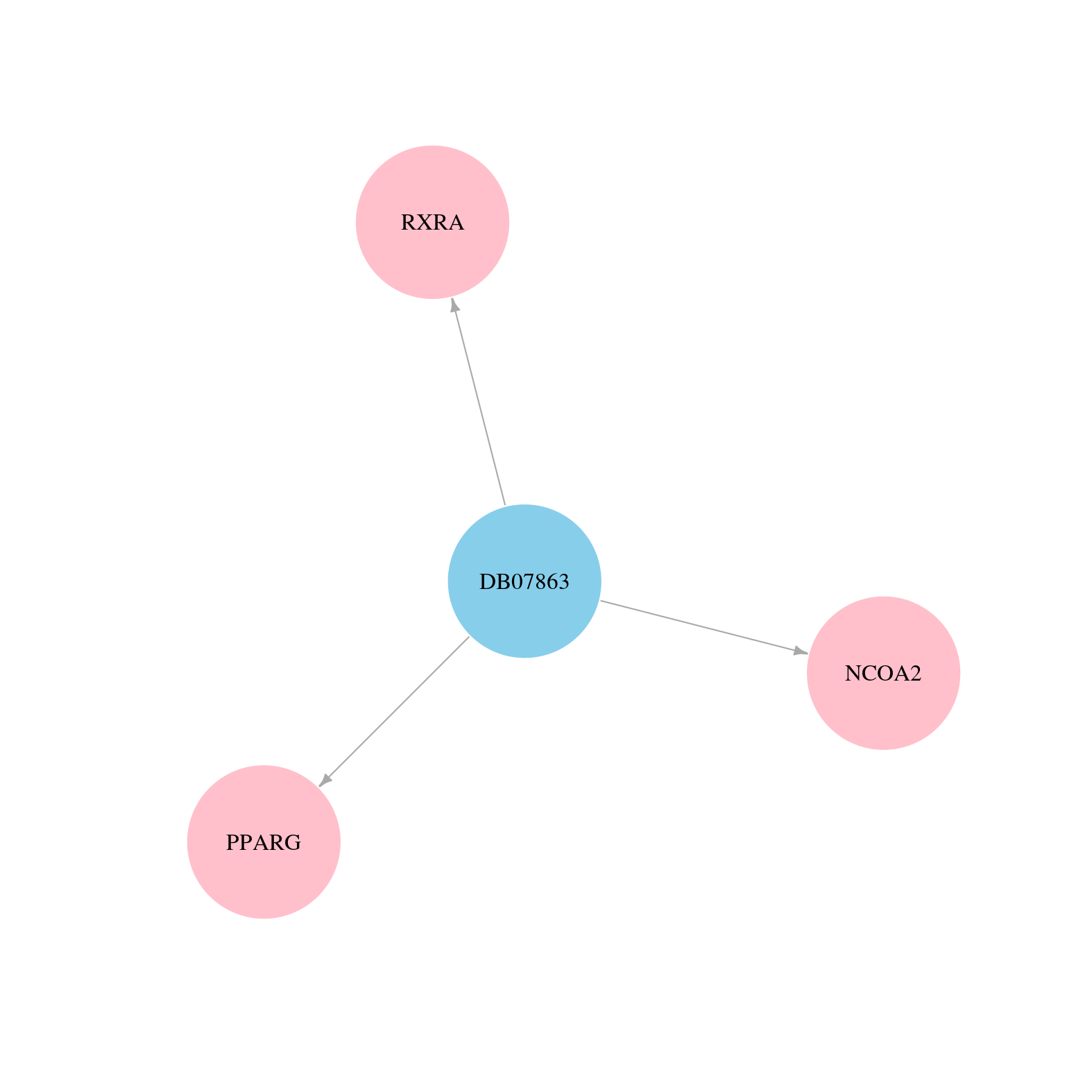
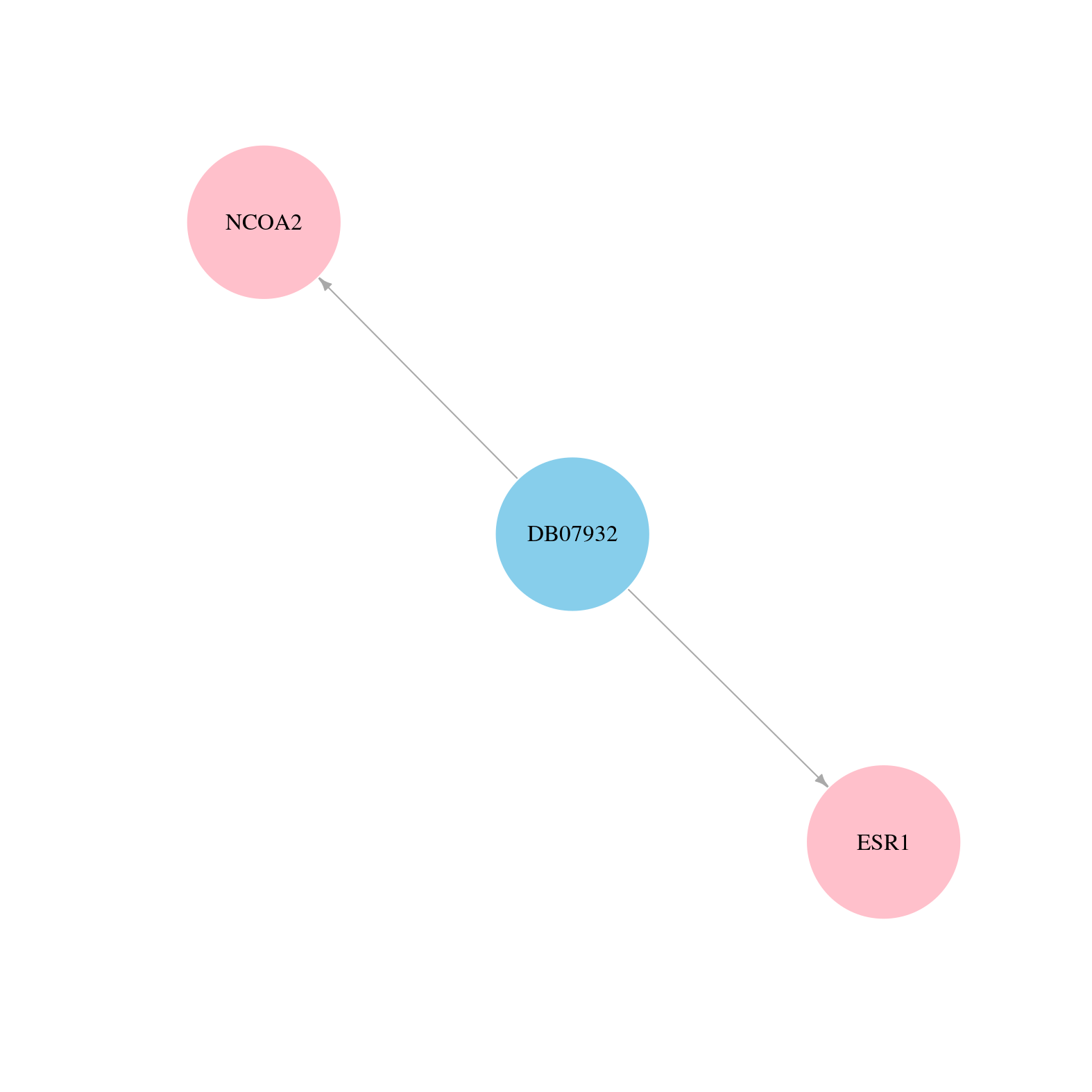
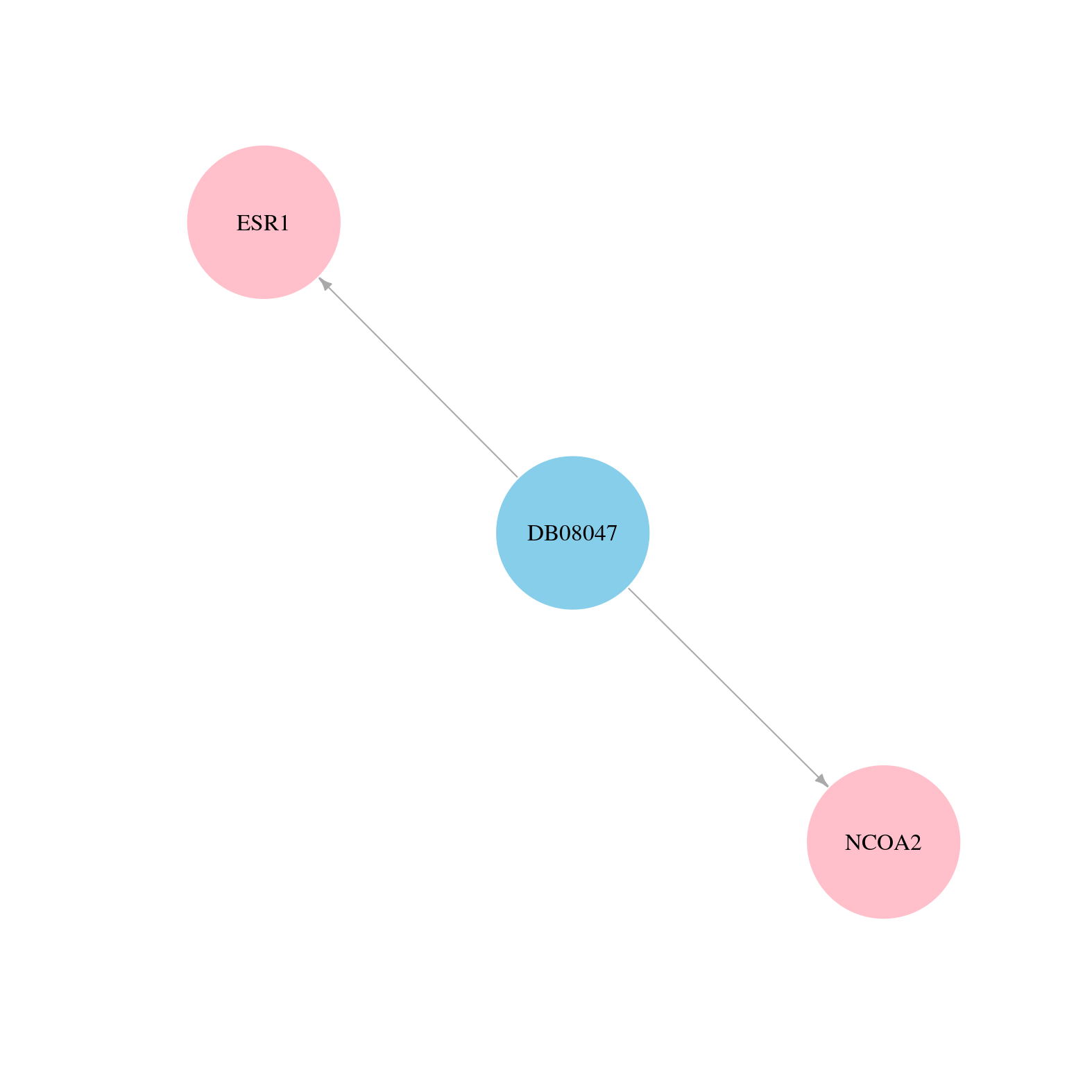
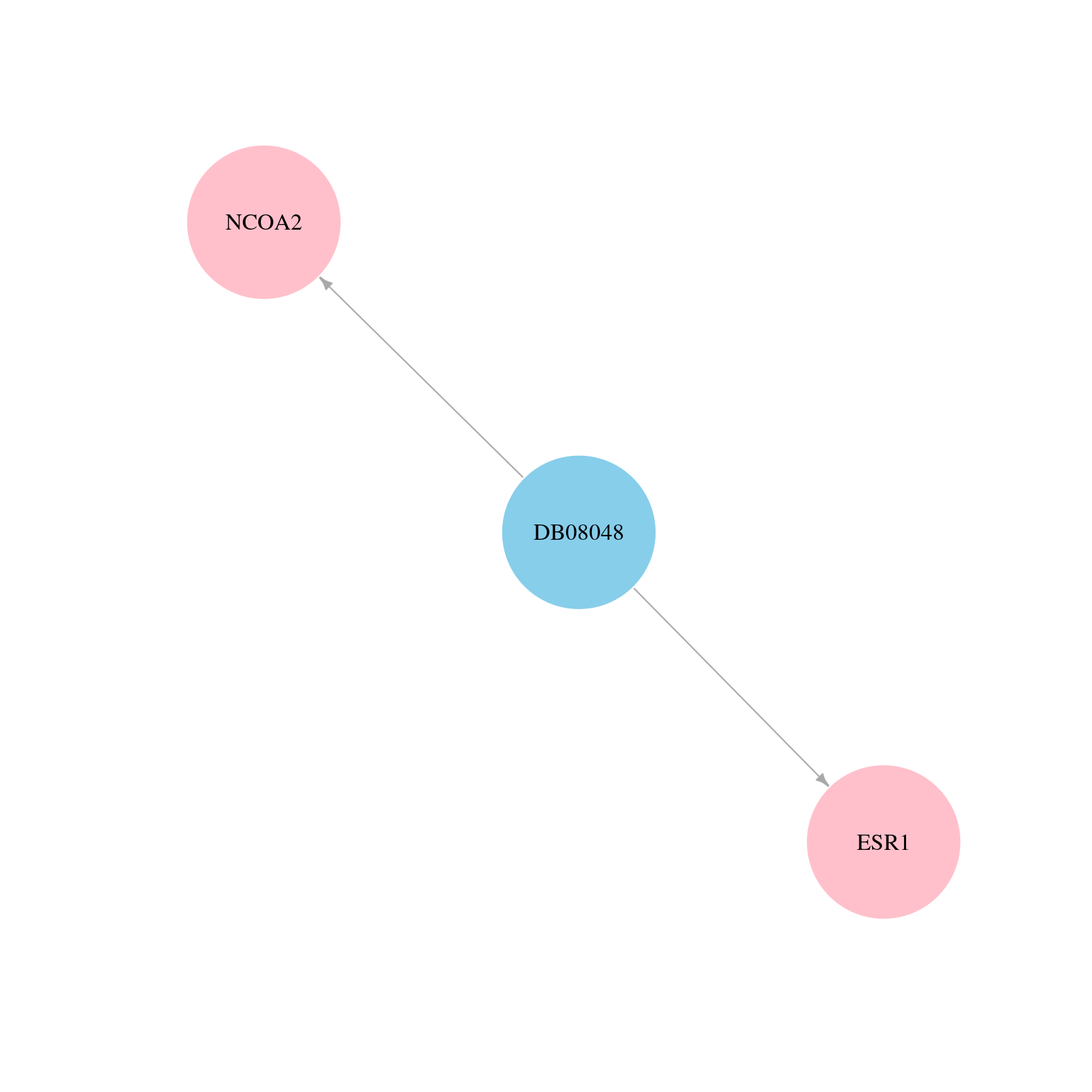
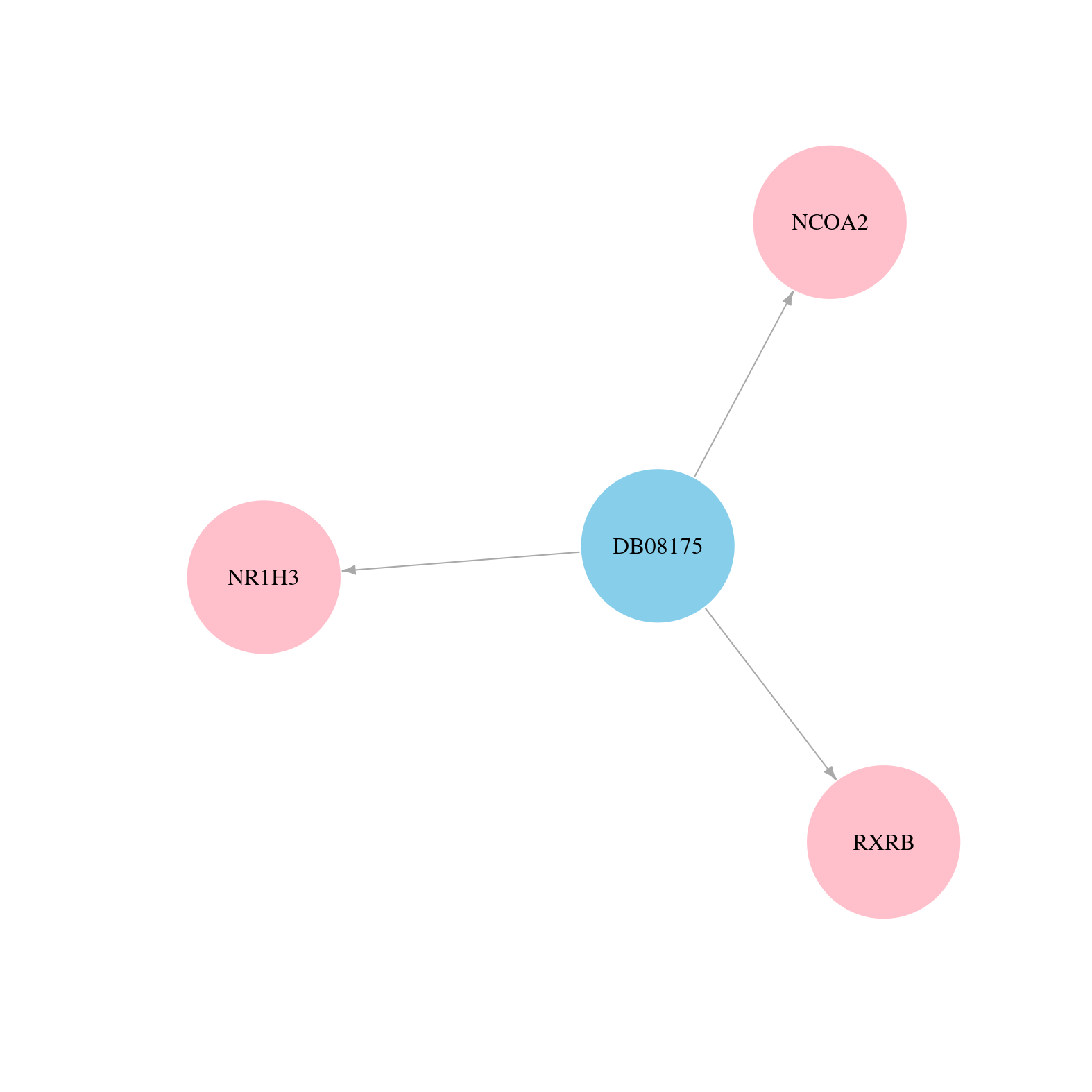
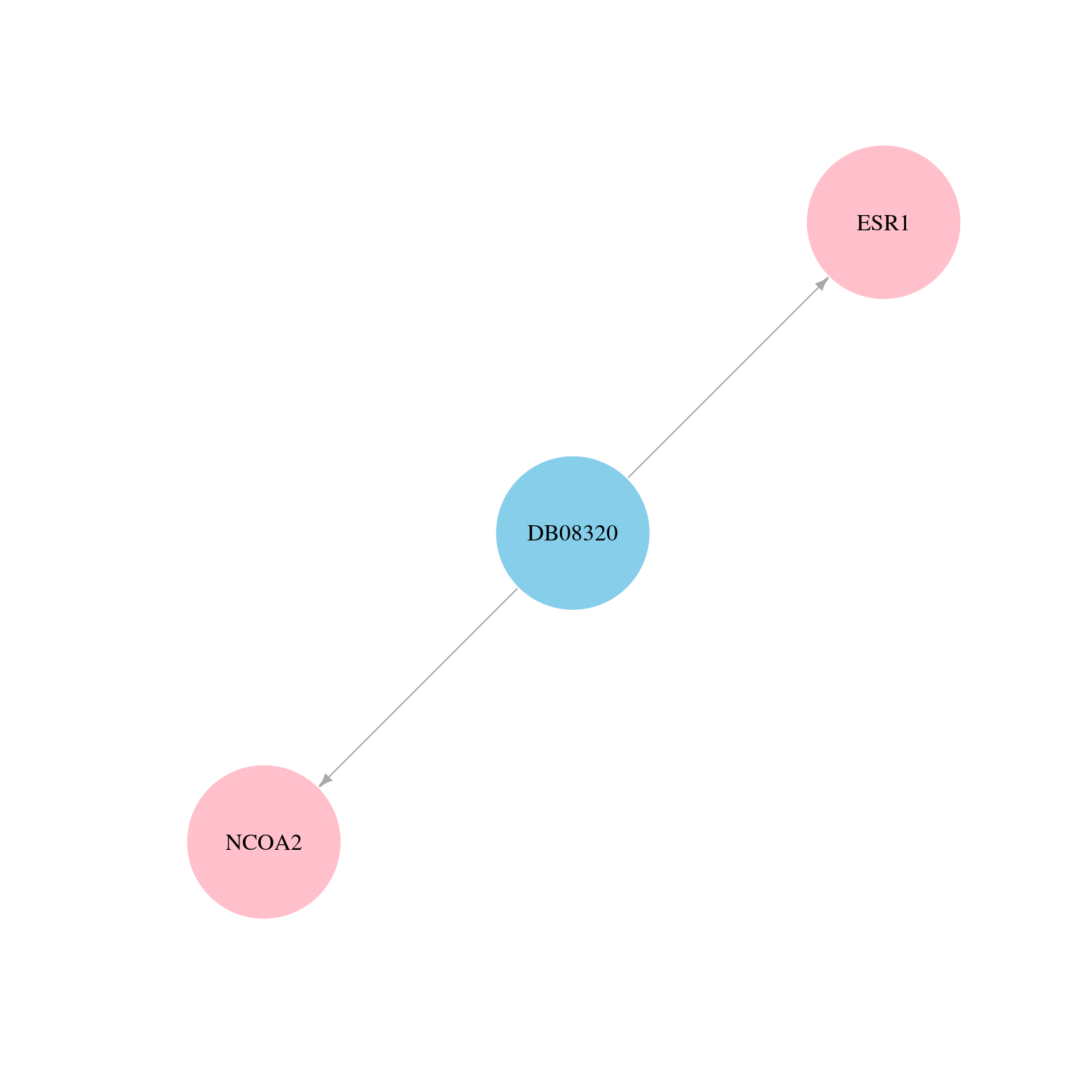
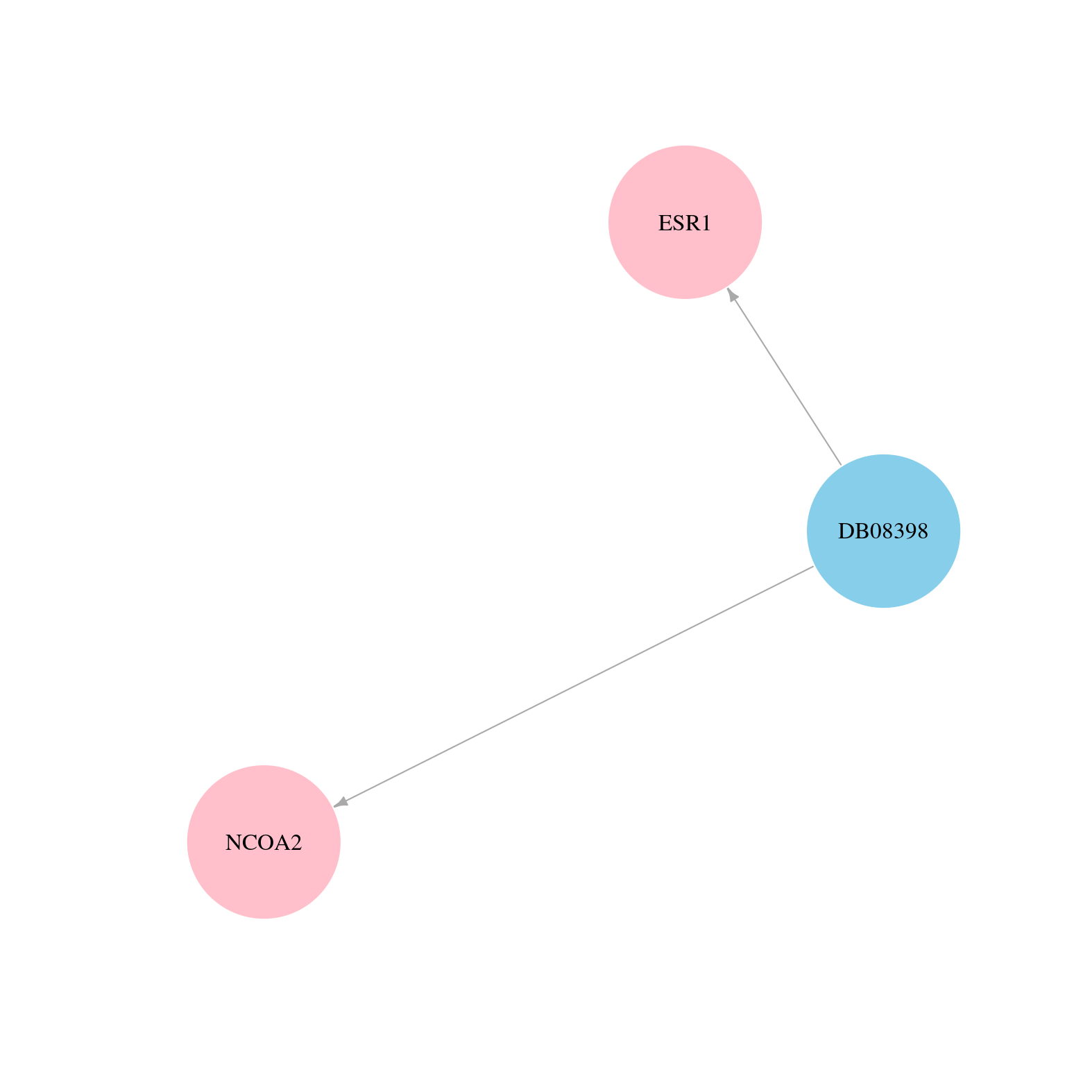
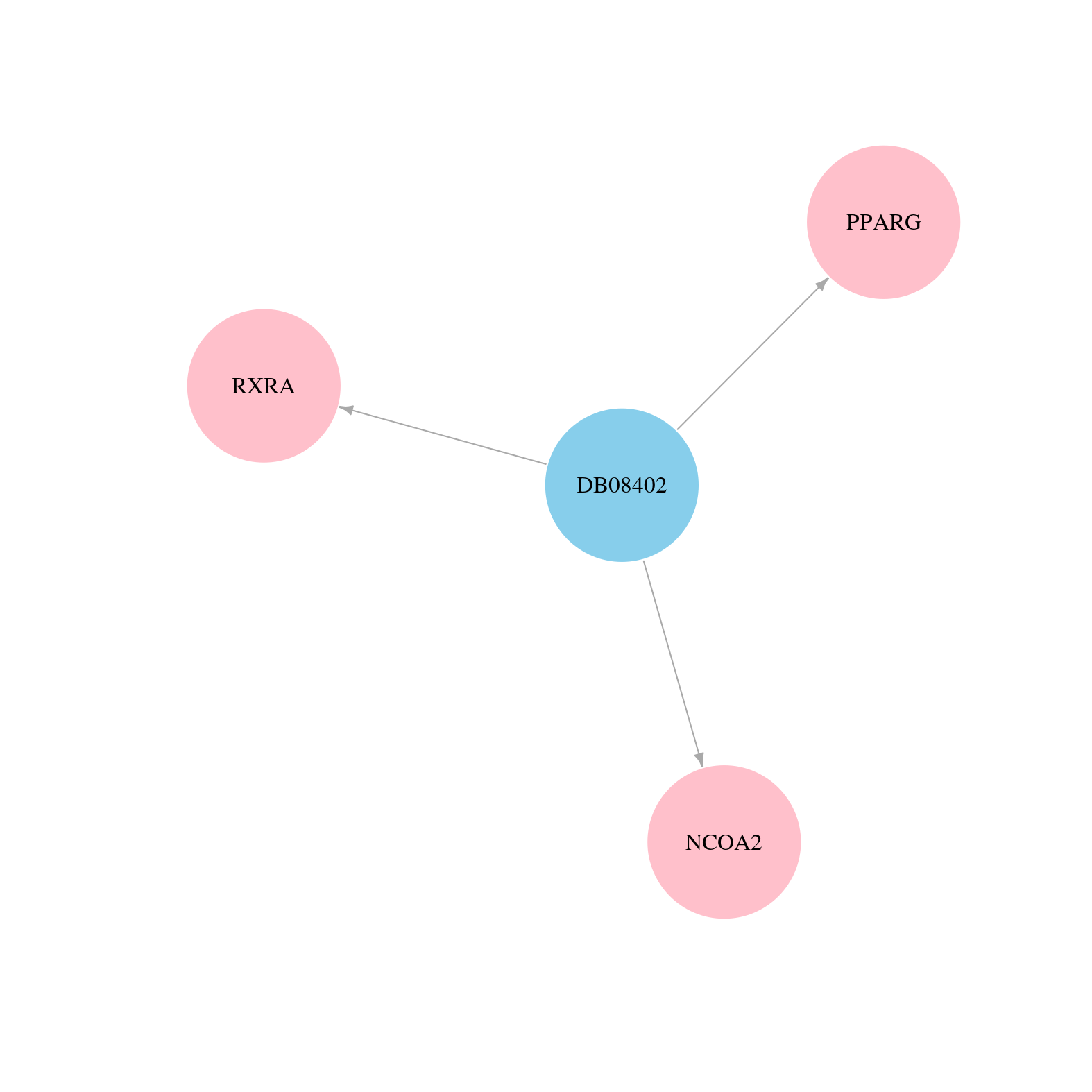
> Drugs from DrugBank database |
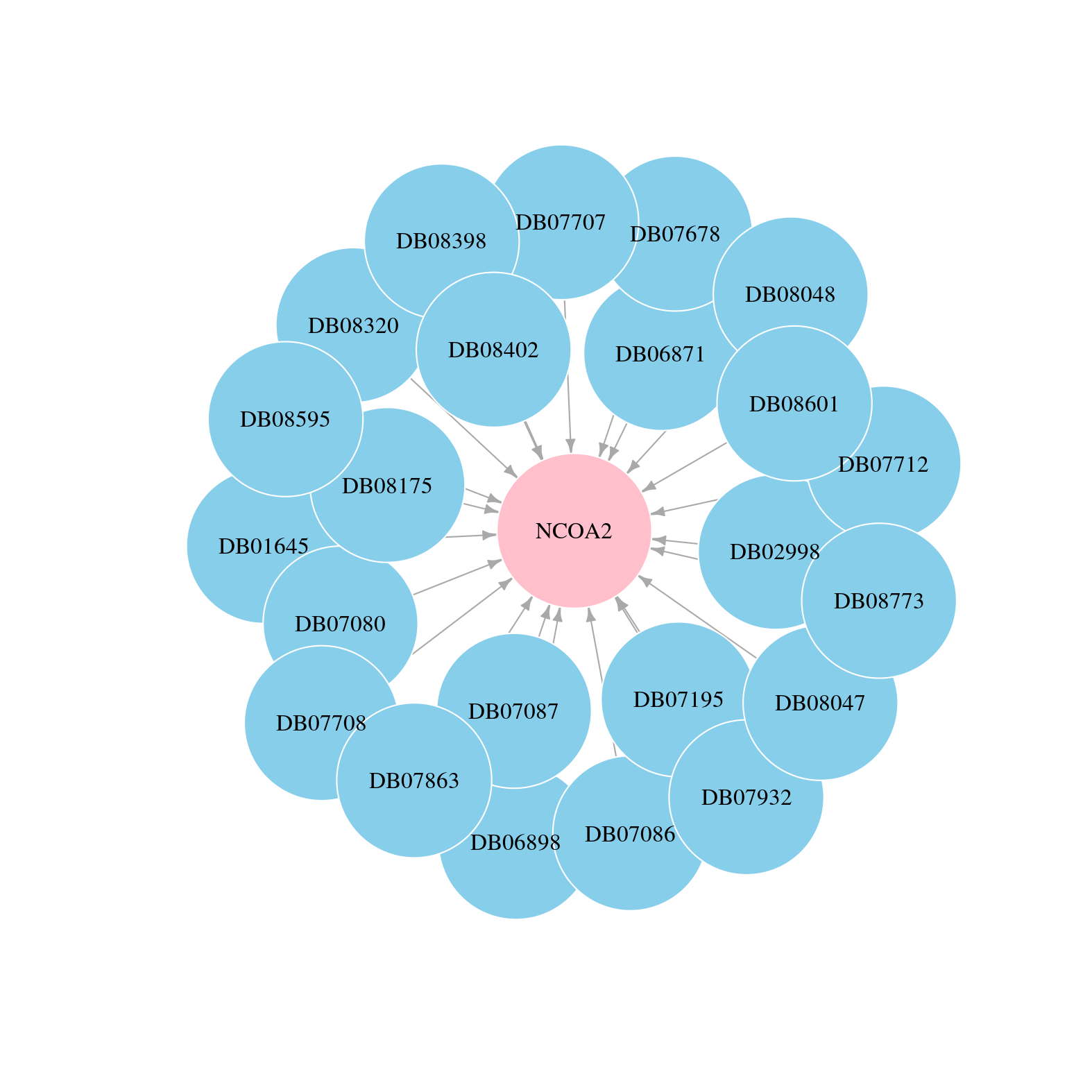
|
| Summary | |
|---|---|
| Symbol | NCOA2 |
| Name | nuclear receptor coactivator 2 |
| Aliases | TIF2; NCoA-2; KAT13C; bHLHe75; class E basic helix-loop-helix protein 75; glucocorticoid receptor-interactin ...... |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|