Browse NSD2 in pancancer
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00505 HMG (high mobility group) box PF00855 PWWP domain PF00856 SET domain |
||||||||||
| Function |
Histone methyltransferase with histone H3 'Lys-27' (H3K27me) methyltransferase activity. Isoform 2 may act as a transcription regulator that binds DNA and suppresses IL5 transcription through HDAC recruitment. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000018 regulation of DNA recombination GO:0000726 non-recombinational repair GO:0001501 skeletal system development GO:0002200 somatic diversification of immune receptors GO:0002204 somatic recombination of immunoglobulin genes involved in immune response GO:0002208 somatic diversification of immunoglobulins involved in immune response GO:0002250 adaptive immune response GO:0002263 cell activation involved in immune response GO:0002285 lymphocyte activation involved in immune response GO:0002312 B cell activation involved in immune response GO:0002366 leukocyte activation involved in immune response GO:0002377 immunoglobulin production GO:0002381 immunoglobulin production involved in immunoglobulin mediated immune response GO:0002440 production of molecular mediator of immune response GO:0002443 leukocyte mediated immunity GO:0002449 lymphocyte mediated immunity GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002562 somatic diversification of immune receptors via germline recombination within a single locus GO:0002637 regulation of immunoglobulin production GO:0002639 positive regulation of immunoglobulin production GO:0002694 regulation of leukocyte activation GO:0002696 positive regulation of leukocyte activation GO:0002697 regulation of immune effector process GO:0002699 positive regulation of immune effector process GO:0002700 regulation of production of molecular mediator of immune response GO:0002702 positive regulation of production of molecular mediator of immune response GO:0002703 regulation of leukocyte mediated immunity GO:0002705 positive regulation of leukocyte mediated immunity GO:0002706 regulation of lymphocyte mediated immunity GO:0002708 positive regulation of lymphocyte mediated immunity GO:0002712 regulation of B cell mediated immunity GO:0002714 positive regulation of B cell mediated immunity GO:0002819 regulation of adaptive immune response GO:0002821 positive regulation of adaptive immune response GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002824 positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002889 regulation of immunoglobulin mediated immune response GO:0002891 positive regulation of immunoglobulin mediated immune response GO:0003007 heart morphogenesis GO:0003149 membranous septum morphogenesis GO:0003205 cardiac chamber development GO:0003206 cardiac chamber morphogenesis GO:0003209 cardiac atrium morphogenesis GO:0003230 cardiac atrium development GO:0003231 cardiac ventricle development GO:0003279 cardiac septum development GO:0003281 ventricular septum development GO:0003283 atrial septum development GO:0003284 septum primum development GO:0003285 septum secundum development GO:0003289 atrial septum primum morphogenesis GO:0003290 atrial septum secundum morphogenesis GO:0006282 regulation of DNA repair GO:0006302 double-strand break repair GO:0006303 double-strand break repair via nonhomologous end joining GO:0006310 DNA recombination GO:0006479 protein methylation GO:0007507 heart development GO:0008213 protein alkylation GO:0010452 histone H3-K36 methylation GO:0016064 immunoglobulin mediated immune response GO:0016444 somatic cell DNA recombination GO:0016445 somatic diversification of immunoglobulins GO:0016447 somatic recombination of immunoglobulin gene segments GO:0016570 histone modification GO:0016571 histone methylation GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0019724 B cell mediated immunity GO:0032259 methylation GO:0034770 histone H4-K20 methylation GO:0034968 histone lysine methylation GO:0042113 B cell activation GO:0043414 macromolecule methylation GO:0045190 isotype switching GO:0045191 regulation of isotype switching GO:0045830 positive regulation of isotype switching GO:0045911 positive regulation of DNA recombination GO:0048290 isotype switching to IgA isotypes GO:0048296 regulation of isotype switching to IgA isotypes GO:0048298 positive regulation of isotype switching to IgA isotypes GO:0050864 regulation of B cell activation GO:0050865 regulation of cell activation GO:0050867 positive regulation of cell activation GO:0050871 positive regulation of B cell activation GO:0051052 regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051249 regulation of lymphocyte activation GO:0051251 positive regulation of lymphocyte activation GO:0060348 bone development GO:0060411 cardiac septum morphogenesis GO:0060412 ventricular septum morphogenesis GO:0060413 atrial septum morphogenesis GO:2000779 regulation of double-strand break repair GO:2001020 regulation of response to DNA damage stimulus GO:2001032 regulation of double-strand break repair via nonhomologous end joining |
| Molecular Function |
GO:0003682 chromatin binding GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018024 histone-lysine N-methyltransferase activity GO:0042054 histone methyltransferase activity GO:0042799 histone methyltransferase activity (H4-K20 specific) |
| Cellular Component | - |
| KEGG |
hsa00310 Lysine degradation |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for NSD2. |
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
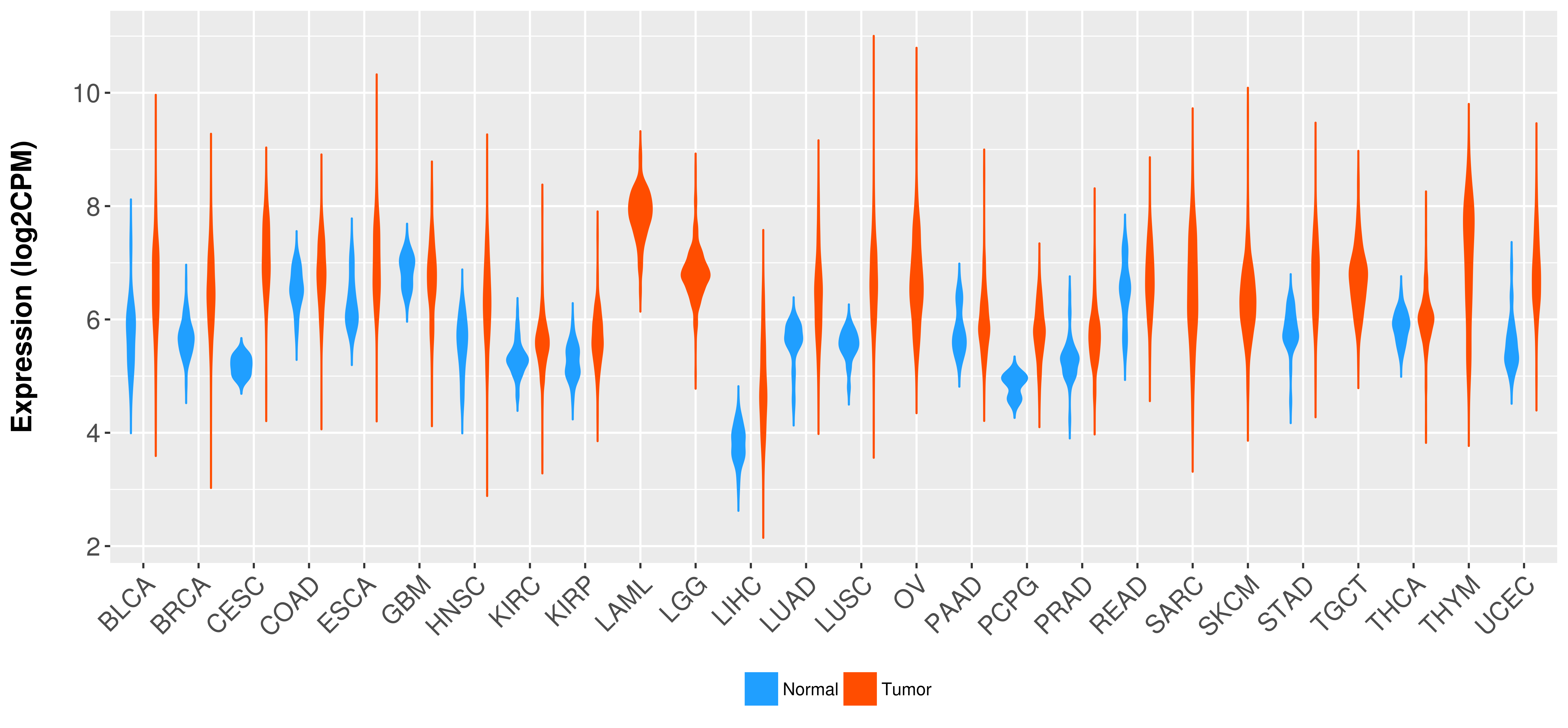
Differential expression analysis for cancers with more than 10 normal samples
|
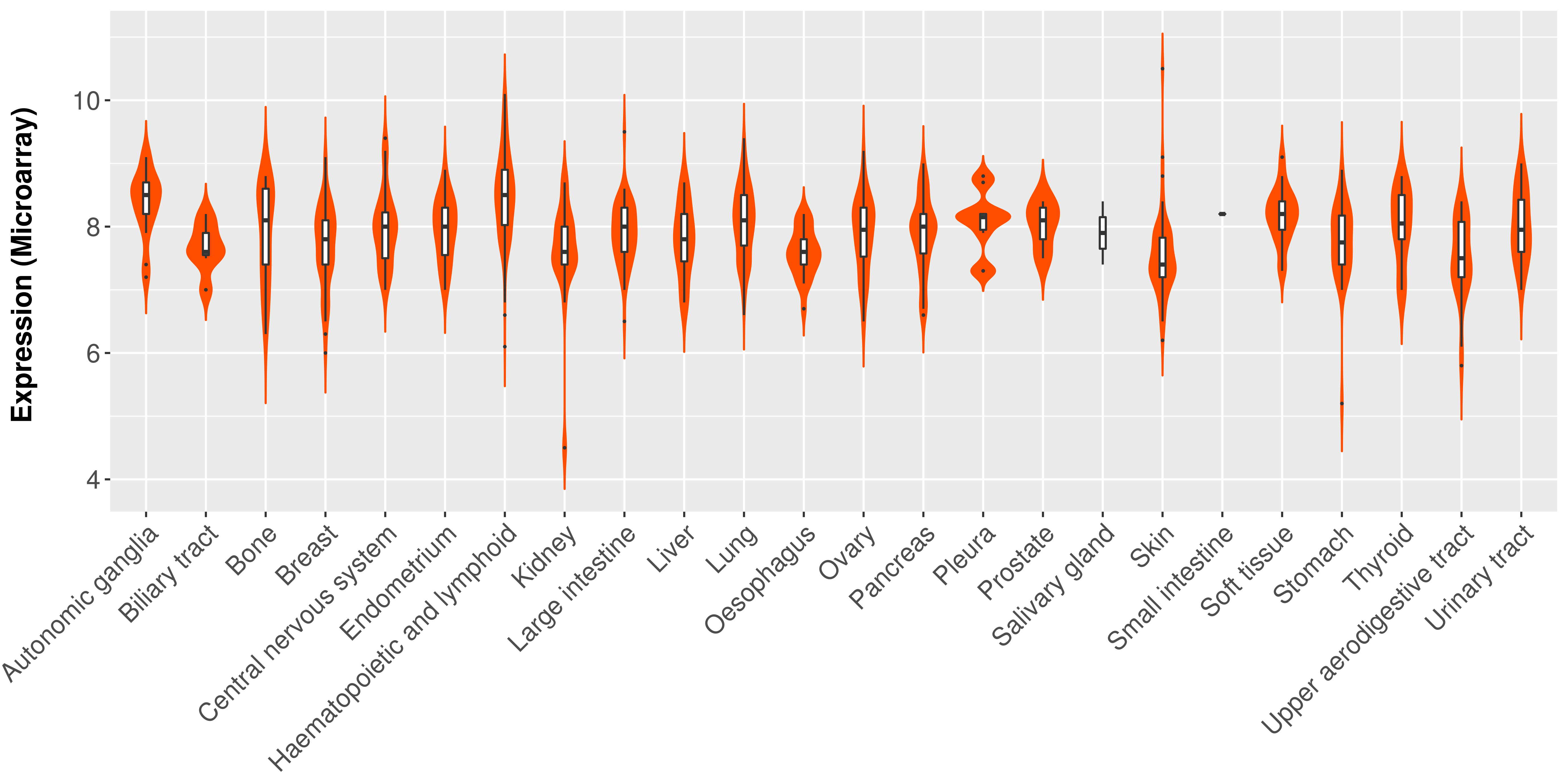
|
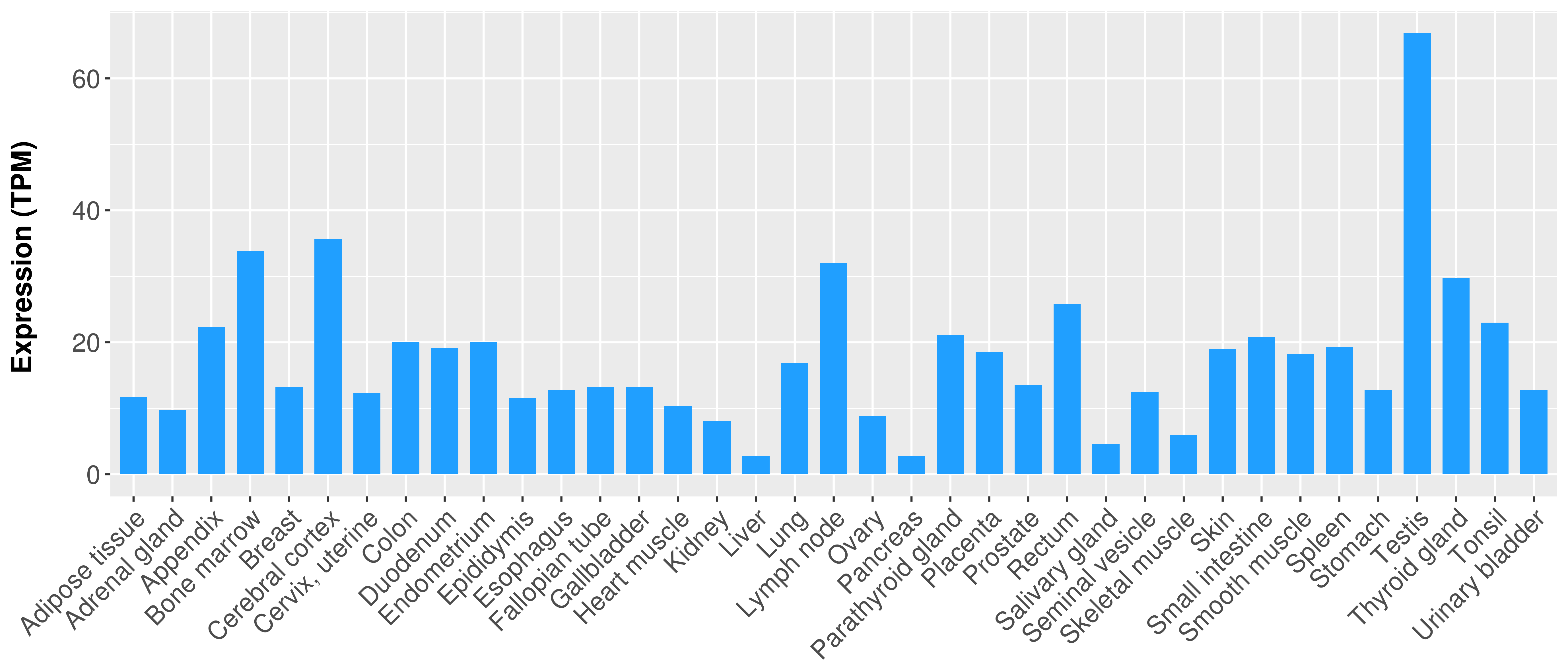
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
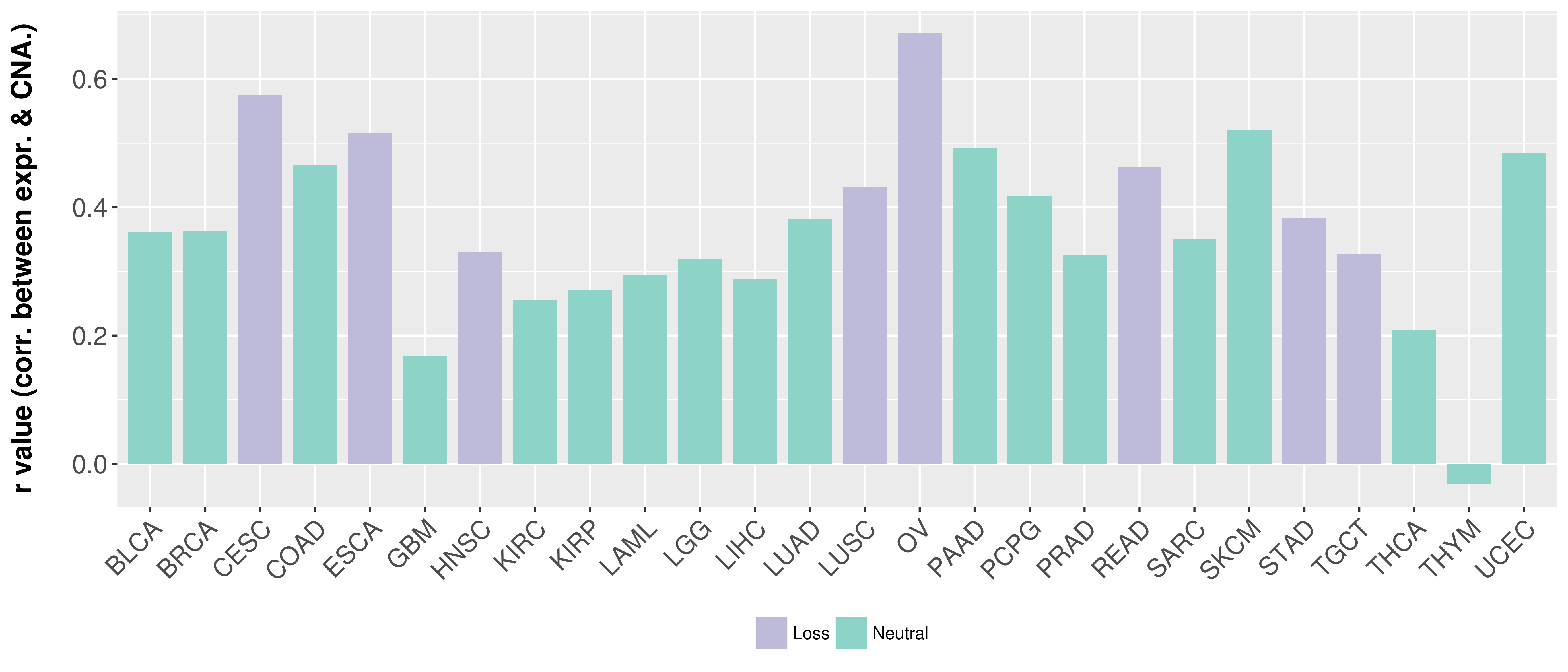
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
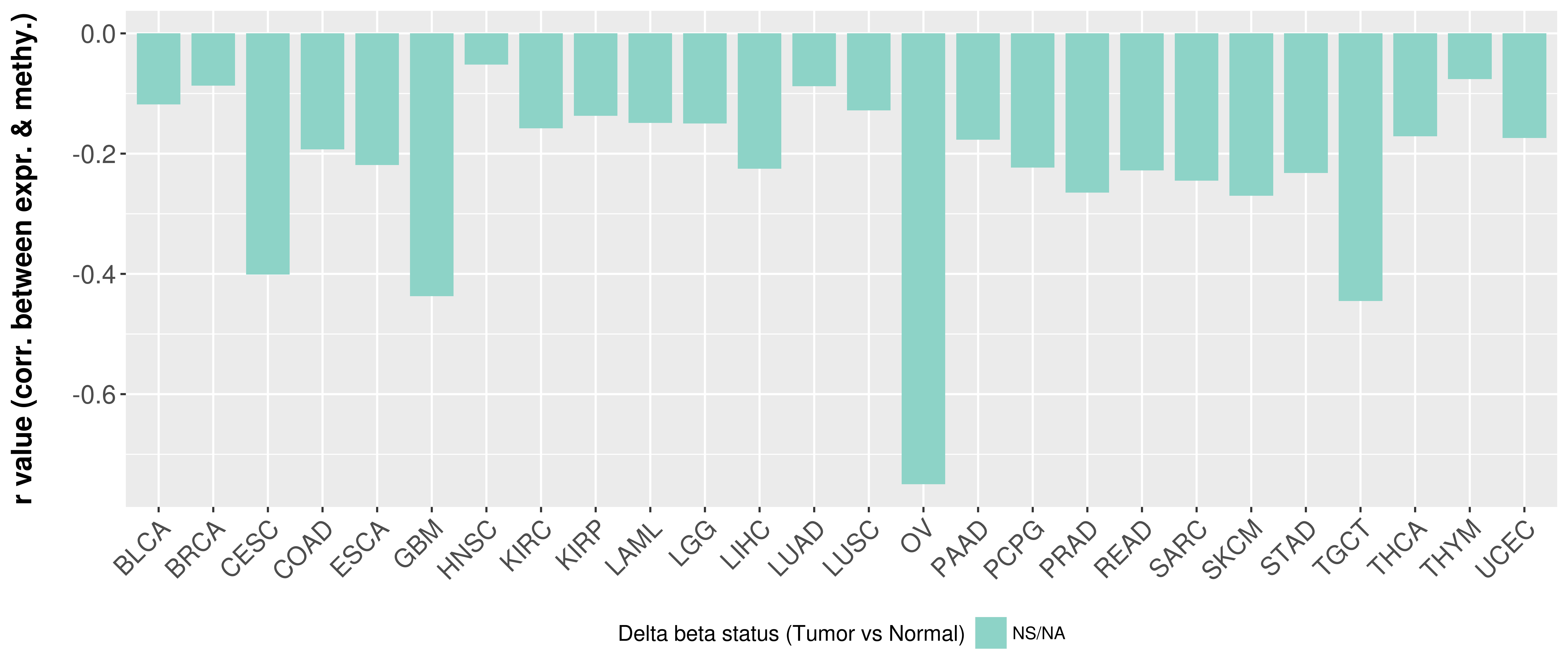
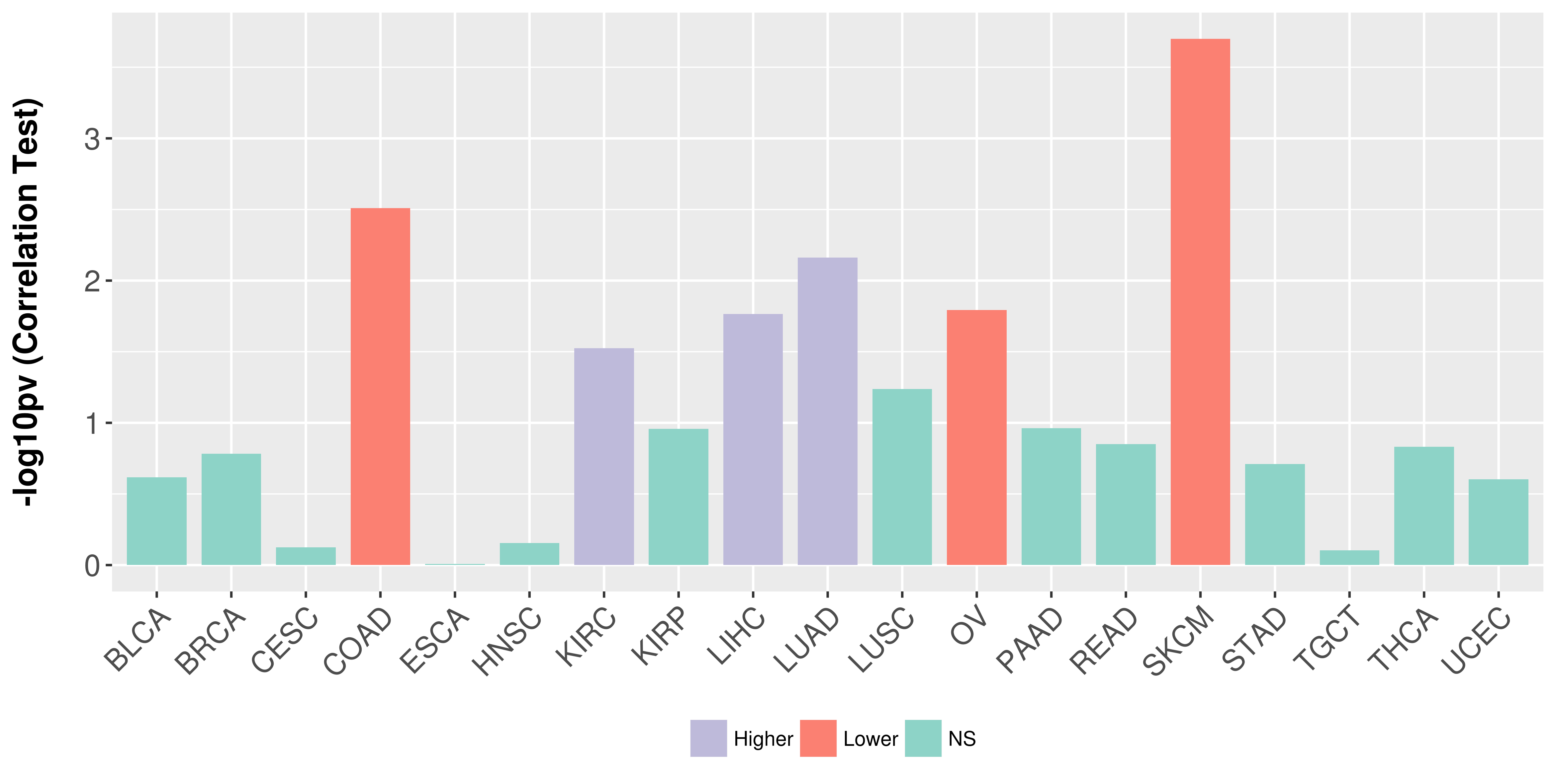
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |

|
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
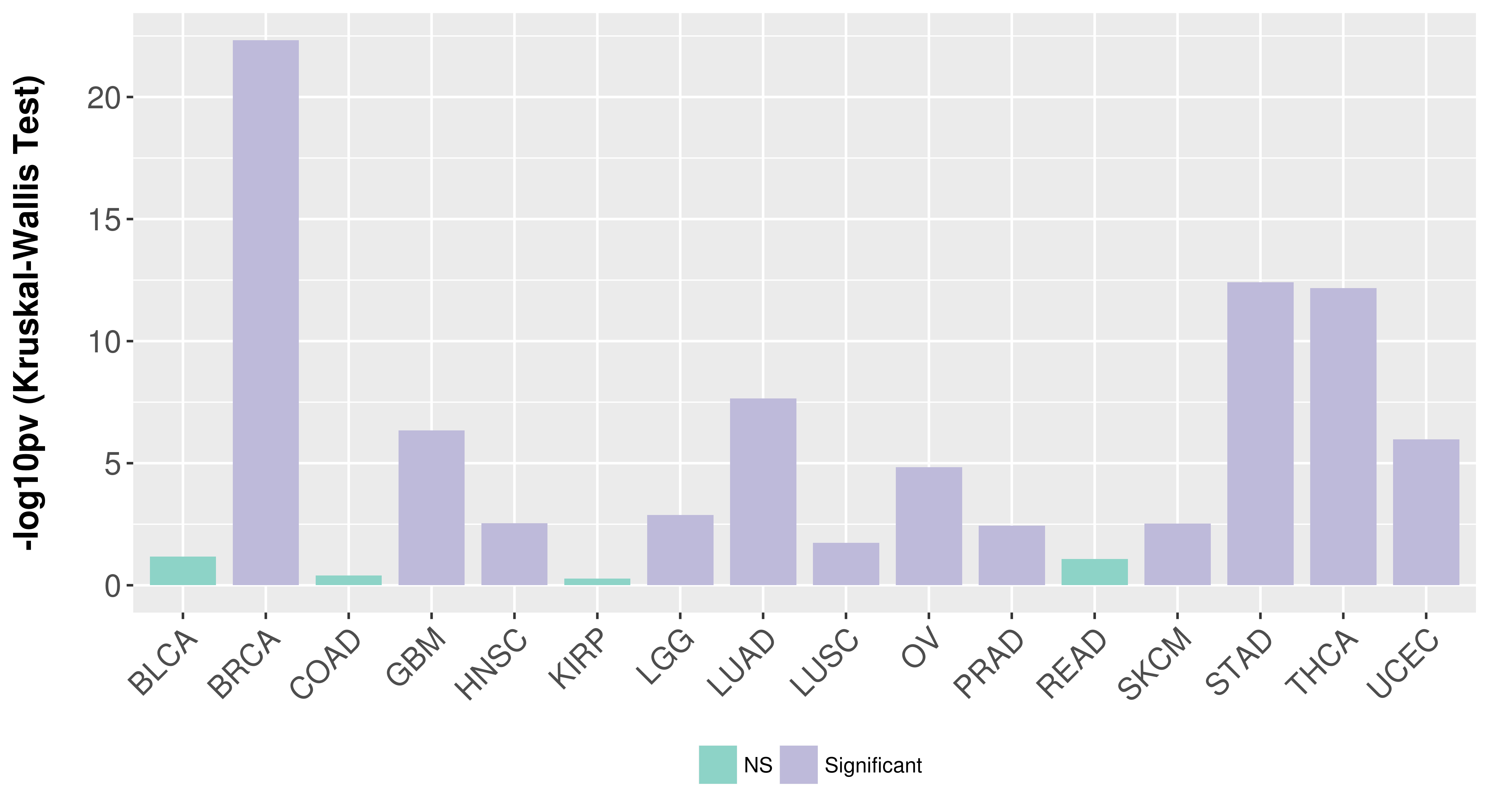
Association between expresson and subtype.
|
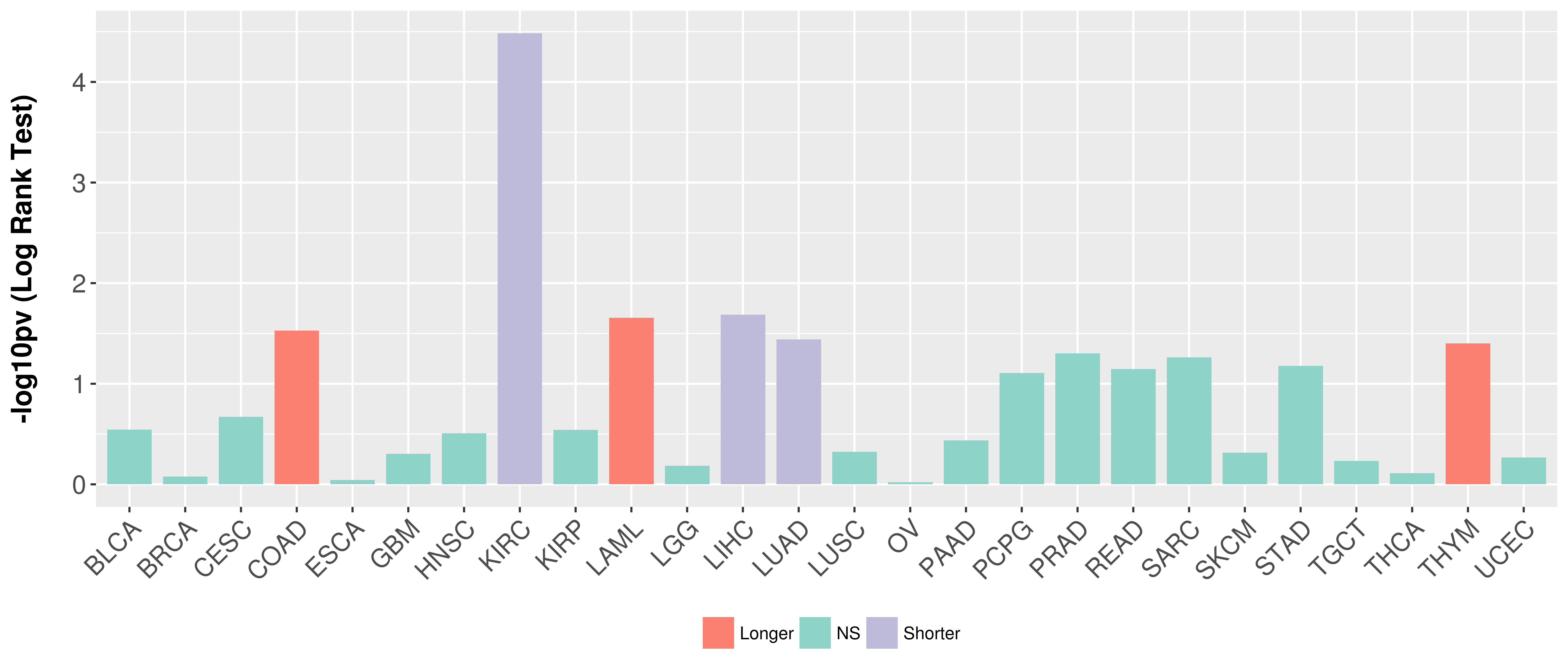
Overall survival analysis based on expression.
|
Association between expresson and stage.
|
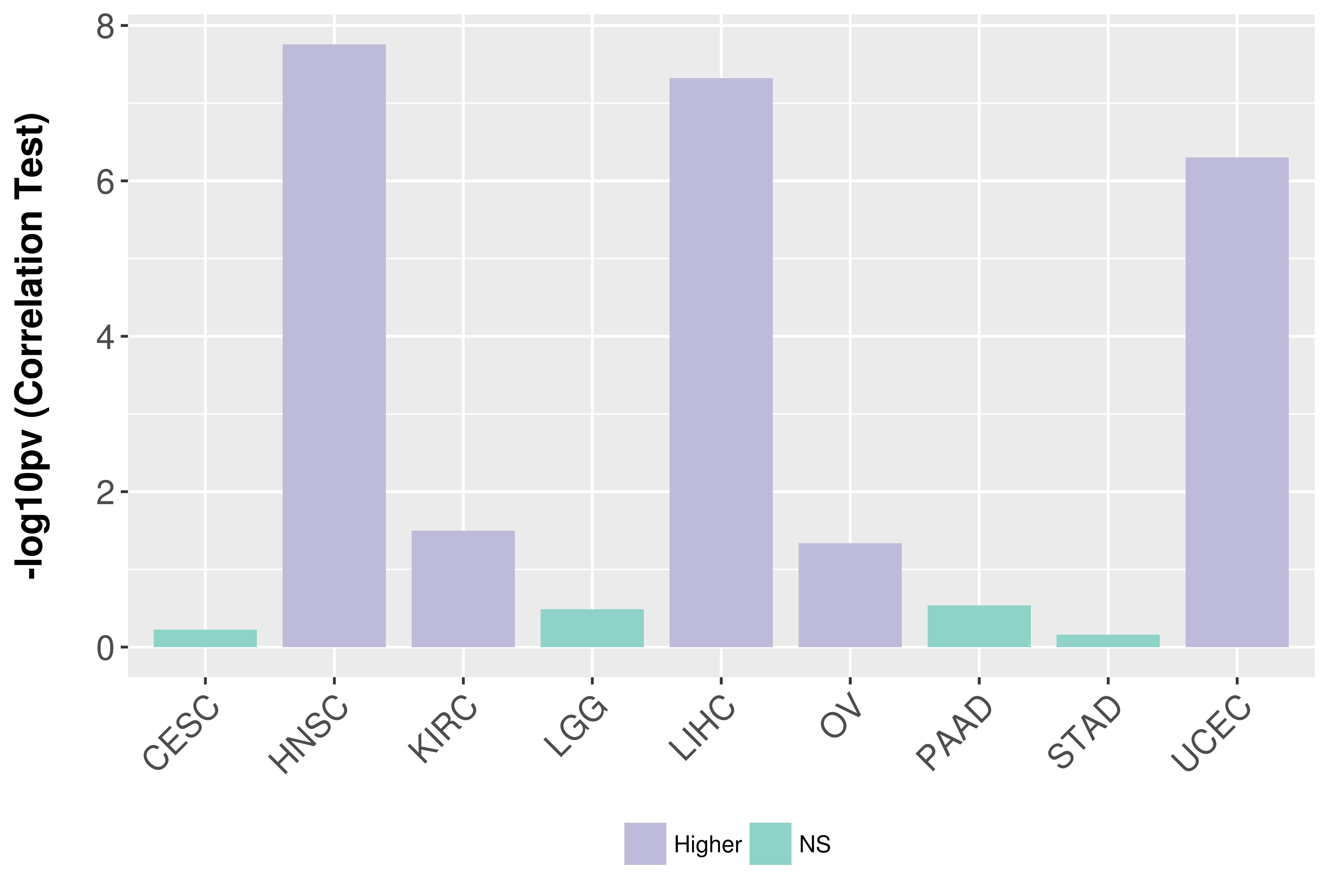
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for NSD2. |
| Summary | |
|---|---|
| Symbol | NSD2 |
| Name | nuclear receptor binding SET domain protein 2 |
| Aliases | |
| Location | 4p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for NSD2. |
| There is no record for NSD2. |