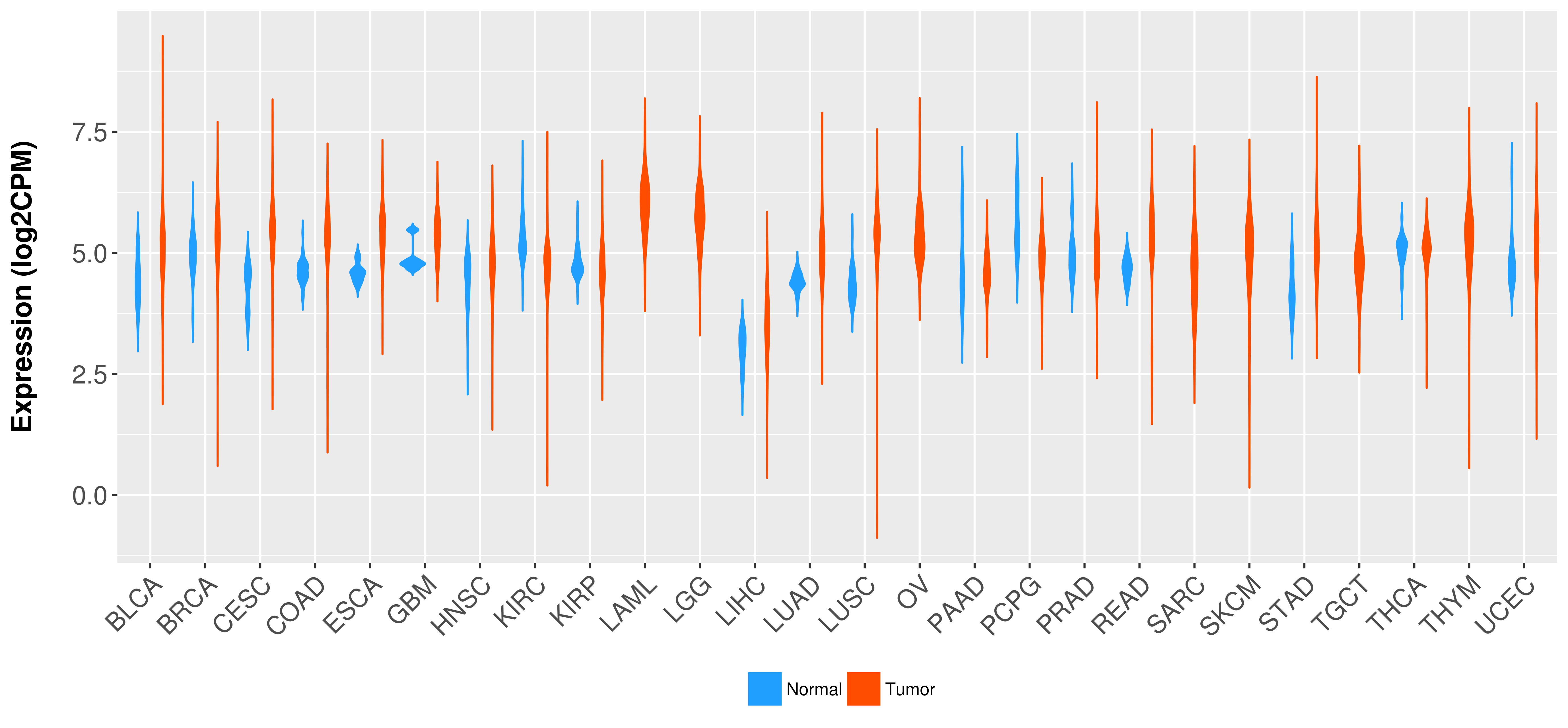
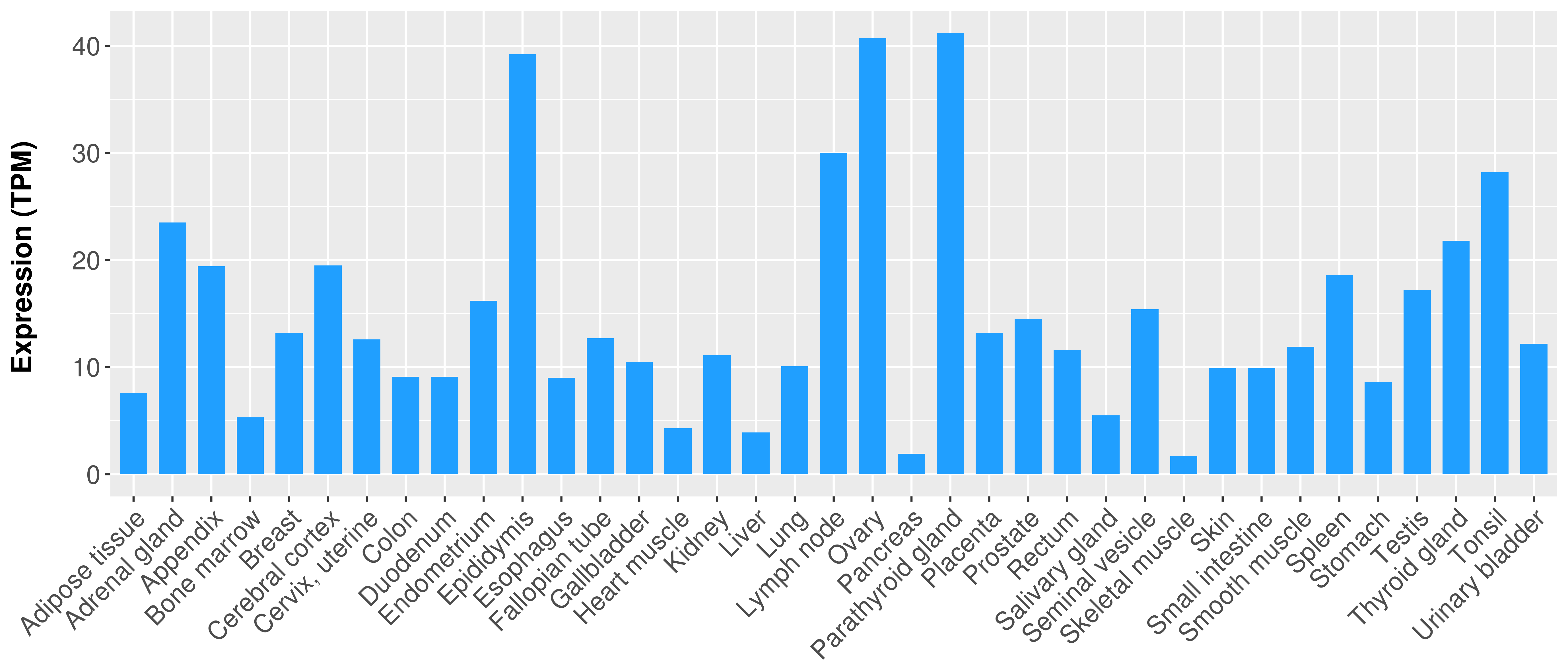
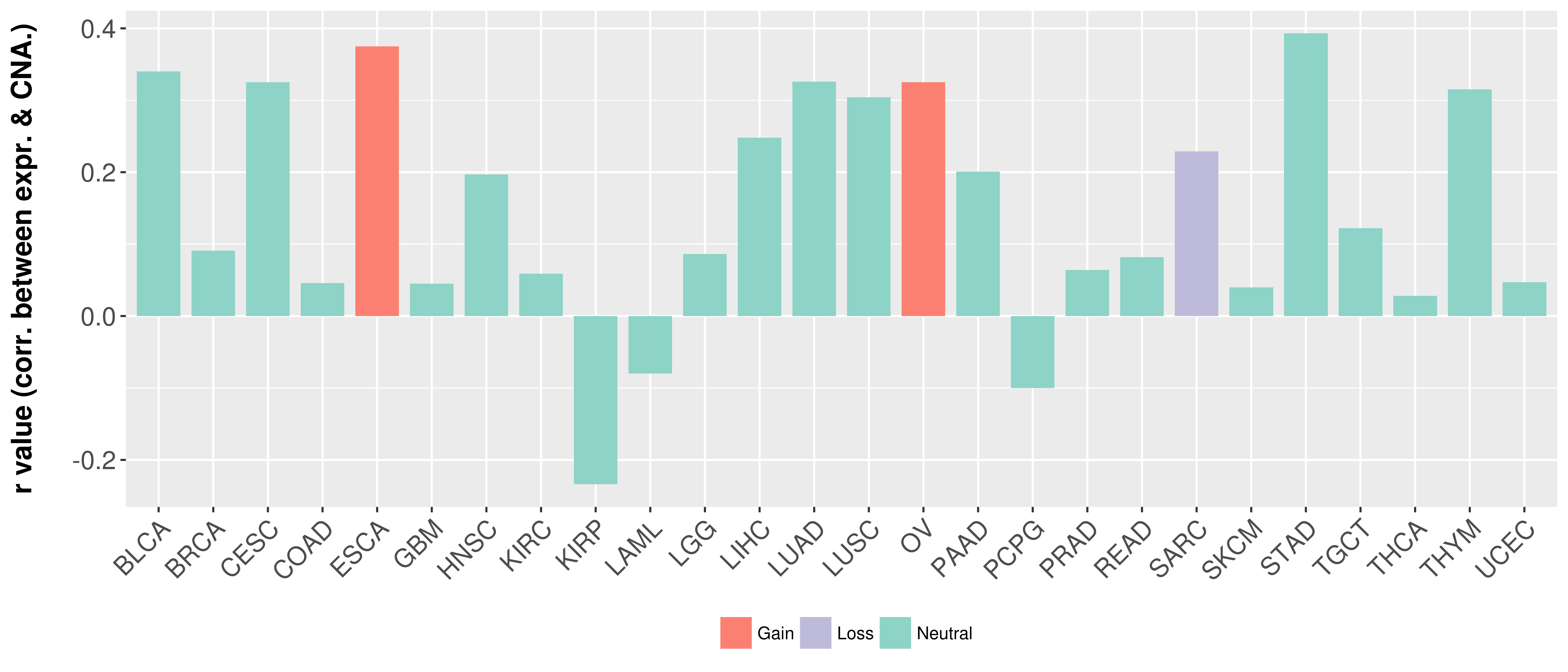
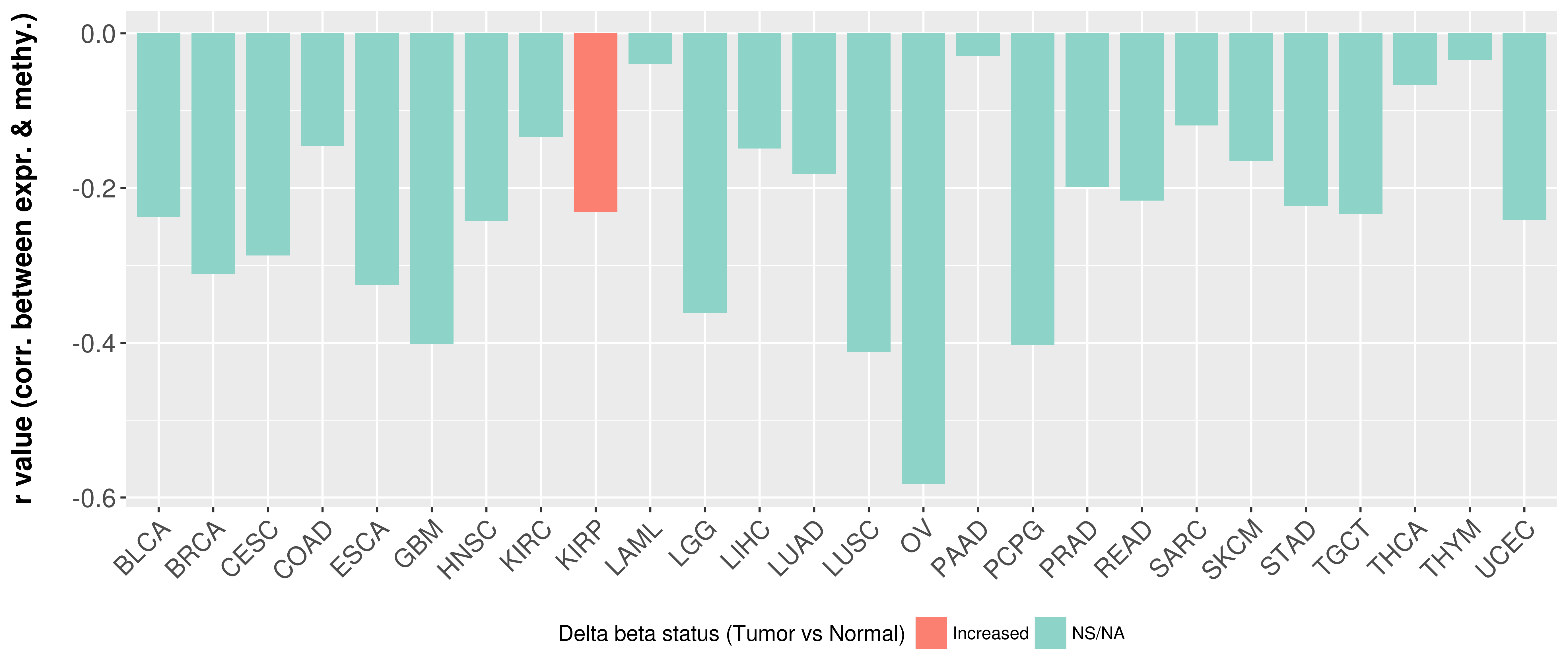
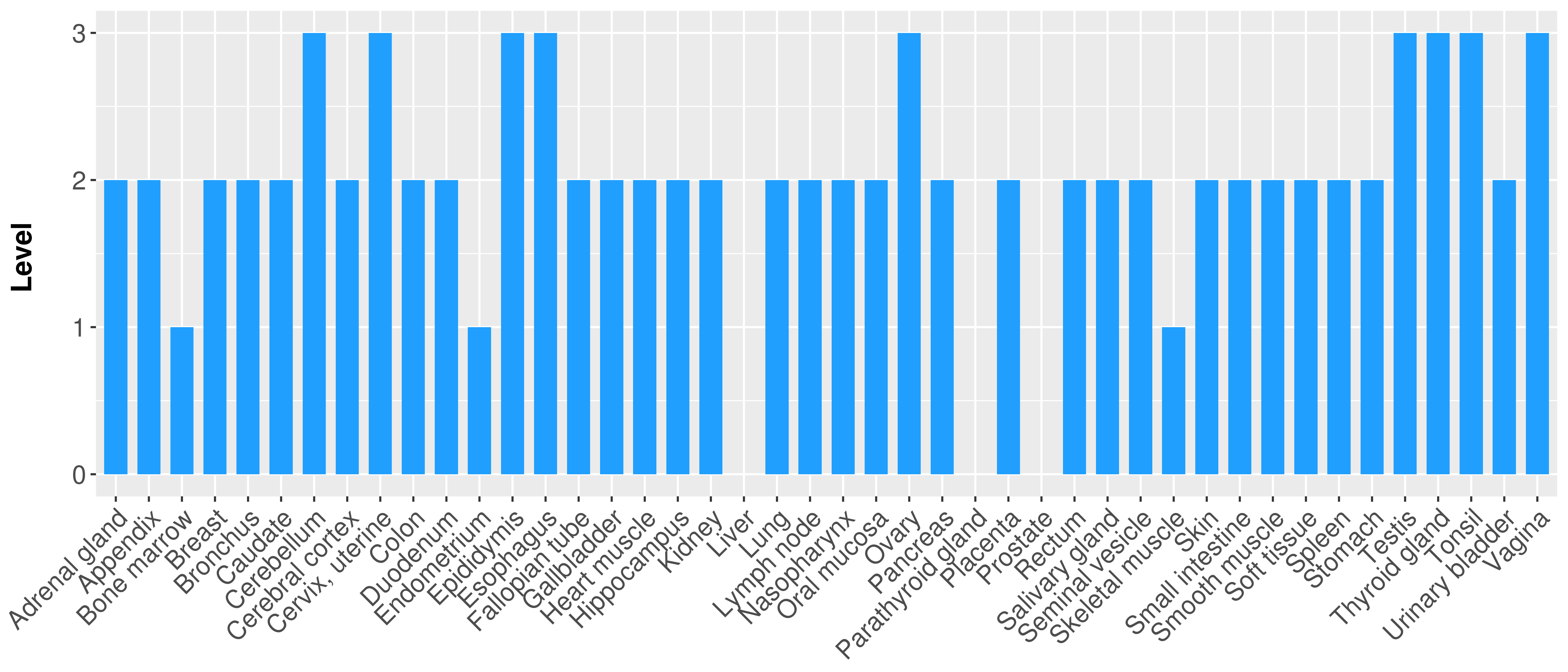
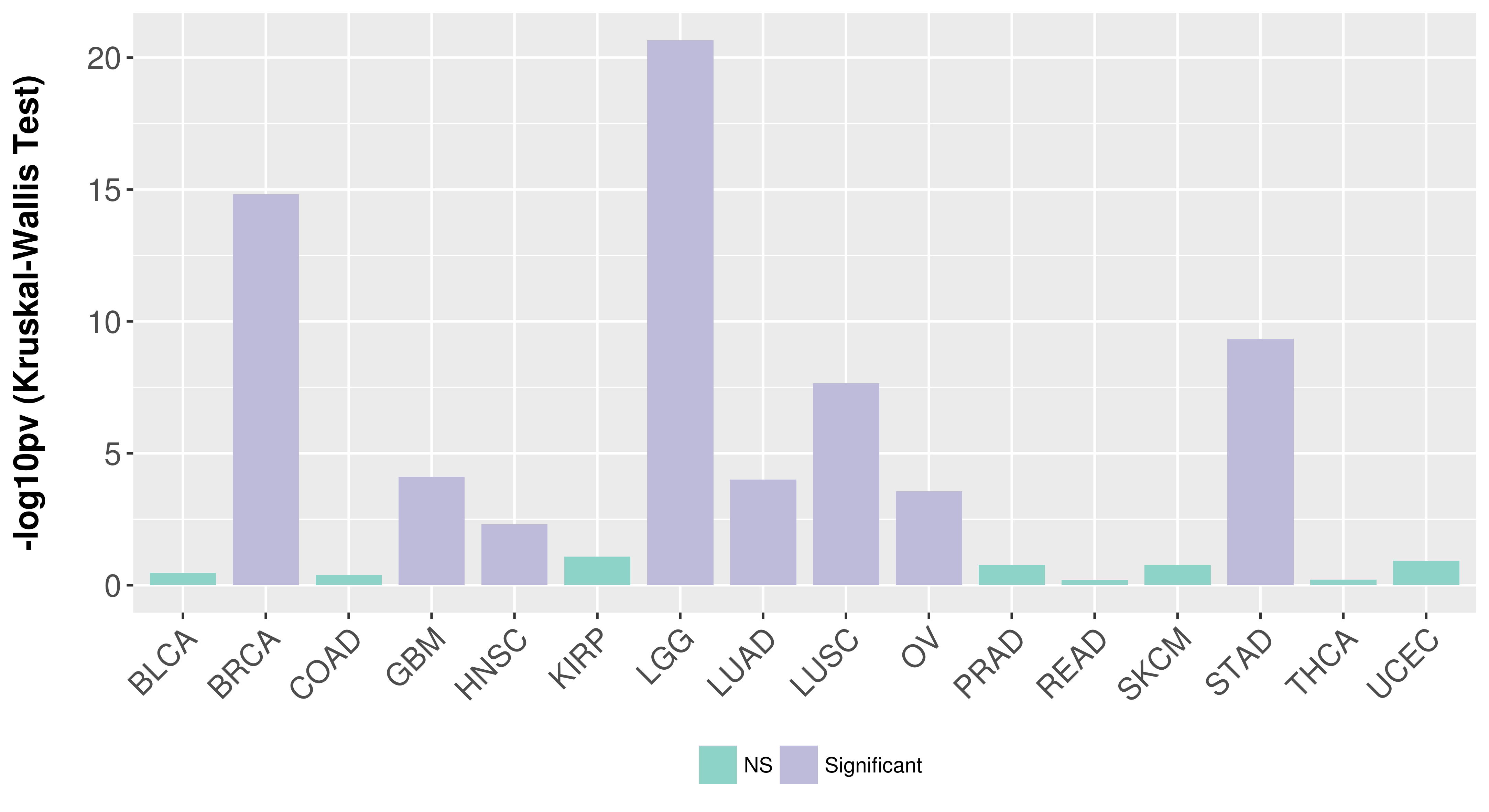
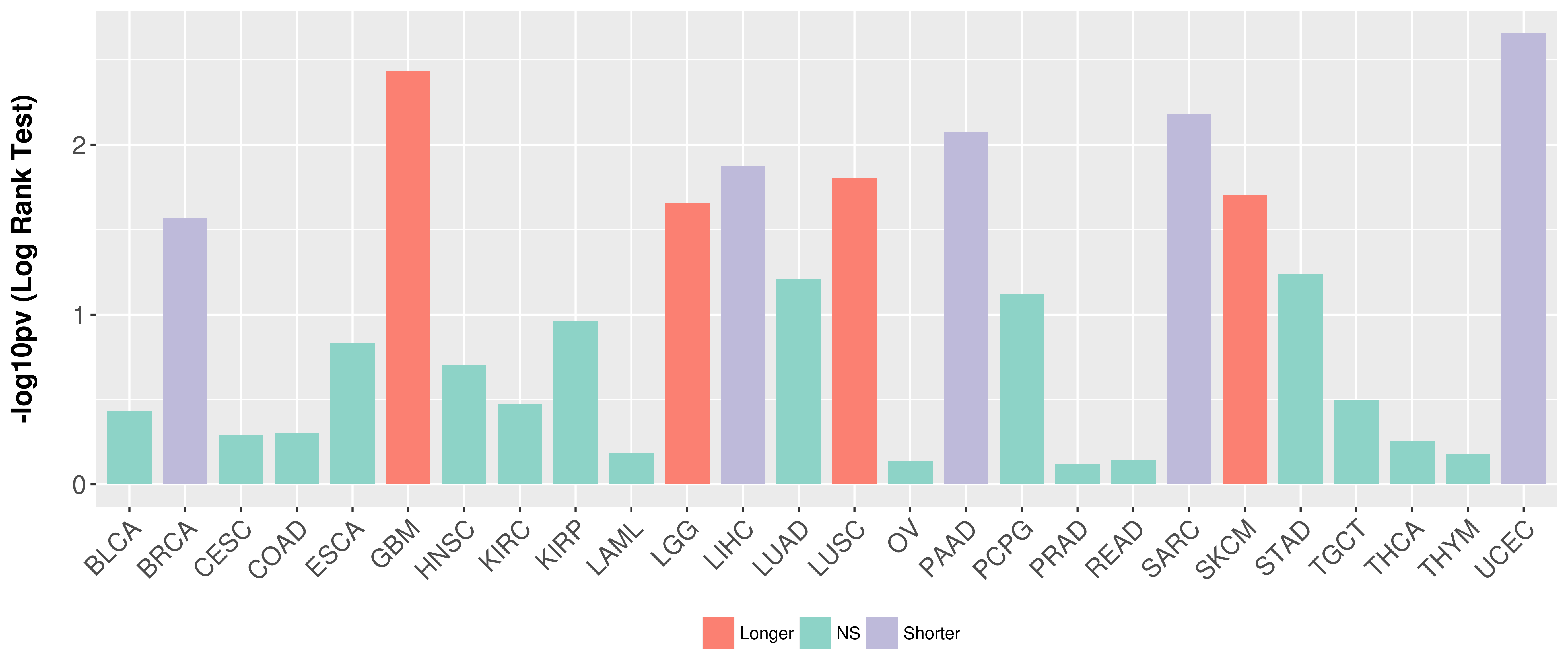
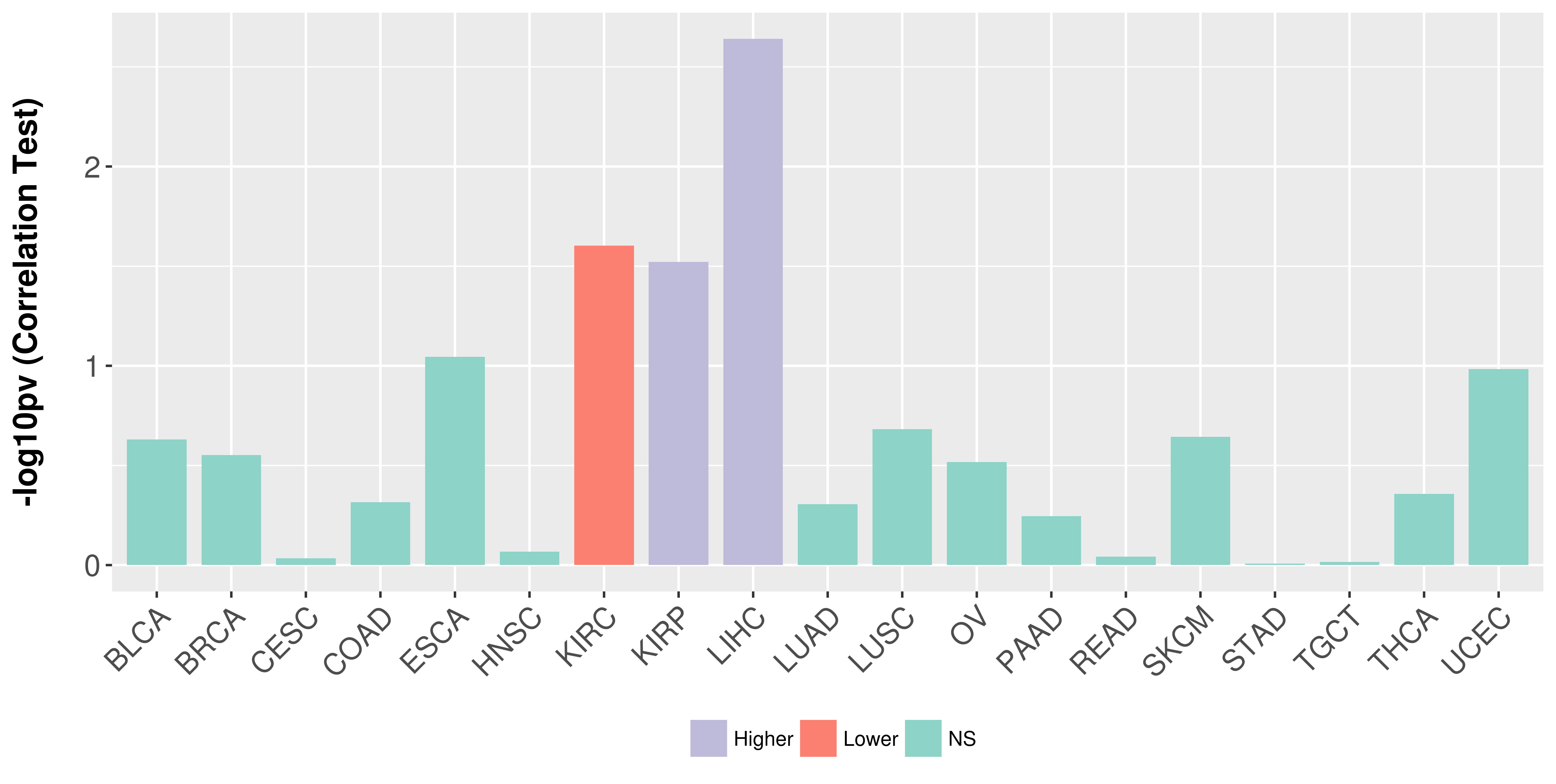
| COSM5880077 | c.879_880insAT | p.K295fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144700 | c.? | p.D333fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5752076 | c.721_722insCA | p.K241fs*39 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5752502 | c.223delA | p.R76fs*5 | Deletion - Frameshift | Breast
|
| COSM5880060 | c.374+2T>C | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144669 | c.898A>G | p.T300A | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385774 | c.308_309insA | p.Y103fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1465737 | c.537C>T | p.T179T | Substitution - coding silent | Large_intestine
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327311 | c.882A>G | p.I294M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1220385 | c.351G>T | p.E117D | Substitution - Missense | Large_intestine
|
| COSM5546652 | c.97_113>G | p.I33fs*43 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4914860 | c.395A>G | p.K132R | Substitution - Missense | Liver
|
| COSM5751890 | c.881T>C | p.I294T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4774973 | c.986A>C | p.H329P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751649 | c.302_303insT | p.R101fs*11 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM144709 | c.? | p.C215F | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115438 | c.897G>T | p.K299N | Substitution - Missense | Endometrium
|
| COSM144707 | c.? | p.S158fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5546654 | c.341_342insGAGGTCCA | p.I115fs*31 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM158743 | c.860G>T | p.G287V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM422226 | c.202G>T | p.E68* | Substitution - Nonsense | Urinary_tract
|
| COSM5880063 | c.435delT | p.F146fs*72 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5991598 | c.225_226delAA | p.G77fs*18 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1235023 | c.554_580>CCCGGGGA | p.H185fs*27 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5880303 | c.479_480insA | p.K161fs*11 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM255183 | c.736_737insTT | p.S246fs*34 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM327311 | c.882A>G | p.I294M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4154847 | c.720T>G | p.Y240* | Substitution - Nonsense | Kidney
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115431 | c.5C>G | p.S2* | Substitution - Nonsense | Endometrium
|
| COSM5944860 | c.187_188insAAGC | p.G63fs*7 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144712 | c.? | p.C280Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4923692 | c.675A>G | p.R225R | Substitution - coding silent | Liver
|
| COSM255188 | c.27_28insA | p.G10fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM255182 | c.653_667>13 | p.G218fs*3 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945630 | c.969-2A>G | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144575 | c.76_95>TTGG | p.K26fs*50 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM3719014 | c.866C>A | p.T289N | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1159590 | c.2T>A | p.M1K | Substitution - Missense | Pancreas
|
| COSM144633 | c.? | p.R335fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5944872 | c.788_789insCCGTC | p.D264fs*17 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5880066 | c.476_480delAAAAG | p.K161fs*9 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385506 | c.674G>A | p.R225Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM69277 | c.730-1G>C | p.? | Unknown | Ovary
|
| COSM158743 | c.860G>T | p.G287V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM5945587 | c.487C>T | p.R163C | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5546648 | c.716A>G | p.H239R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5944863 | c.395_396insGAAAACT | p.A135fs*5 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5904086 | c.307T>A | p.Y103N | Substitution - Missense | Skin
|
| COSM4606362 | c.?_?insGGCA | p.N23fs | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5751973 | c.375-1G>AG | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144631 | c.? | p.H302Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945584 | c.375-2A>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM3405981 | c.943G>C | p.E315Q | Substitution - Missense | Central_nervous_system
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945575 | c.241-1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4106766 | c.512A>T | p.N171I | Substitution - Missense | Stomach
|
| COSM5945590 | c.585+5G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1756397 | c.956G>T | p.R319L | Substitution - Missense | Urinary_tract
|
| COSM5427493 | c.785A>T | p.D262V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3558101 | c.888C>T | p.A296A | Substitution - coding silent | Skin
|
| COSM4775141 | c.93_94insC | p.L32fs*4 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5752028 | c.986_987insG | p.H329fs*2 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM158731 | c.725G>A | p.C242Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385554 | c.643T>C | p.C215R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144715 | c.? | p.Y303* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144573 | c.986A>G | p.H329R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5546651 | c.384C>G | p.C128W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5752502 | c.223delA | p.R76fs*5 | Deletion - Frameshift | Breast
|
| COSM5880075 | c.834G>A | p.M278I | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1319506 | c.636_637insGGATA | p.F214fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751599 | c.76_77insGTATGGAG | p.K26fs*10 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5880309 | c.905A>C | p.H302P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5427493 | c.785A>T | p.D262V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606366 | c.?_?insCC | p.G213fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880054 | c.79_80insA | p.C28fs*8 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115434 | c.415G>A | p.E139K | Substitution - Missense | Endometrium
|
| COSM5944854 | c.75_76insAAGG | p.C28fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880311 | c.94_94C>GCCTA | p.L32fs*5 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4168277 | c.941T>A | p.I314N | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM6008821 | c.93_94delAC | p.L31fs*4 | Deletion - Frameshift | Prostate
|
| COSM144628 | c.986A>T | p.H329L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4775025 | c.789_790insGC | p.D264fs*16 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1115432 | c.256C>T | p.H86Y | Substitution - Missense | Endometrium
|
| COSM1235025 | c.461_462delCA | p.S154fs*3 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM158731 | c.725G>A | p.C242Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880057 | c.128_129insT | p.K44fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144703 | c.? | p.C28fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4385492 | c.130A>T | p.K44* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606374 | c.890G>A | p.C297Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1194694 | c.70A>G | p.R24G | Substitution - Missense | Lung
|
| COSM5752056 | c.295_296insA | p.C99fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3357385 | c.374+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1724607 | c.585+6_585+7insA | p.? | Unknown | Skin
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144570 | c.903C>A | p.Y301* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144697 | c.? | p.K235* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751375 | c.125_126insCC | p.H43fs*39 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4002259 | c.585+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5546647 | c.222_224TAA>CCCC | p.K75fs*21 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4172512 | c.? | p.A41V | Substitution - Missense | Skin
|
| COSM144568 | c.823G>A | p.G275R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5366922 | c.978T>C | p.C326C | Substitution - coding silent | Large_intestine
|
| COSM4994825 | c.619G>T | p.D207Y | Substitution - Missense | Large_intestine
|
| COSM1235291 | c.409_418del10 | p.N137fs*3 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1133045 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144708 | c.? | p.C215Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM219084 | c.? | p.K44E | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3357388 | c.418+2T>C | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4385489 | c.92T>G | p.L31* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3973273 | c.410A>G | p.N137S | Substitution - Missense | Central_nervous_system
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751236 | c.821G>AG | p.R274fs*20 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945618 | c.824G>T | p.G275V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3372072 | c.277G>T | p.G93C | Substitution - Missense | Thyroid
|
| COSM4385518 | c.941T>C | p.I314T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM306075 | c.374+1G>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM255185 | c.267_268insTTAGGACC | p.A90fs*10 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144574 | c.779_780ins11 | p.D262fs*21 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM255190 | c.968+2_968+5delTAAG | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5945593 | c.586-2A>G | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM305985 | c.730-1G>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM305985 | c.730-1G>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM3747319 | c.313T>C | p.Y105H | Substitution - Missense | Liver
|
| COSM4606372 | c.?_?insG | p.F263fs | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1319510 | c.415G>T | p.E139* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1636387 | c.1088A>G | p.N363S | Substitution - Missense | Liver
|
| COSM5945575 | c.241-1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5752669 | c.1042G>T | p.G348* | Substitution - Nonsense | Breast
|
| COSM4774987 | c.407_408insT | p.N137fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945628 | c.903C>G | p.Y301* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3357385 | c.374+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4385483 | c.3G>C | p.M1I | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5944857 | c.127_128insG | p.H43fs*2 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Liver
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1262069 | c.378C>G | p.V126V | Substitution - coding silent | Oesophagus
|
| COSM5880069 | c.639_640insCC | p.F214fs*5 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4606370 | c.?_?insT | p.C215fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM144695 | c.? | p.F172fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4775910 | c.91_101del11 | p.L31fs*1 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144568 | c.823G>A | p.G275R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144705 | c.? | p.T98fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4408226 | c.17A>C | p.E6A | Substitution - Missense | Pancreas
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385509 | c.719A>G | p.Y240C | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5427493 | c.785A>T | p.D262V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1319508 | c.586-1G>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4170005 | c.868A>T | p.I290F | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168272 | c.847T>A | p.C283S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606364 | c.?_?ins11 | p.C242fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4168269 | c.806T>C | p.L269P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255187 | c.83_101>AT | p.C28fs*2 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945607 | c.725G>T | p.C242F | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945610 | c.729G>A | p.M243I | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5944866 | c.585+4_585+5insAGTAA | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144570 | c.903C>A | p.Y301* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM5991599 | c.904_904delC | p.H302fs*49 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM3363819 | c.638G>T | p.G213V | Substitution - Missense | Kidney
|
| COSM5944469 | c.70_71delAG | p.D25fs*10 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5095008 | c.677G>A | p.G226E | Substitution - Missense | Thymus
|
| COSM1235292 | c.634_635delTG | p.C212fs*11 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5944851 | c.58_59insT | p.C20fs*2 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1319512 | c.65_66insA | p.N23fs*2 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5944869 | c.591_592insAGGTCTC | p.P200fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144629 | c.? | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144698 | c.? | p.A41fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM255181 | c.90_91insCCCG | p.L31fs*6 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144634 | c.? | p.C20fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144569 | c.835_835delA | p.K279fs*16 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM2726751 | c.506A>C | p.K169T | Substitution - Missense | Large_intestine
|
| COSM5880302 | c.631A>T | p.K211* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1756397 | c.956G>T | p.R319L | Substitution - Missense | Urinary_tract
|
| COSM5546650 | c.839_840ins13 | p.C280fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM755295 | c.742G>T | p.G248C | Substitution - Missense | Lung
|
| COSM5762915 | c.661G>T | p.E221* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM6005414 | c.210C>T | p.V70V | Substitution - coding silent | Prostate
|
| COSM211020 | c.871G>T | p.G291* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144709 | c.? | p.C215F | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255188 | c.27_28insA | p.G10fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5752669 | c.1042G>T | p.G348* | Substitution - Nonsense | Breast
|
| COSM1262067 | c.570G>T | p.M190I | Substitution - Missense | Oesophagus
|
| COSM4385771 | c.69_70insA | p.R24fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1235026 | c.834+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5945596 | c.637G>A | p.G213R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM2726745 | c.414C>T | p.S138S | Substitution - coding silent | Upper_aerodigestive_tract
|
| COSM2726745 | c.414C>T | p.S138S | Substitution - coding silent | Upper_aerodigestive_tract
|
| COSM306075 | c.374+1G>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1315227 | c.137+2G>A | p.? | Unknown | Urinary_tract
|
| COSM5752094 | c.780_789del10 | p.F260fs*16 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1315229 | c.790G>C | p.D264H | Substitution - Missense | Urinary_tract
|
| COSM4606368 | c.?_?insGGCCC | p.Y103fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5991601 | c.241_374del134 | p.M81fs*12 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM3558103 | c.903C>T | p.Y301Y | Substitution - coding silent | Skin
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Kidney
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751934 | c.79_80insT | p.E27fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5751501 | c.673_674insT | p.R225fs*8 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4606373 | c.(787_789)T>C | p.F263L | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1319510 | c.415G>T | p.E139* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751283 | c.820_821insT | p.R274fs*20 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5950134 | c.948T>G | p.N316K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1235283 | c.978T>G | p.C326W | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880304 | c.517A>T | p.K173* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168250 | c.22_23insA | p.G10fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1115436 | c.587G>A | p.R196K | Substitution - Missense | Endometrium
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144573 | c.986A>G | p.H329R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880308 | c.957_958insGT | p.G320fs*32 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385498 | c.469delA | p.S158fs*60 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM2726749 | c.457C>T | p.P153S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144629 | c.? | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM755296 | c.187G>T | p.G63C | Substitution - Missense | Lung
|
| COSM5944472 | c.347_348delGA | p.K118fs*19 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385518 | c.941T>C | p.I314T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5991600 | c.434_439GTTTTA>TCAGTTC | p.S145fs*4 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144630 | c.? | p.R342* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751755 | c.28_29insA | p.G10fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144635 | c.? | p.N171fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945604 | c.725G>C | p.C242S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115433 | c.396G>T | p.K132N | Substitution - Missense | Endometrium
|
| COSM5880312 | c.134_134G>CC | p.C45fs*14 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945623 | c.853C>T | p.Q285* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606369 | c.?_?insCA | p.N147fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5546653 | c.128_130ATA>TGGGTTC | p.H43fs*17 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1235290 | c.404C>T | p.A135V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM158731 | c.725G>A | p.C242Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168259 | c.261T>A | p.C87* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1490507 | c.421G>A | p.D141N | Substitution - Missense | Breast
|
| COSM4775145 | c.340_341insA | p.I115fs*23 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385516 | c.865A>G | p.T289A | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945572 | c.68A>G | p.N23S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144708 | c.? | p.C215Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM456777 | c.347G>A | p.R116Q | Substitution - Missense | Endometrium
|
| COSM4168279 | c.953delC | p.S318fs*33 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5546655 | c.442G>T | p.E148* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144701 | c.? | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM255186 | c.902_903insA | p.Y301fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5945626 | c.877G>A | p.E293K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5546648 | c.716A>G | p.H239R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM456778 | c.982A>T | p.N328Y | Substitution - Missense | Breast
|
| COSM5546649 | c.604A>T | p.R202* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168274 | c.874T>C | p.C292R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM256007 | c.? | p.H136fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144568 | c.823G>A | p.G275R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144563 | c.1024C>T | p.R342* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751501 | c.673_674insT | p.R225fs*8 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1115437 | c.786_787insT | p.D264fs*1 | Insertion - Frameshift | Endometrium
|
| COSM4168256 | c.129T>A | p.H43Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144564 | c.289A>T | p.K97* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168253 | c.128_129insGAGTC | p.H43fs*40 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5880306 | c.586_587insA | p.R196fs*3 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4606371 | c.?_?insATCCT | p.S246fs*? | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5880307 | c.971T>C | p.L324P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945581 | c.323C>A | p.A108E | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945578 | c.322G>A | p.A108T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM256008 | c.1_1098del1098 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM137286 | c.811G>A | p.E271K | Substitution - Missense | Skin
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115439 | c.954A>G | p.S318S | Substitution - coding silent | Endometrium
|
| COSM144714 | c.? | p.R257* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327314 | c.916G>T | p.G306* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5702604 | c.119C>A | p.A40E | Substitution - Missense | Autonomic_ganglia
|
| COSM4774967 | c.375-1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144565 | c.525_526delGT | p.S176fs*13 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144572 | c.973T>C | p.Y325H | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606365 | c.737C>A | p.S246Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255218 | c.123_124insAGGCA | p.H42fs*41 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385495 | c.418G>A | p.A140T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945621 | c.834+2T>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4168281 | c.968+1_968+2delGT | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144629 | c.? | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385501 | c.596C>G | p.S199C | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255184 | c.673C>T | p.R225* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327312 | c.631A>G | p.K211E | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1115431 | c.5C>G | p.S2* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1636387 | c.1088A>G | p.N363S | Substitution - Missense | Liver
|
| COSM255189 | c.968+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144627 | c.600_600delA | p.H201fs*17 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144694 | c.? | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4775163 | c.66_69delAAAT | p.N23fs*9 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144711 | c.? | p.A311P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM487987 | c.356C>A | p.P119H | Substitution - Missense | Kidney
|
| COSM456777 | c.347G>A | p.R116Q | Substitution - Missense | Breast
|
| COSM487988 | c.875G>C | p.C292S | Substitution - Missense | Kidney
|
| COSM5752102 | c.729+1_729+2insACCCATG | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5880072 | c.711_712insTC | p.A238fs*42 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4168263 | c.407delA | p.H136fs*7 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM158743 | c.860G>T | p.G287V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1636385 | c.418+8A>G | p.? | Unknown | Liver
|
| COSM144702 | c.? | p.C283R | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385486 | c.61delA | p.K21fs*12 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM256006 | c.? | p.S193fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM385771 | c.565G>A | p.E189K | Substitution - Missense | Lung
|
| COSM5945613 | c.730-2A>G | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM144696 | c.? | p.Y303fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144632 | c.? | p.A40G | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4606374 | c.890G>A | p.C297Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144567 | c.820C>T | p.R274* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5991599 | c.904_904delC | p.H302fs*49 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1235024 | c.807_808insGGCCCCTC | p.Q270fs*12 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4606367 | c.385C>T | p.R129* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5945599 | c.709G>A | p.A237T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751924 | c.839G>A | p.C280Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880079 | c.914G>A | p.C305Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880079 | c.914G>A | p.C305Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144566 | c.808C>T | p.Q270* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1159590 | c.2T>A | p.M1K | Substitution - Missense | Pancreas
|
| COSM5751883 | c.955_956insT | p.R319fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4168279 | c.953delC | p.S318fs*33 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM144706 | c.? | p.Y105fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1235026 | c.834+1G>A | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM306061 | c.821G>A | p.R274Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144626 | c.955C>T | p.R319* | Substitution - Nonsense | Stomach
|
| COSM144704 | c.? | p.H43fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5751150 | c.385_386insT | p.R129fs*9 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4169632 | c.? | p.A296fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4761565 | c.892G>A | p.V298I | Substitution - Missense | Stomach
|
| COSM4606361 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5751236 | c.821G>AG | p.R274fs*20 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM3747319 | c.313T>C | p.Y105H | Substitution - Missense | Breast
|
| COSM1133045 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4606363 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1115435 | c.567A>T | p.E189D | Substitution - Missense | Endometrium
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3558099 | c.491A>G | p.K164R | Substitution - Missense | Skin
|
| COSM144571 | c.933_934insT | p.K312fs*1 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4385518 | c.941T>C | p.I314T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4168266 | c.513delT | p.F172fs*46 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM158731 | c.725G>A | p.C242Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4385514 | c.833T>A | p.M278K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5751440 | c.941T>AT | p.I314fs*2 | Complex - frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4774973 | c.986A>C | p.H329P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM158743 | c.860G>T | p.G287V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM144693 | c.? | p.G122* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM144694 | c.? | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM327313 | c.346C>T | p.R116* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM3719601 | c.1018G>A | p.E340K | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5880305 | c.731_732delTG | p.L244fs*20 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|