Browse PYGO1 in pancancer
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00628 PHD-finger |
||||||||||
| Function |
Involved in signal transduction through the Wnt pathway. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001655 urogenital system development GO:0001822 kidney development GO:0002244 hematopoietic progenitor cell differentiation GO:0006997 nucleus organization GO:0007281 germ cell development GO:0007283 spermatogenesis GO:0007286 spermatid development GO:0007289 spermatid nucleus differentiation GO:0009791 post-embryonic development GO:0016055 Wnt signaling pathway GO:0022412 cellular process involved in reproduction in multicellular organism GO:0034504 protein localization to nucleus GO:0048232 male gamete generation GO:0048515 spermatid differentiation GO:0072001 renal system development GO:0198738 cell-cell signaling by wnt GO:1904837 beta-catenin-TCF complex assembly |
| Molecular Function | - |
| Cellular Component | - |
| KEGG | - |
| Reactome |
R-HSA-3769402: Deactivation of the beta-catenin transactivating complex R-HSA-201722: Formation of the beta-catenin R-HSA-162582: Signal Transduction R-HSA-195721: Signaling by Wnt R-HSA-201681: TCF dependent signaling in response to WNT |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for PYGO1. |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
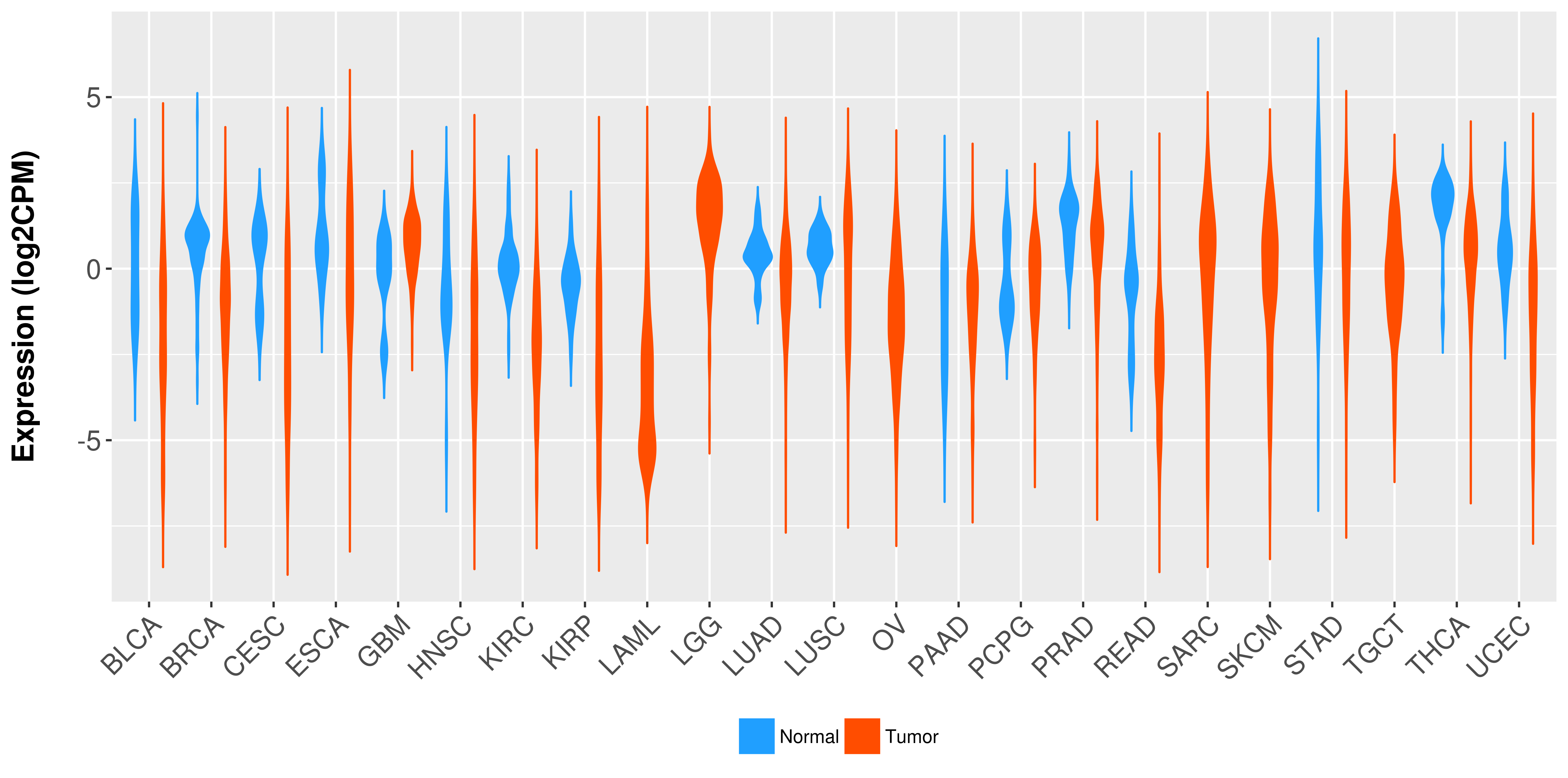
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
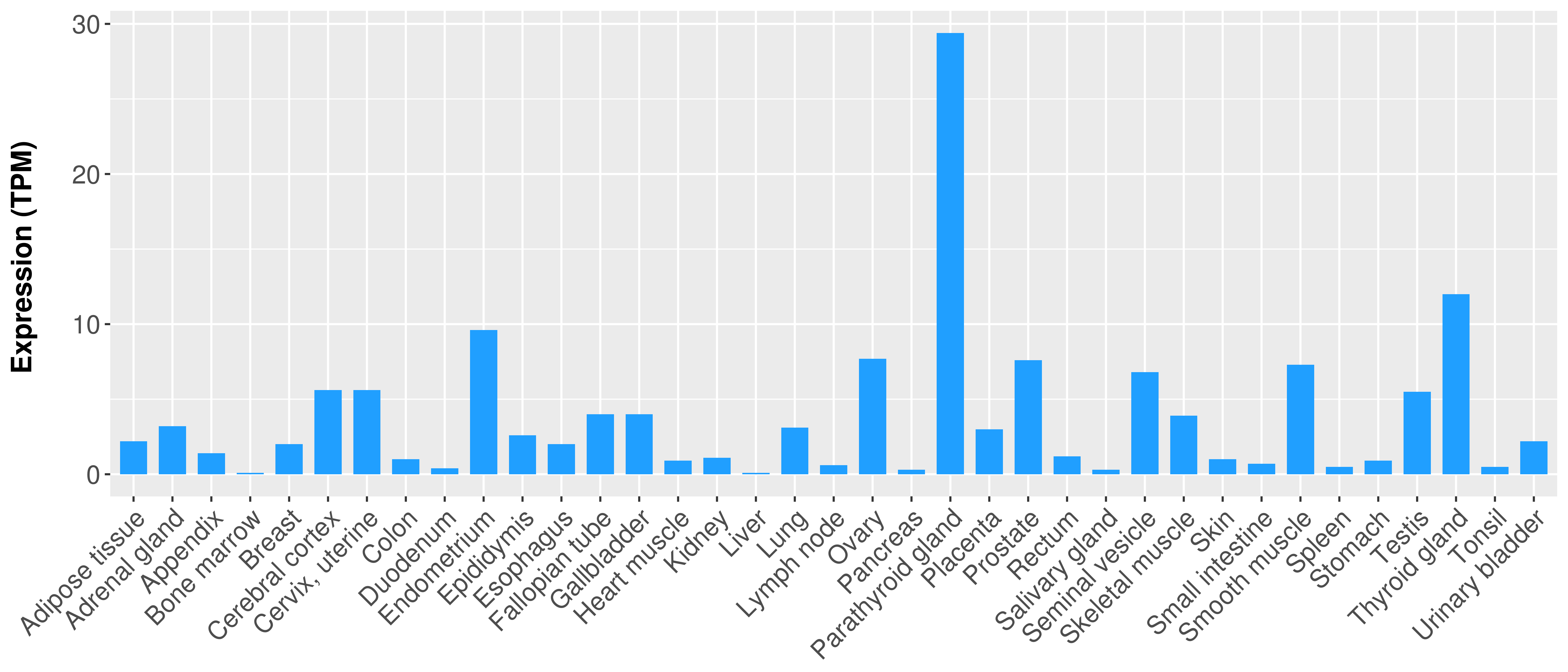
|
|
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
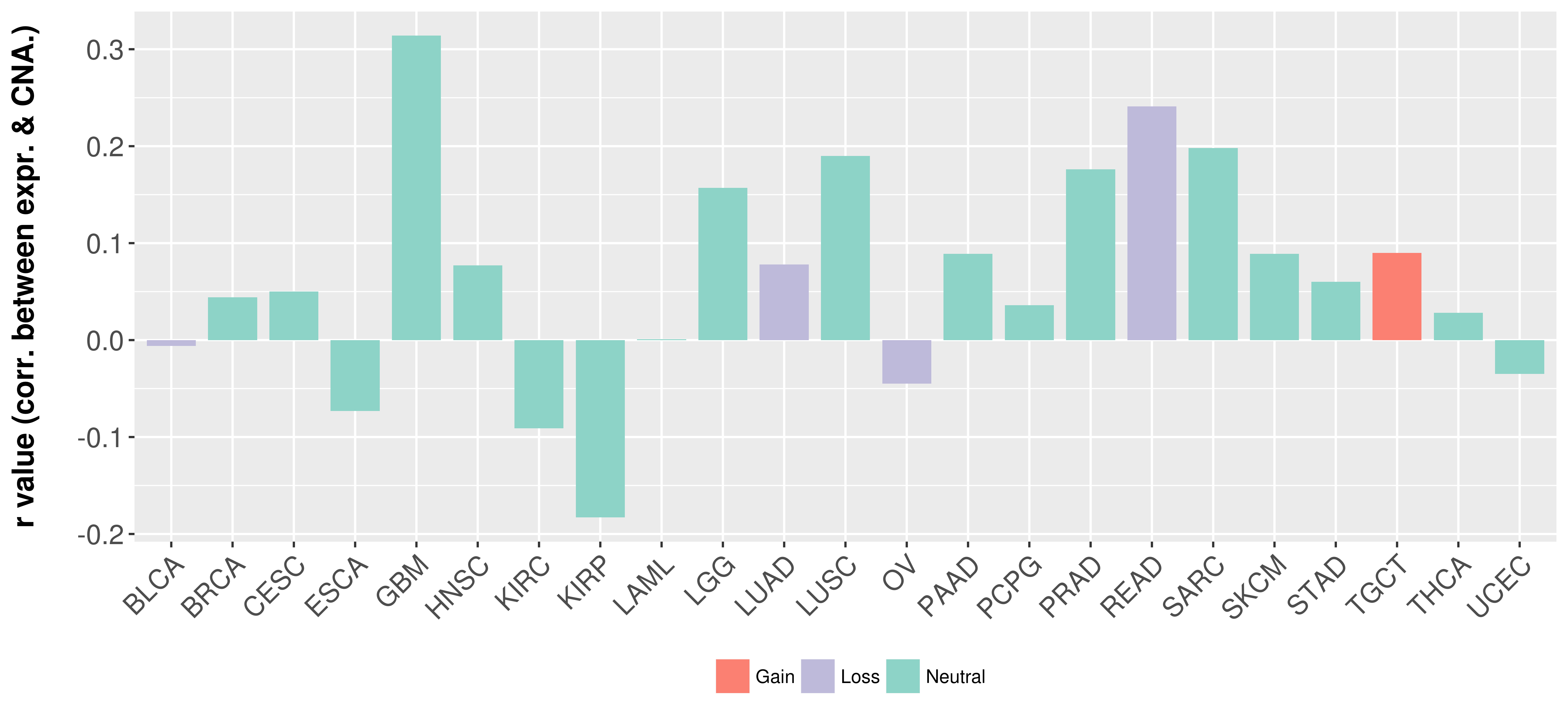
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
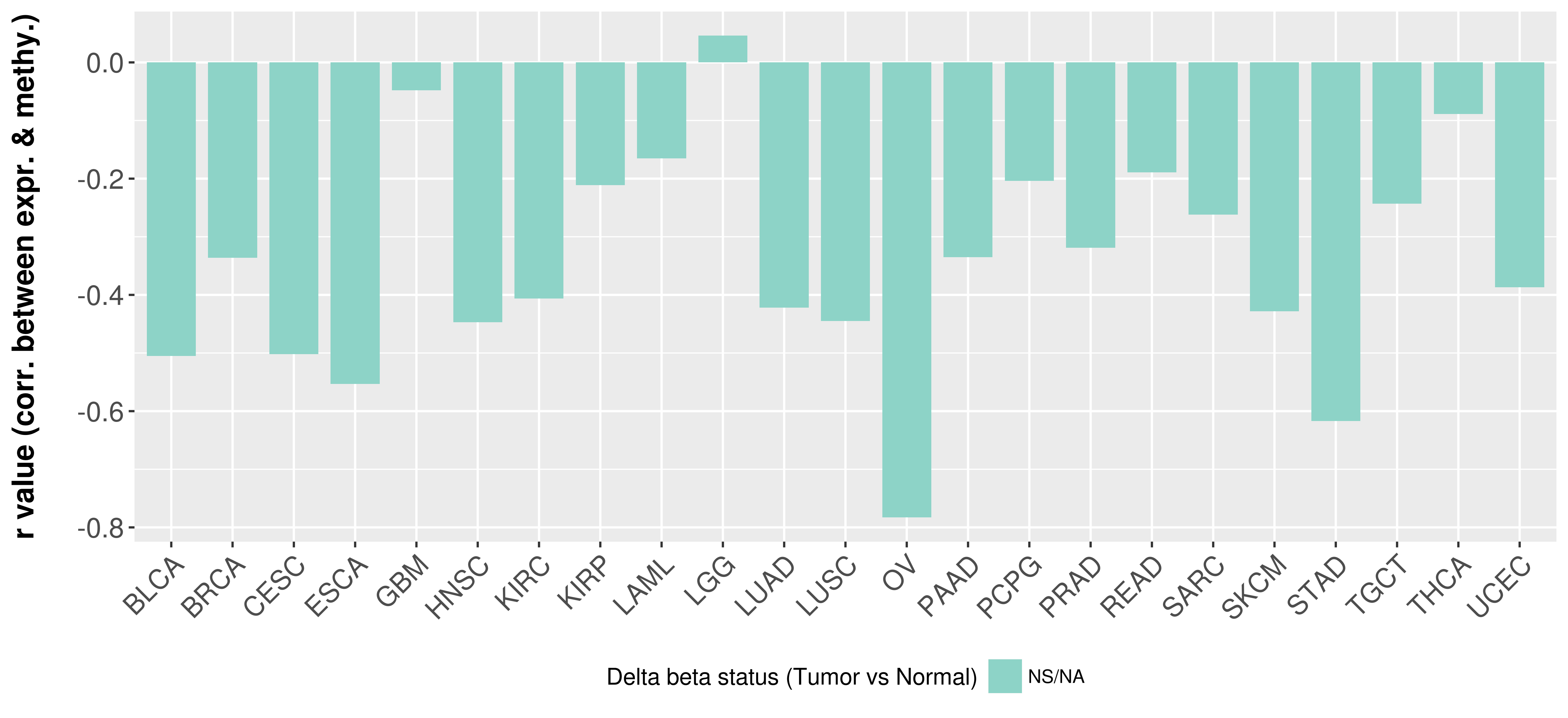
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
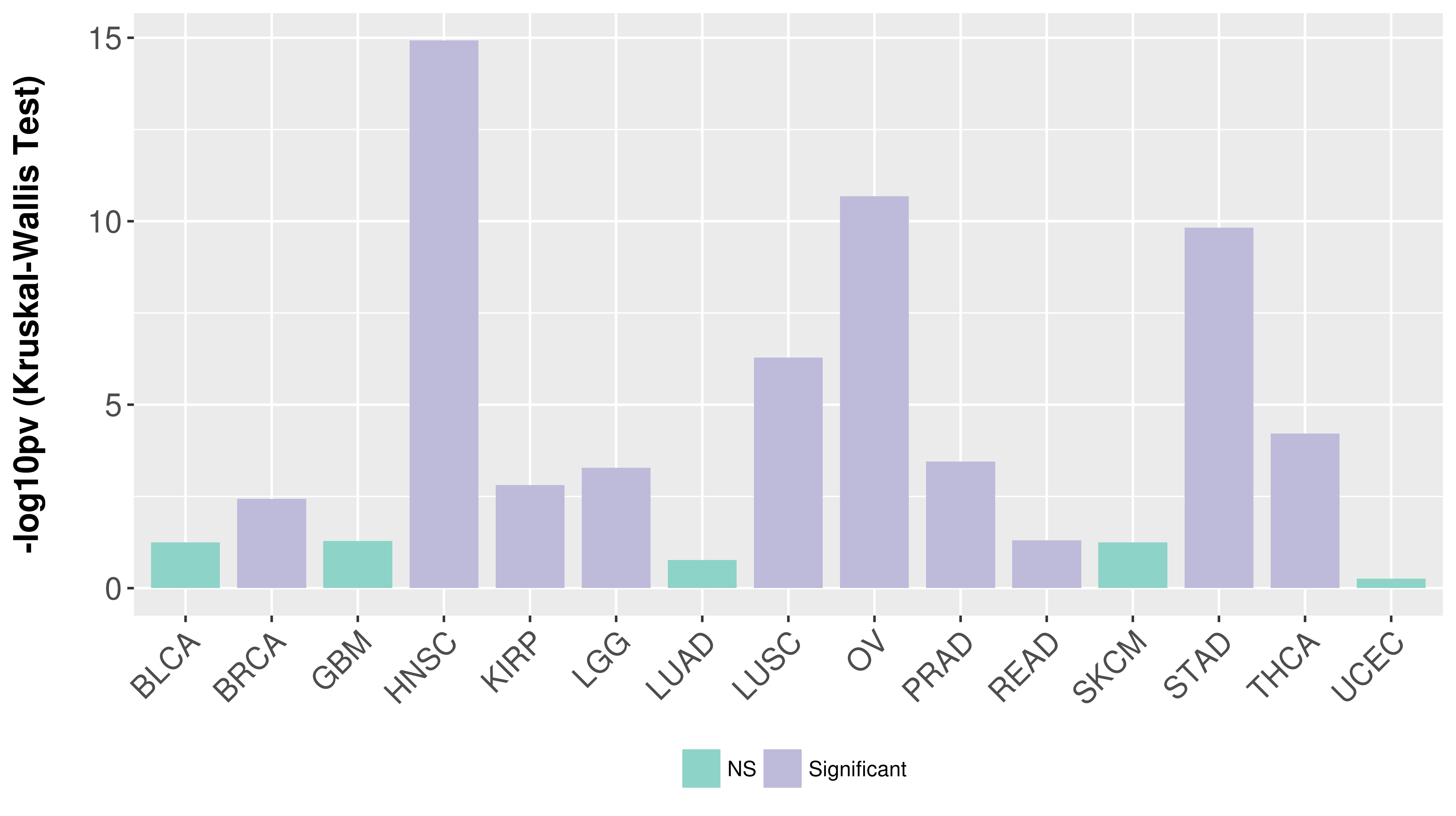
Association between expresson and subtype.
|
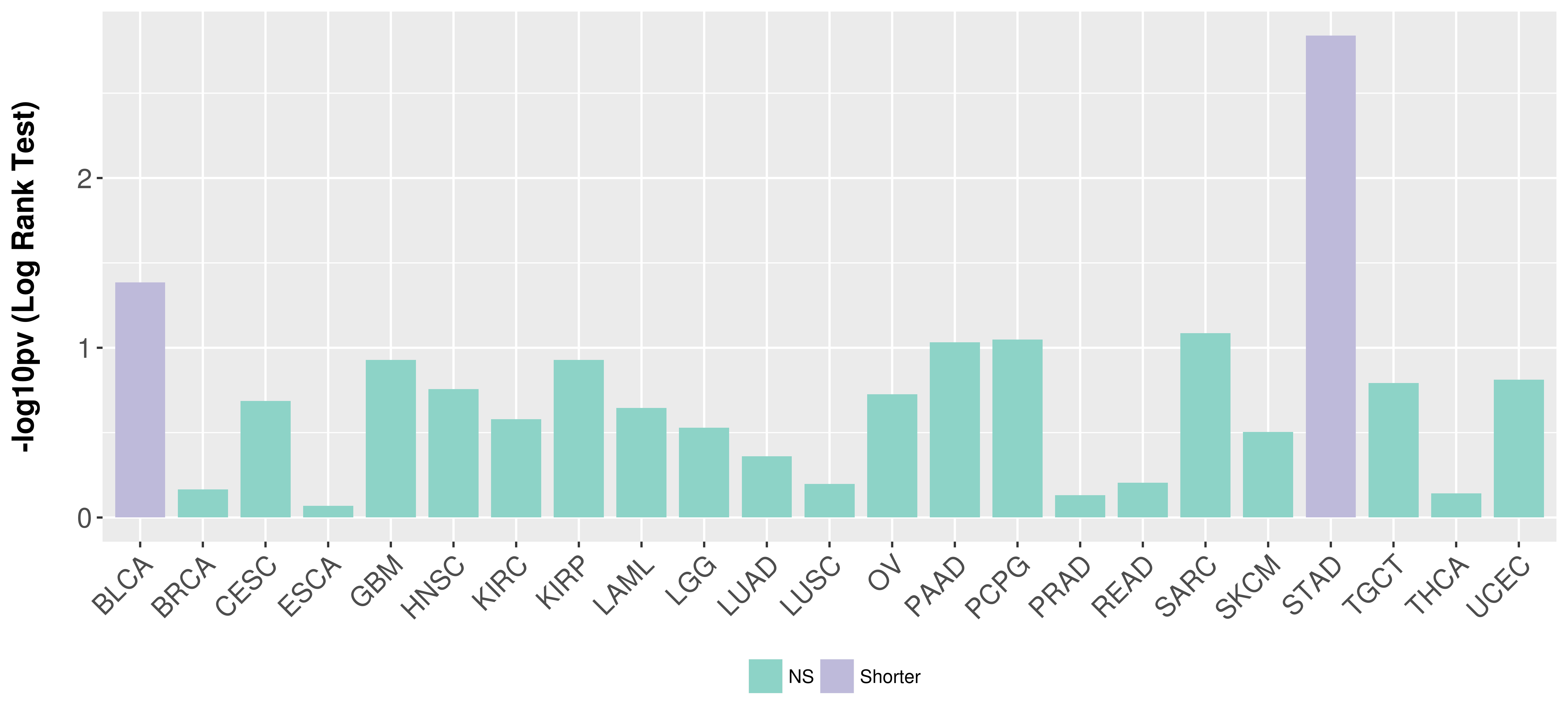
Overall survival analysis based on expression.
|
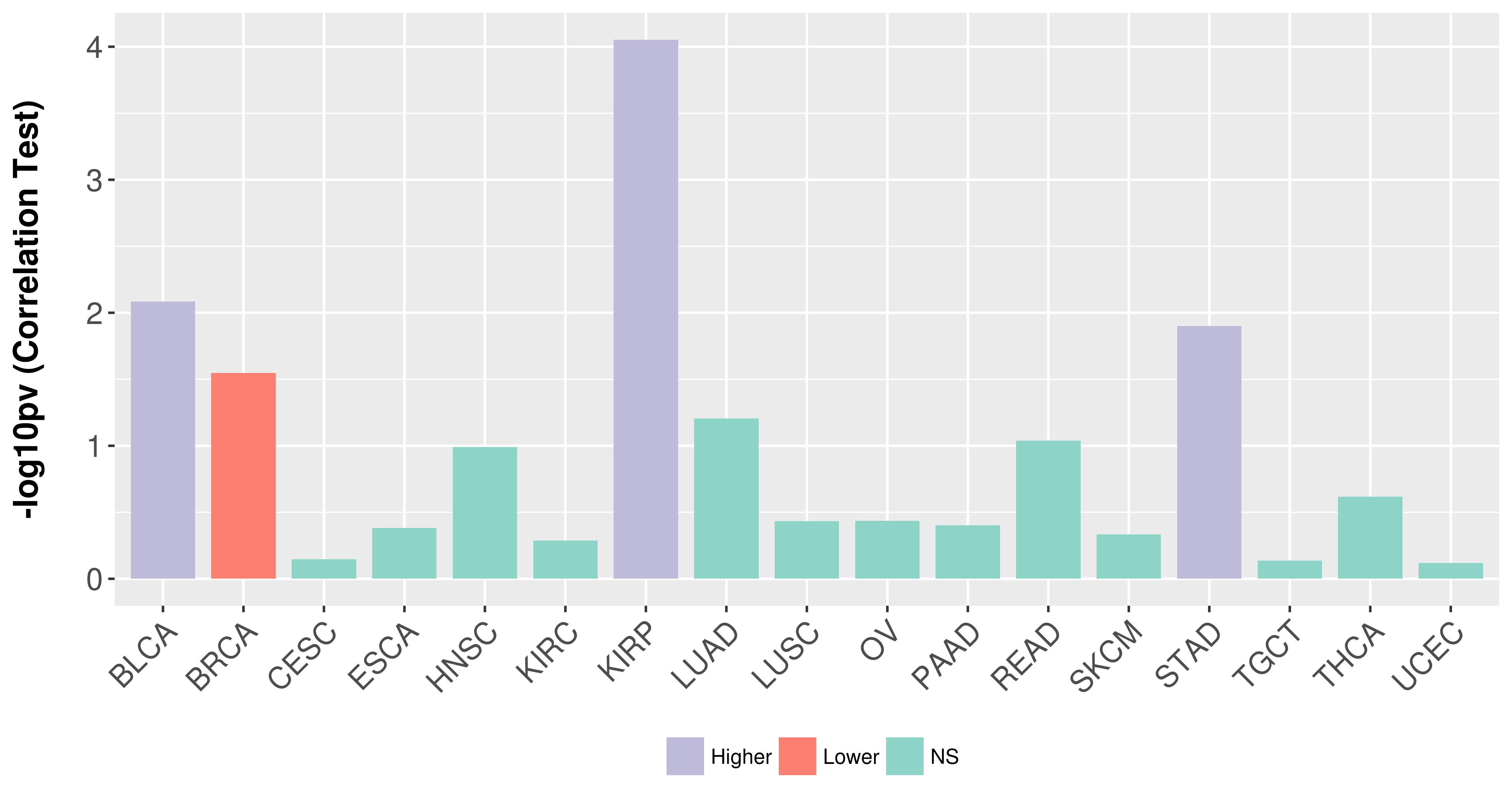
Association between expresson and stage.
|
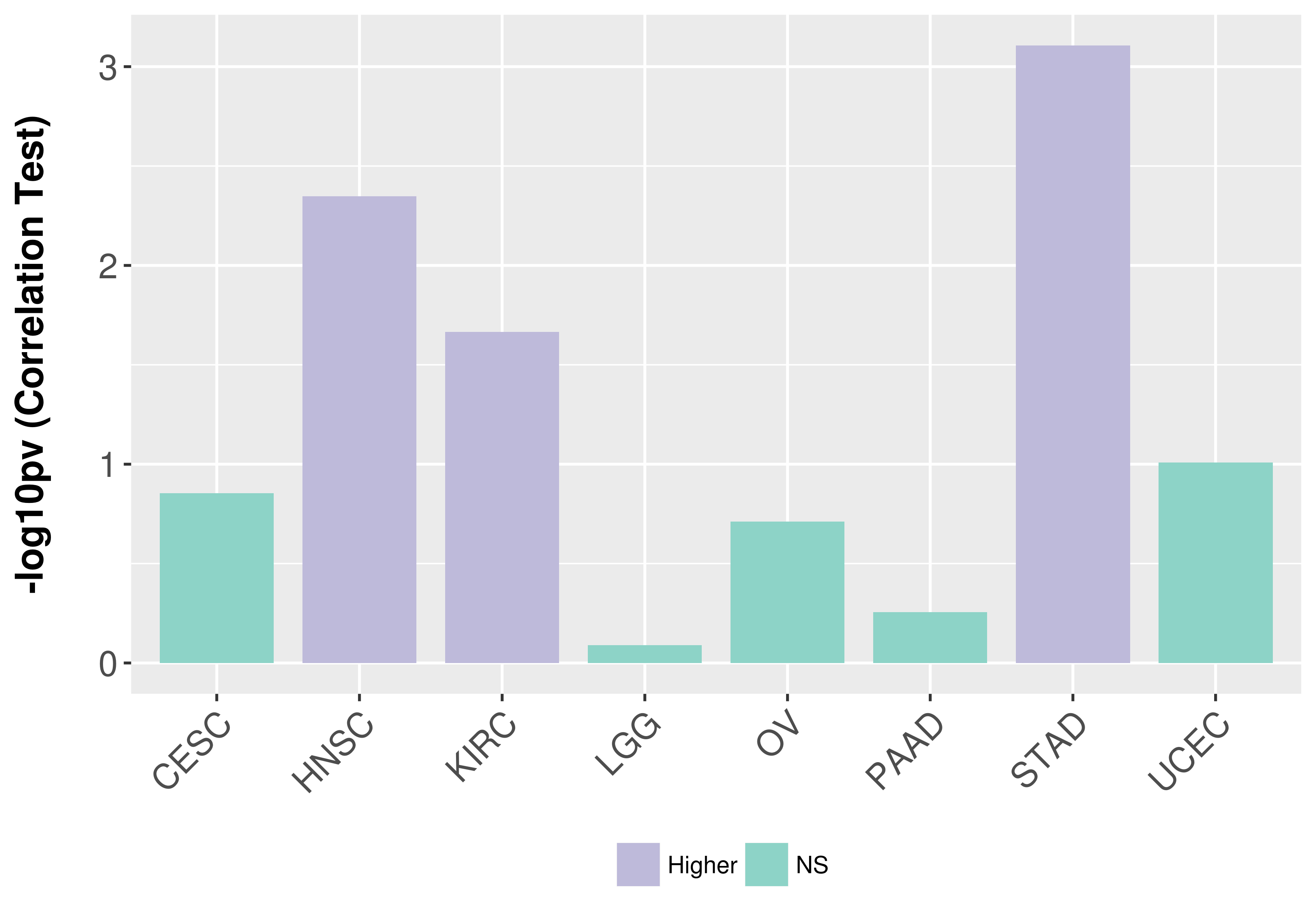
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for PYGO1. |
| Summary | |
|---|---|
| Symbol | PYGO1 |
| Name | pygopus family PHD finger 1 |
| Aliases | pygopus homolog 1 (Drosophila); pygopus-like protein 1; Pygopus homolog 1 |
| Location | 15q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for PYGO1. |