Browse SIRT2 in pancancer
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02146 Sir2 family |
||||||||||
| Function |
NAD-dependent protein deacetylase, which deacetylates internal lysines on histone and alpha-tubulin as well as many other proteins such as key transcription factors. Participates in the modulation of multiple and diverse biological processes such as cell cycle control, genomic integrity, microtubule dynamics, cell differentiation, metabolic networks, and autophagy. Plays a major role in the control of cell cycle progression and genomic stability. Functions in the antephase checkpoint preventing precocious mitotic entry in response to microtubule stress agents, and hence allowing proper inheritance of chromosomes. Positively regulates the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase complex activity by deacetylating CDC20 and FZR1, then allowing progression through mitosis. Associates both with chromatin at transcriptional start sites (TSSs) and enhancers of active genes. Plays a role in cell cycle and chromatin compaction through epigenetic modulation of the regulation of histone H4 'Lys-20' methylation (H4K20me1) during early mitosis. Specifically deacetylates histone H4 at 'Lys-16' (H4K16ac) between the G2/M transition and metaphase enabling H4K20me1 deposition by KMT5A leading to ulterior levels of H4K20me2 and H4K20me3 deposition throughout cell cycle, and mitotic S-phase progression. Deacetylates KMT5A modulating KMT5A chromatin localization during the mitotic stress response. Deacetylates also histone H3 at 'Lys-57' (H3K56ac) during the mitotic G2/M transition. Upon bacterium Listeria monocytogenes infection, deacetylates 'Lys-18' of histone H3 in a receptor tyrosine kinase MET- and PI3K/Akt-dependent manner, thereby inhibiting transcriptional activity and promoting late stages of listeria infection. During oocyte meiosis progression, may deacetylate histone H4 at 'Lys-16' (H4K16ac) and alpha-tubulin, regulating spindle assembly and chromosome alignment by influencing microtubule dynamics and kinetochore function. Deacetylates alpha-tubulin at 'Lys-40' and hence controls neuronal motility, oligodendroglial cell arbor projection processes and proliferation of non-neuronal cells. Phosphorylation at Ser-368 by a G1/S-specific cyclin E-CDK2 complex inactivates SIRT2-mediated alpha-tubulin deacetylation, negatively regulating cell adhesion, cell migration and neurite outgrowth during neuronal differentiation. Deacetylates PARD3 and participates in the regulation of Schwann cell peripheral myelination formation during early postnatal development and during postinjury remyelination. Involved in several cellular metabolic pathways. Plays a role in the regulation of blood glucose homeostasis by deacetylating and stabilizing phosphoenolpyruvate carboxykinase PCK1 activity in response to low nutrient availability. Acts as a key regulator in the pentose phosphate pathway (PPP) by deacetylating and activating the glucose-6-phosphate G6PD enzyme, and therefore, stimulates the production of cytosolic NADPH to counteract oxidative damage. Maintains energy homeostasis in response to nutrient deprivation as well as energy expenditure by inhibiting adipogenesis and promoting lipolysis. Attenuates adipocyte differentiation by deacetylating and promoting FOXO1 interaction to PPARG and subsequent repression of PPARG-dependent transcriptional activity. Plays a role in the regulation of lysosome-mediated degradation of protein aggregates by autophagy in neuronal cells. Deacetylates FOXO1 in response to oxidative stress or serum deprivation, thereby negatively regulating FOXO1-mediated autophagy. Deacetylates a broad range of transcription factors and co-regulators regulating target gene expression. Deacetylates transcriptional factor FOXO3 stimulating the ubiquitin ligase SCF(SKP2)-mediated FOXO3 ubiquitination and degradation. Deacetylates HIF1A and therefore promotes HIF1A degradation and inhibition of HIF1A transcriptional activity in tumor cells in response to hypoxia. Deacetylates RELA in the cytoplasm inhibiting NF-kappaB-dependent transcription activation upon TNF-alpha stimulation. Inhibits transcriptional activation by deacetylating p53/TP53 and EP300. Deacetylates also EIF5A. Functions as a negative regulator on oxidative stress-tolerance in response to anoxia-reoxygenation conditions. Plays a role as tumor suppressor.; FUNCTION: Isoform 1: Deacetylates EP300, alpha-tubulin and histone H3 and H4.; FUNCTION: Isoform 2: Deacetylates EP300, alpha-tubulin and histone H3 and H4.; FUNCTION: Isoform 5: Lacks deacetylation activity. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000183 chromatin silencing at rDNA GO:0001556 oocyte maturation GO:0001666 response to hypoxia GO:0001933 negative regulation of protein phosphorylation GO:0002237 response to molecule of bacterial origin GO:0002831 regulation of response to biotic stimulus GO:0002832 negative regulation of response to biotic stimulus GO:0006342 chromatin silencing GO:0006348 chromatin silencing at telomere GO:0006471 protein ADP-ribosylation GO:0006476 protein deacetylation GO:0006486 protein glycosylation GO:0006914 autophagy GO:0006979 response to oxidative stress GO:0007059 chromosome segregation GO:0007067 mitotic nuclear division GO:0007088 regulation of mitotic nuclear division GO:0007096 regulation of exit from mitosis GO:0007126 meiotic nuclear division GO:0007272 ensheathment of neurons GO:0007281 germ cell development GO:0007292 female gamete generation GO:0007346 regulation of mitotic cell cycle GO:0007422 peripheral nervous system development GO:0007517 muscle organ development GO:0008366 axon ensheathment GO:0008608 attachment of spindle microtubules to kinetochore GO:0009100 glycoprotein metabolic process GO:0009101 glycoprotein biosynthetic process GO:0009894 regulation of catabolic process GO:0009895 negative regulation of catabolic process GO:0009896 positive regulation of catabolic process GO:0009991 response to extracellular stimulus GO:0009994 oocyte differentiation GO:0010001 glial cell differentiation GO:0010458 exit from mitosis GO:0010498 proteasomal protein catabolic process GO:0010506 regulation of autophagy GO:0010507 negative regulation of autophagy GO:0010720 positive regulation of cell development GO:0010721 negative regulation of cell development GO:0010799 regulation of peptidyl-threonine phosphorylation GO:0010801 negative regulation of peptidyl-threonine phosphorylation GO:0014013 regulation of gliogenesis GO:0014014 negative regulation of gliogenesis GO:0014037 Schwann cell differentiation GO:0014044 Schwann cell development GO:0014065 phosphatidylinositol 3-kinase signaling GO:0014706 striated muscle tissue development GO:0016042 lipid catabolic process GO:0016202 regulation of striated muscle tissue development GO:0016458 gene silencing GO:0016570 histone modification GO:0016575 histone deacetylation GO:0018107 peptidyl-threonine phosphorylation GO:0018205 peptidyl-lysine modification GO:0018210 peptidyl-threonine modification GO:0021700 developmental maturation GO:0021762 substantia nigra development GO:0021782 glial cell development GO:0022011 myelination in peripheral nervous system GO:0022412 cellular process involved in reproduction in multicellular organism GO:0030901 midbrain development GO:0031329 regulation of cellular catabolic process GO:0031331 positive regulation of cellular catabolic process GO:0031333 negative regulation of protein complex assembly GO:0031348 negative regulation of defense response GO:0031641 regulation of myelination GO:0031667 response to nutrient levels GO:0032102 negative regulation of response to external stimulus GO:0032292 peripheral nervous system axon ensheathment GO:0032434 regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0032436 positive regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0034599 cellular response to oxidative stress GO:0034983 peptidyl-lysine deacetylation GO:0035601 protein deacylation GO:0035728 response to hepatocyte growth factor GO:0035729 cellular response to hepatocyte growth factor stimulus GO:0036293 response to decreased oxygen levels GO:0036294 cellular response to decreased oxygen levels GO:0040020 regulation of meiotic nuclear division GO:0040029 regulation of gene expression, epigenetic GO:0042063 gliogenesis GO:0042176 regulation of protein catabolic process GO:0042177 negative regulation of protein catabolic process GO:0042326 negative regulation of phosphorylation GO:0042552 myelination GO:0042742 defense response to bacterium GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043254 regulation of protein complex assembly GO:0043388 positive regulation of DNA binding GO:0043413 macromolecule glycosylation GO:0043491 protein kinase B signaling GO:0043618 regulation of transcription from RNA polymerase II promoter in response to stress GO:0043620 regulation of DNA-templated transcription in response to stress GO:0043900 regulation of multi-organism process GO:0043901 negative regulation of multi-organism process GO:0043902 positive regulation of multi-organism process GO:0044242 cellular lipid catabolic process GO:0044546 NLRP3 inflammasome complex assembly GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0045444 fat cell differentiation GO:0045598 regulation of fat cell differentiation GO:0045599 negative regulation of fat cell differentiation GO:0045732 positive regulation of protein catabolic process GO:0045787 positive regulation of cell cycle GO:0045814 negative regulation of gene expression, epigenetic GO:0045836 positive regulation of meiotic nuclear division GO:0045843 negative regulation of striated muscle tissue development GO:0045862 positive regulation of proteolysis GO:0048012 hepatocyte growth factor receptor signaling pathway GO:0048015 phosphatidylinositol-mediated signaling GO:0048017 inositol lipid-mediated signaling GO:0048469 cell maturation GO:0048477 oogenesis GO:0048599 oocyte development GO:0048634 regulation of muscle organ development GO:0048635 negative regulation of muscle organ development GO:0048857 neural nucleus development GO:0050768 negative regulation of neurogenesis GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051101 regulation of DNA binding GO:0051302 regulation of cell division GO:0051321 meiotic cell cycle GO:0051445 regulation of meiotic cell cycle GO:0051446 positive regulation of meiotic cell cycle GO:0051775 response to redox state GO:0051781 positive regulation of cell division GO:0051783 regulation of nuclear division GO:0051785 positive regulation of nuclear division GO:0051961 negative regulation of nervous system development GO:0051983 regulation of chromosome segregation GO:0051984 positive regulation of chromosome segregation GO:0051987 positive regulation of attachment of spindle microtubules to kinetochore GO:0051988 regulation of attachment of spindle microtubules to kinetochore GO:0060281 regulation of oocyte development GO:0060282 positive regulation of oocyte development GO:0060537 muscle tissue development GO:0061136 regulation of proteasomal protein catabolic process GO:0061351 neural precursor cell proliferation GO:0061418 regulation of transcription from RNA polymerase II promoter in response to hypoxia GO:0061428 negative regulation of transcription from RNA polymerase II promoter in response to hypoxia GO:0061433 cellular response to caloric restriction GO:0061771 response to caloric restriction GO:0070085 glycosylation GO:0070265 necrotic cell death GO:0070266 necroptotic process GO:0070444 oligodendrocyte progenitor proliferation GO:0070445 regulation of oligodendrocyte progenitor proliferation GO:0070446 negative regulation of oligodendrocyte progenitor proliferation GO:0070482 response to oxygen levels GO:0070932 histone H3 deacetylation GO:0070933 histone H4 deacetylation GO:0071216 cellular response to biotic stimulus GO:0071219 cellular response to molecule of bacterial origin GO:0071407 cellular response to organic cyclic compound GO:0071417 cellular response to organonitrogen compound GO:0071453 cellular response to oxygen levels GO:0071456 cellular response to hypoxia GO:0071867 response to monoamine GO:0071868 cellular response to monoamine stimulus GO:0071869 response to catecholamine GO:0071870 cellular response to catecholamine stimulus GO:0071871 response to epinephrine GO:0071872 cellular response to epinephrine stimulus GO:0072593 reactive oxygen species metabolic process GO:0090042 tubulin deacetylation GO:0090068 positive regulation of cell cycle process GO:0097194 execution phase of apoptosis GO:0097201 negative regulation of transcription from RNA polymerase II promoter in response to stress GO:0097300 programmed necrotic cell death GO:0097343 ripoptosome assembly GO:0098542 defense response to other organism GO:0098732 macromolecule deacylation GO:0098813 nuclear chromosome segregation GO:1900117 regulation of execution phase of apoptosis GO:1900119 positive regulation of execution phase of apoptosis GO:1900193 regulation of oocyte maturation GO:1900195 positive regulation of oocyte maturation GO:1900225 regulation of NLRP3 inflammasome complex assembly GO:1900226 negative regulation of NLRP3 inflammasome complex assembly GO:1900424 regulation of defense response to bacterium GO:1900425 negative regulation of defense response to bacterium GO:1901026 ripoptosome assembly involved in necroptotic process GO:1901800 positive regulation of proteasomal protein catabolic process GO:1901861 regulation of muscle tissue development GO:1901862 negative regulation of muscle tissue development GO:1901987 regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1903046 meiotic cell cycle process GO:1903050 regulation of proteolysis involved in cellular protein catabolic process GO:1903052 positive regulation of proteolysis involved in cellular protein catabolic process GO:1903362 regulation of cellular protein catabolic process GO:1903364 positive regulation of cellular protein catabolic process GO:1903429 regulation of cell maturation GO:1903431 positive regulation of cell maturation GO:2000177 regulation of neural precursor cell proliferation GO:2000178 negative regulation of neural precursor cell proliferation GO:2000241 regulation of reproductive process GO:2000243 positive regulation of reproductive process GO:2000377 regulation of reactive oxygen species metabolic process GO:2000378 negative regulation of reactive oxygen species metabolic process GO:2000777 positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia |
| Molecular Function |
GO:0003682 chromatin binding GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0015631 tubulin binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0032182 ubiquitin-like protein binding GO:0033558 protein deacetylase activity GO:0034739 histone deacetylase activity (H4-K16 specific) GO:0034979 NAD-dependent protein deacetylase activity GO:0035035 histone acetyltransferase binding GO:0042826 histone deacetylase binding GO:0042903 tubulin deacetylase activity GO:0043130 ubiquitin binding GO:0046970 NAD-dependent histone deacetylase activity (H4-K16 specific) GO:0048037 cofactor binding GO:0048487 beta-tubulin binding GO:0050662 coenzyme binding GO:0051287 NAD binding GO:0070403 NAD+ binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000792 heterochromatin GO:0005677 chromatin silencing complex GO:0005720 nuclear heterochromatin GO:0005813 centrosome GO:0005814 centriole GO:0005819 spindle GO:0005874 microtubule GO:0017053 transcriptional repressor complex GO:0030424 axon GO:0030426 growth cone GO:0030427 site of polarized growth GO:0030496 midbody GO:0033010 paranodal junction GO:0033267 axon part GO:0033270 paranode region of axon GO:0043025 neuronal cell body GO:0043204 perikaryon GO:0043209 myelin sheath GO:0043218 compact myelin GO:0043219 lateral loop GO:0043220 Schmidt-Lanterman incisure GO:0044224 juxtaparanode region of axon GO:0044297 cell body GO:0044304 main axon GO:0044450 microtubule organizing center part GO:0044454 nuclear chromosome part GO:0072686 mitotic spindle GO:0072687 meiotic spindle GO:0090568 nuclear transcriptional repressor complex GO:0097386 glial cell projection GO:0098687 chromosomal region |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SIRT2. |
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
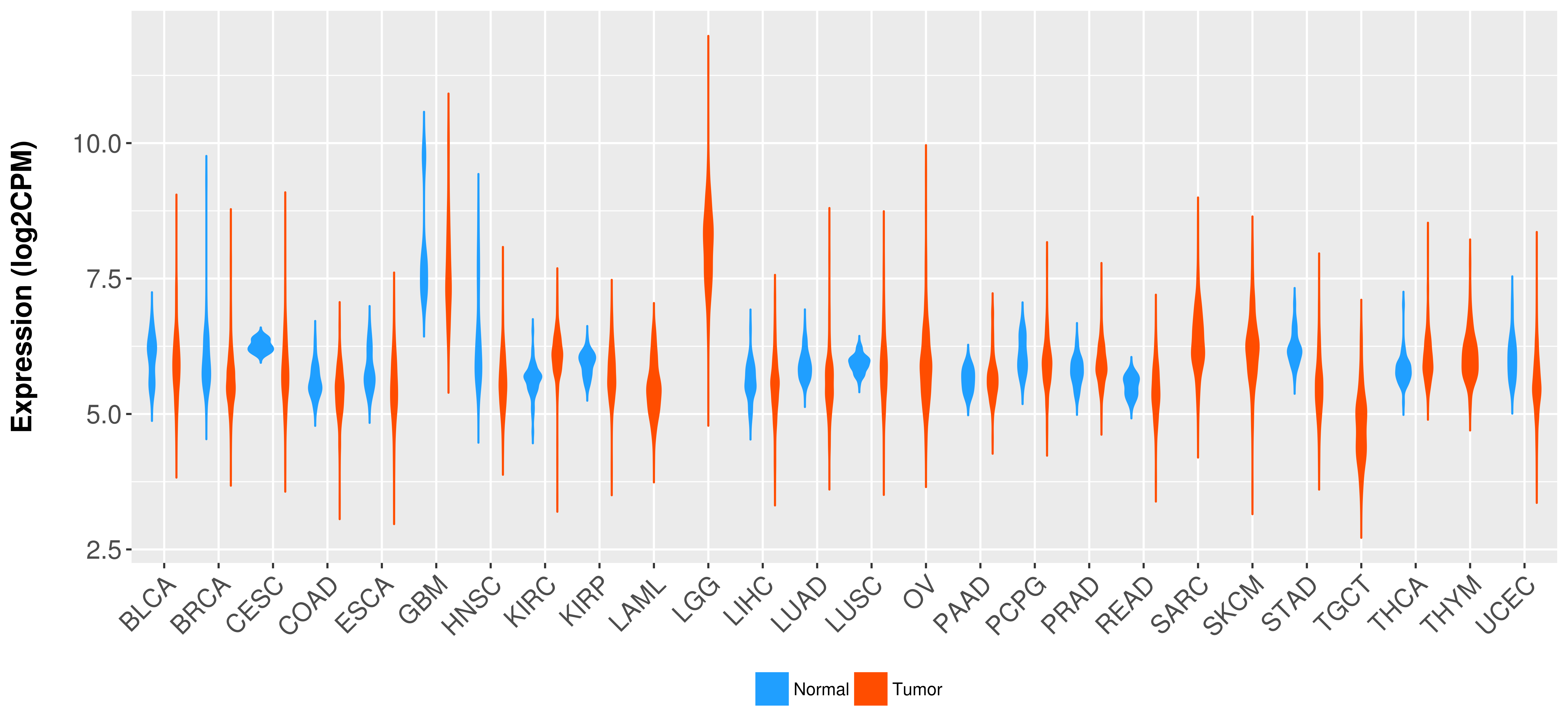
Differential expression analysis for cancers with more than 10 normal samples
|
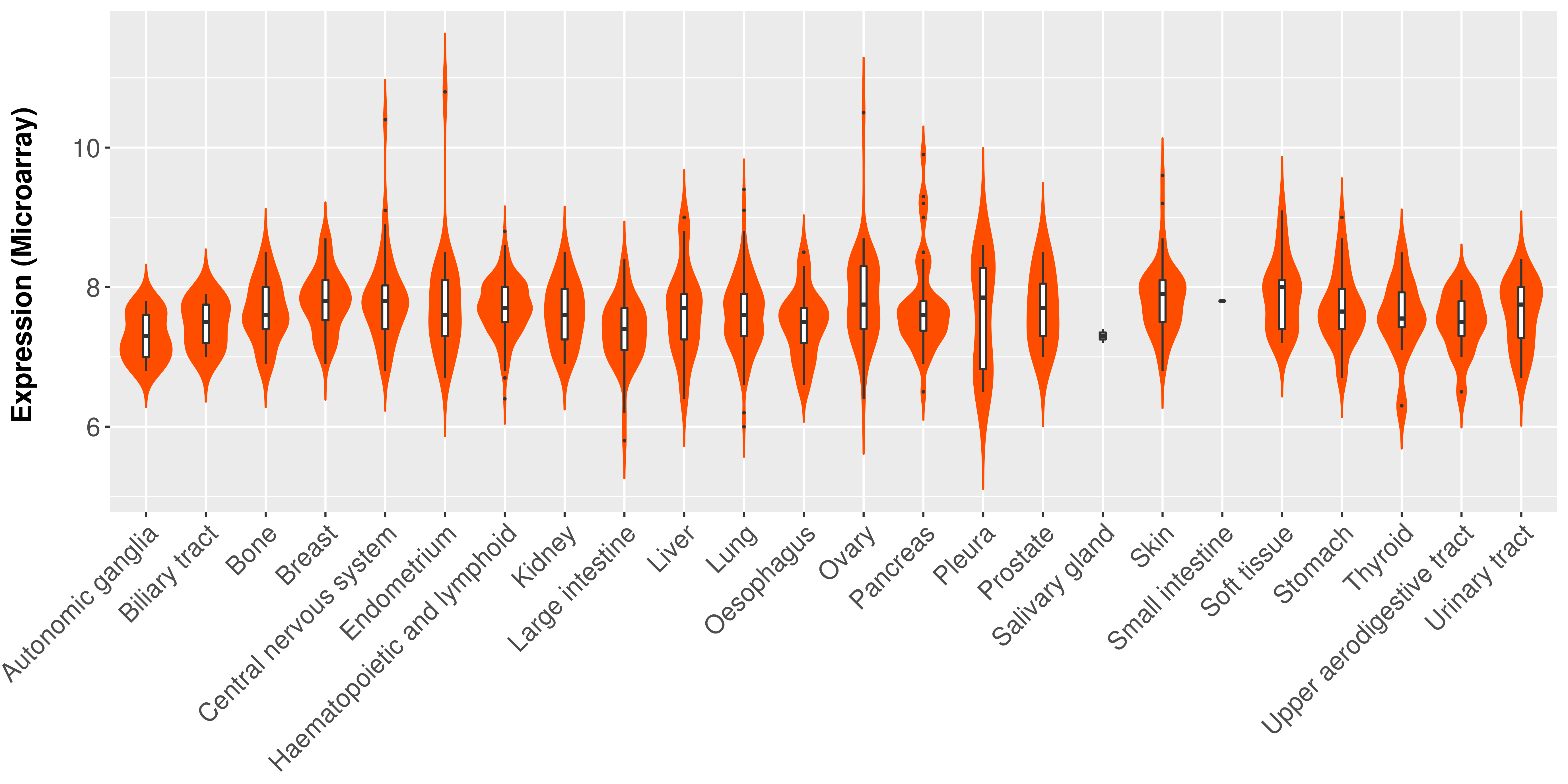
|
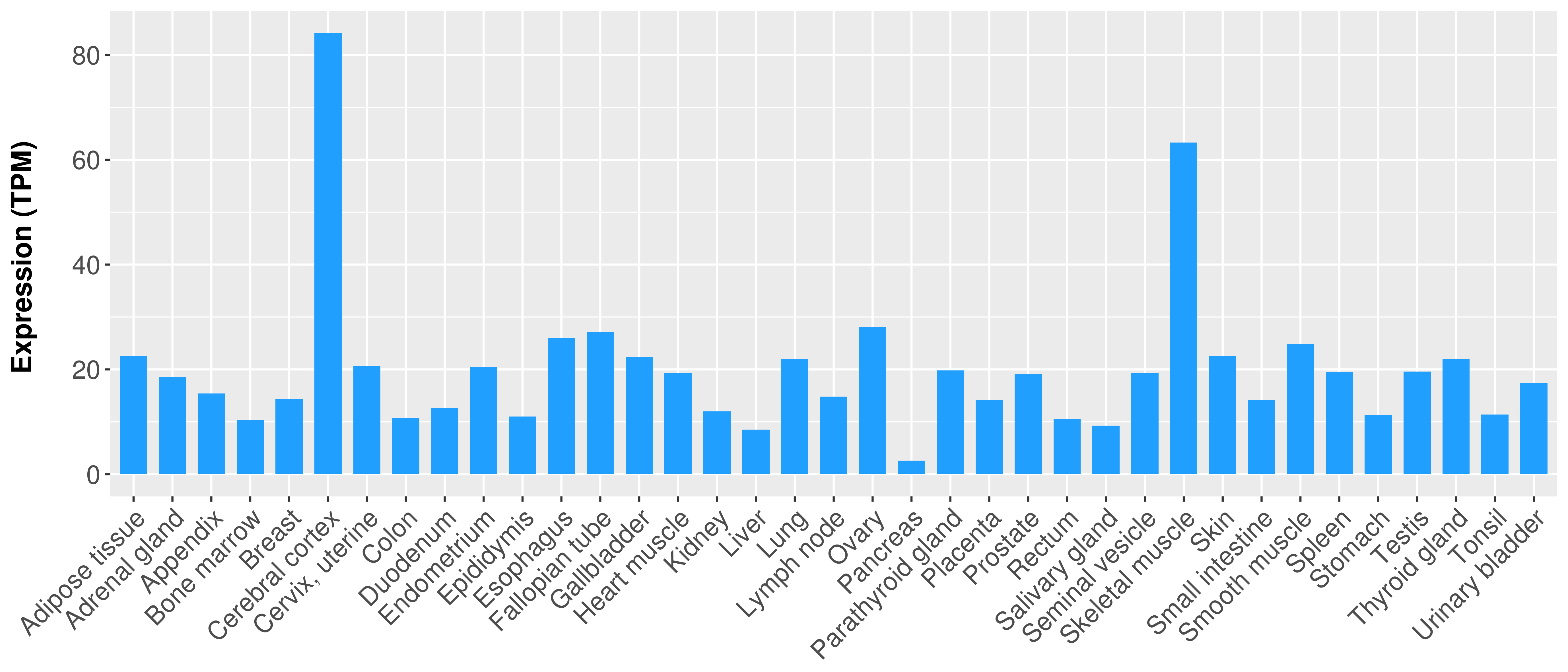
|
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
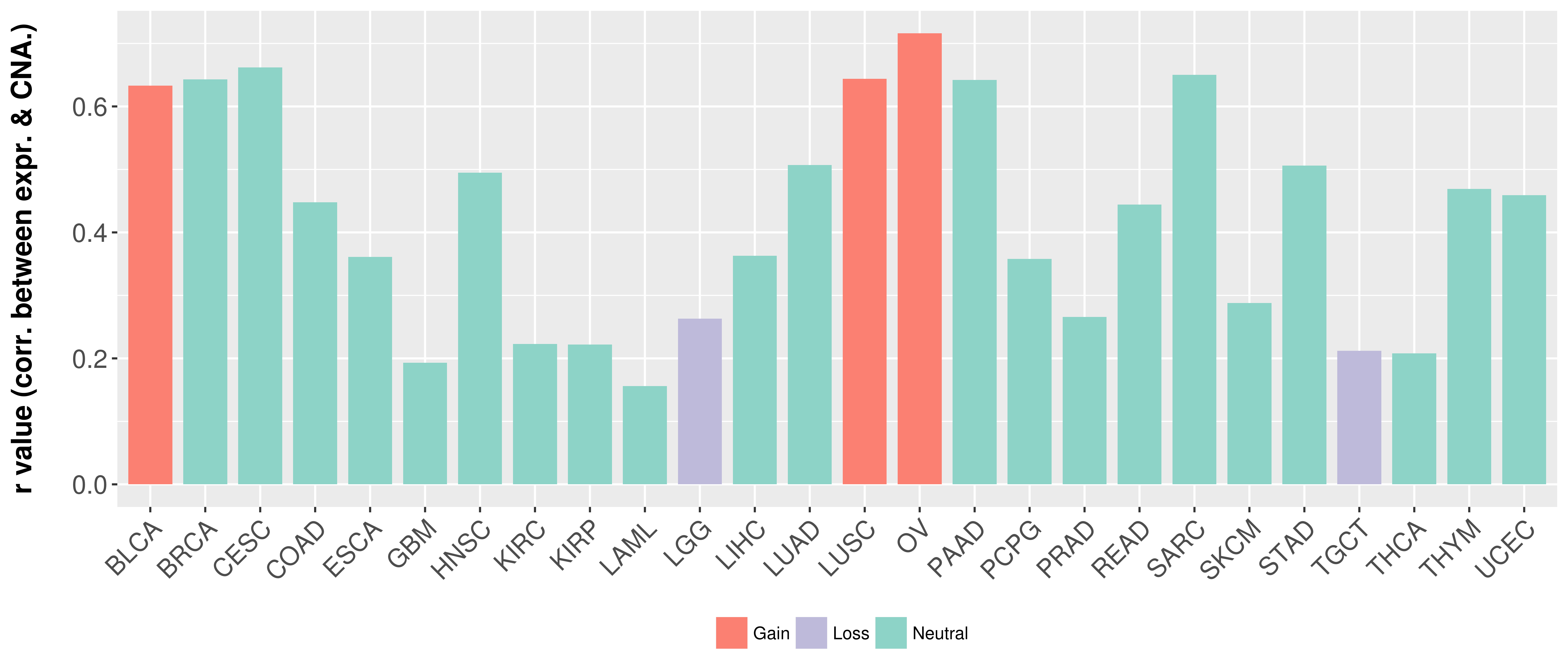
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
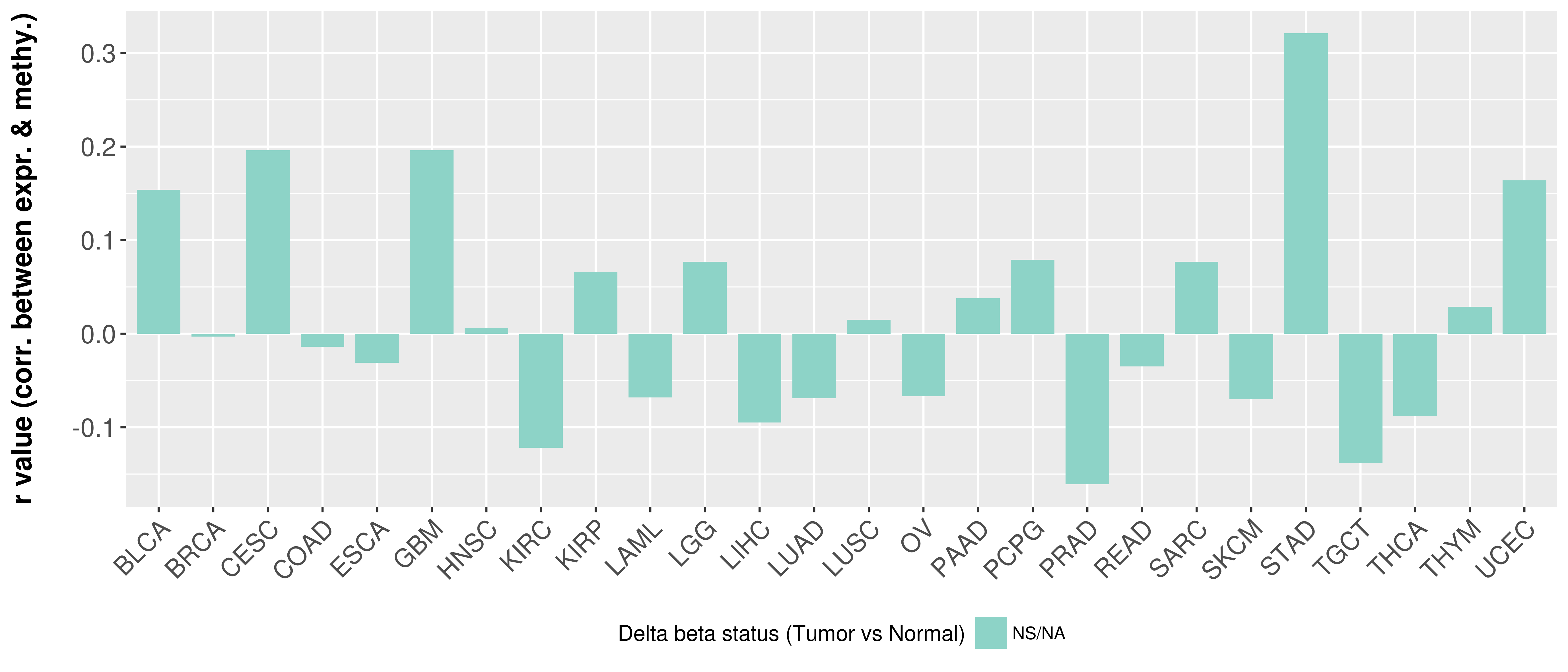
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
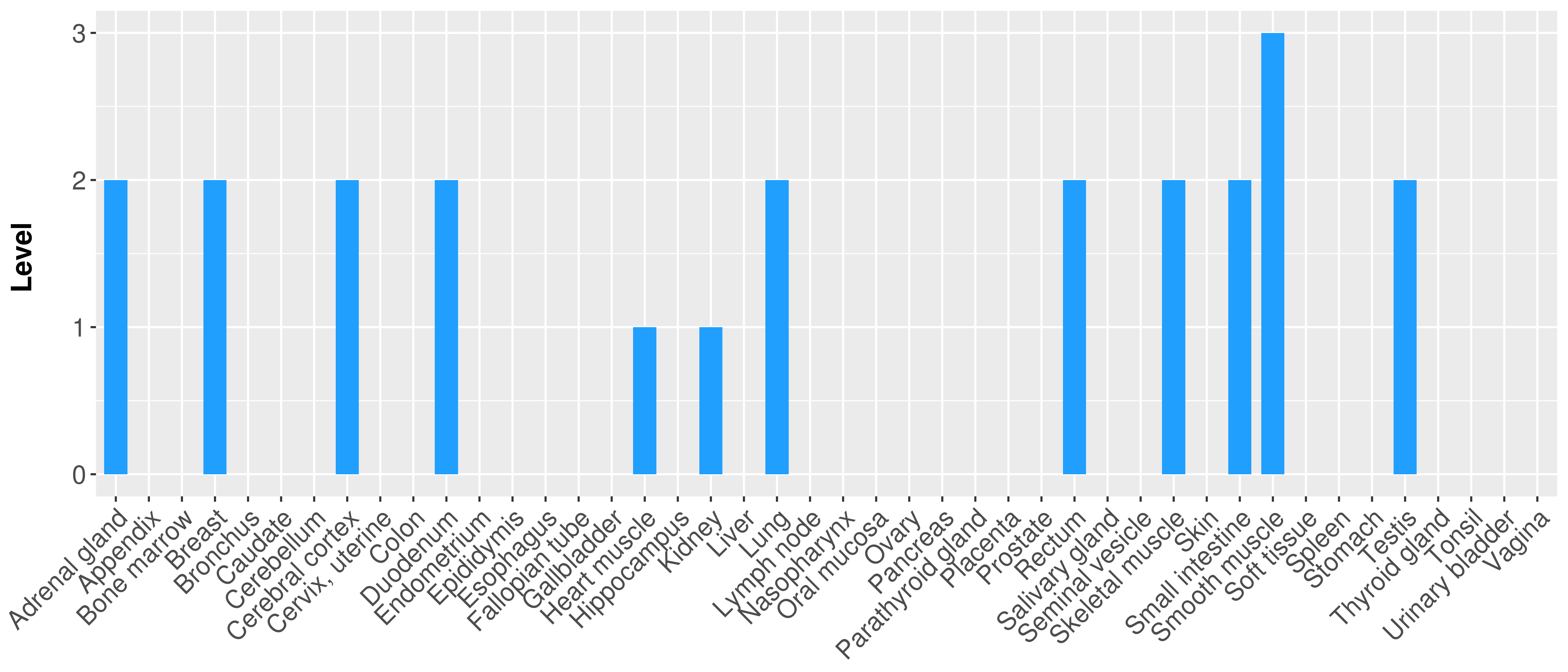
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
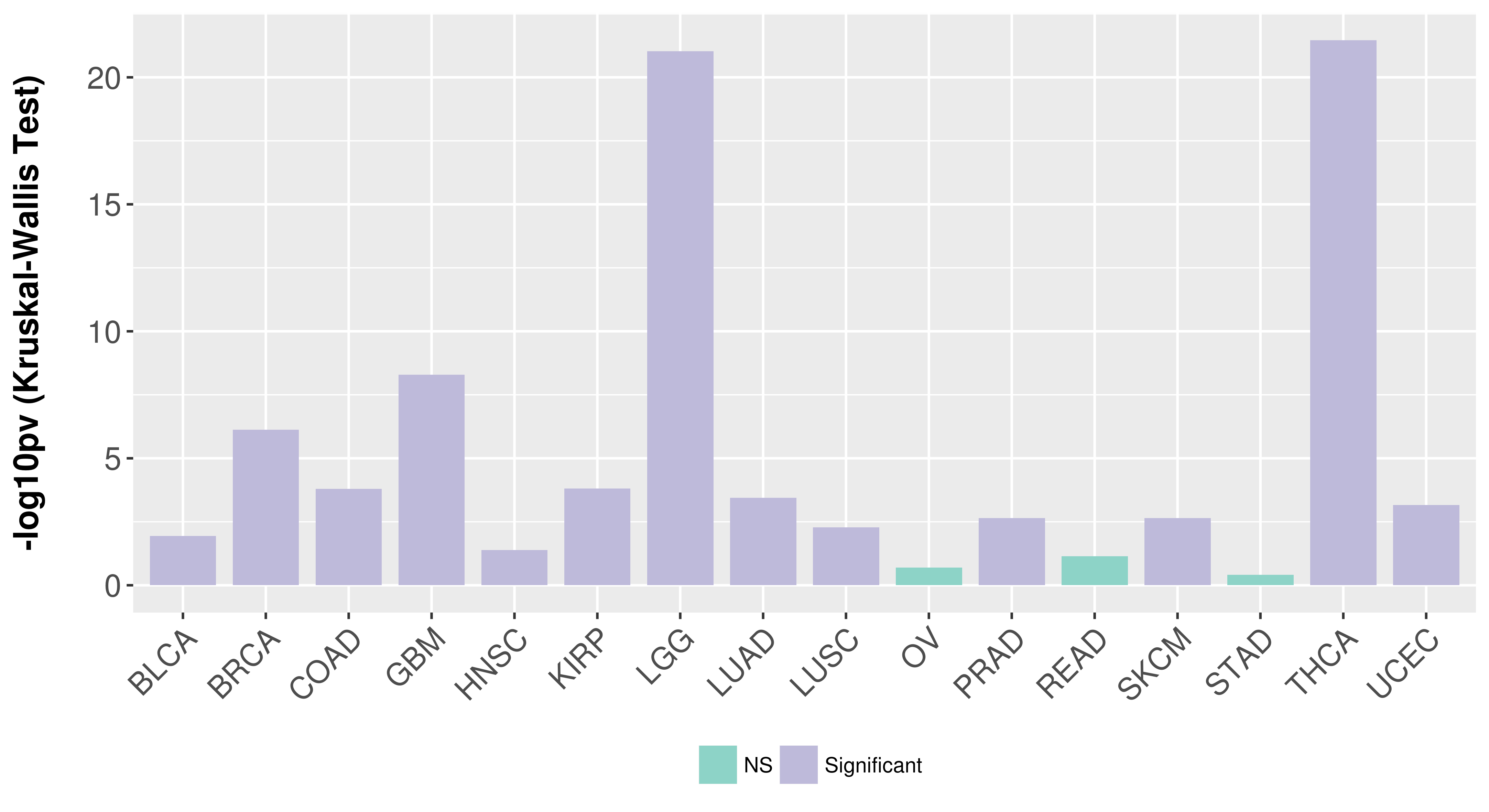
Association between expresson and subtype.
|
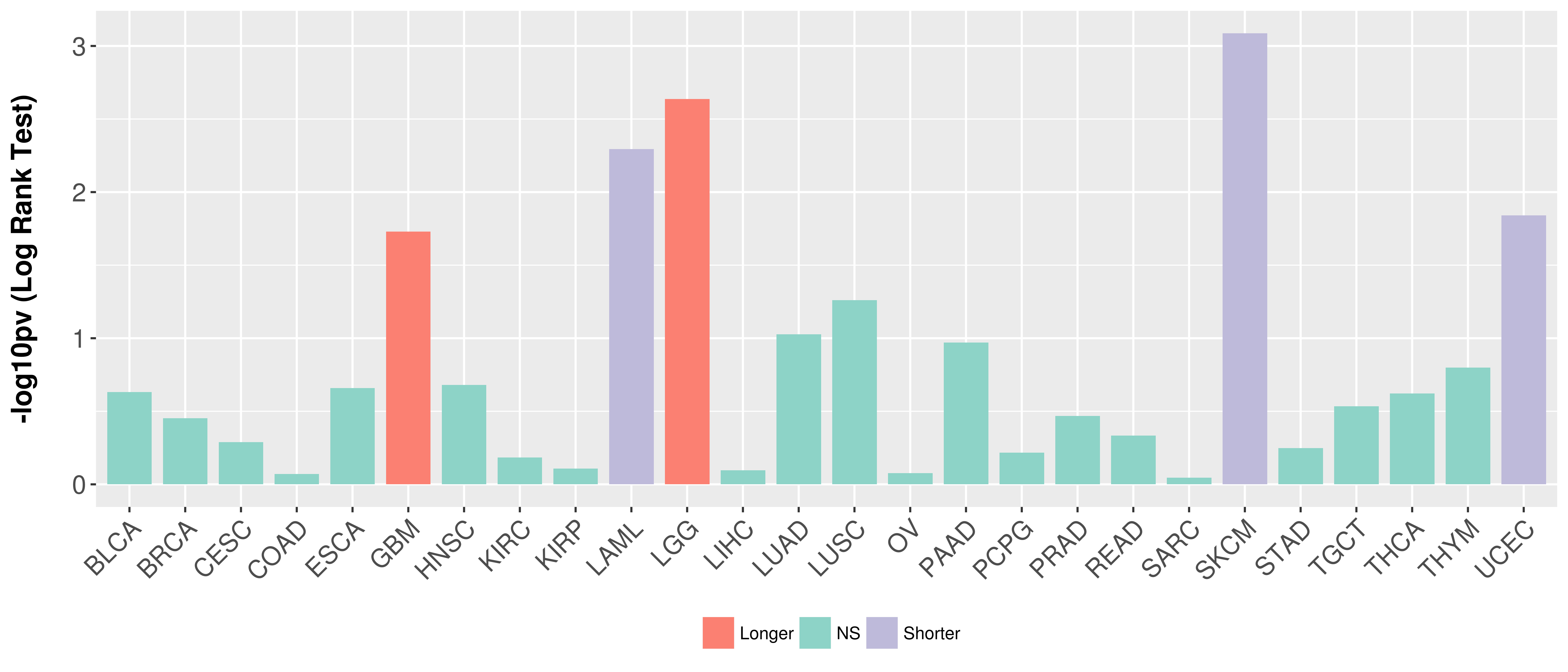
Overall survival analysis based on expression.
|
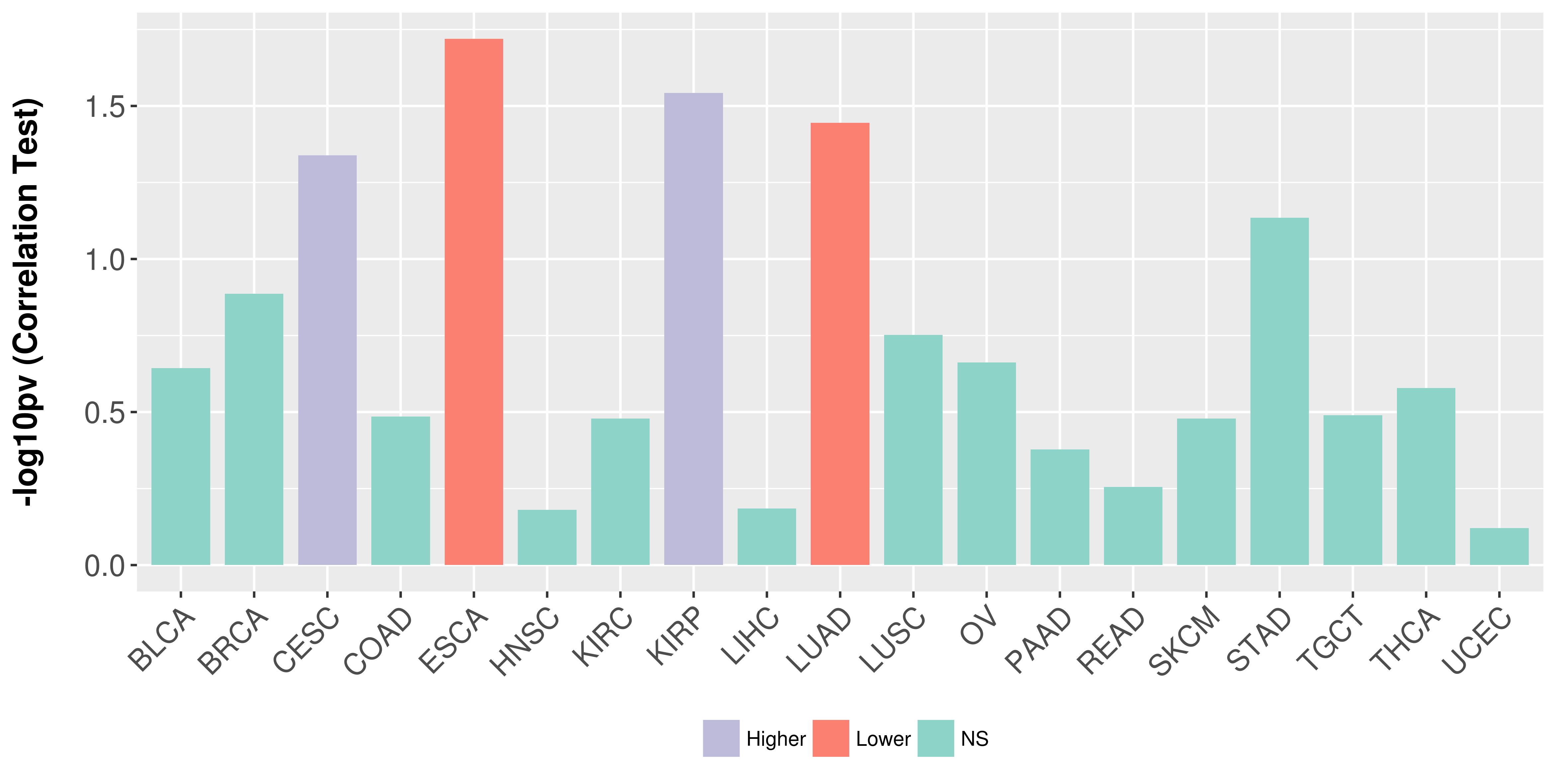
Association between expresson and stage.
|
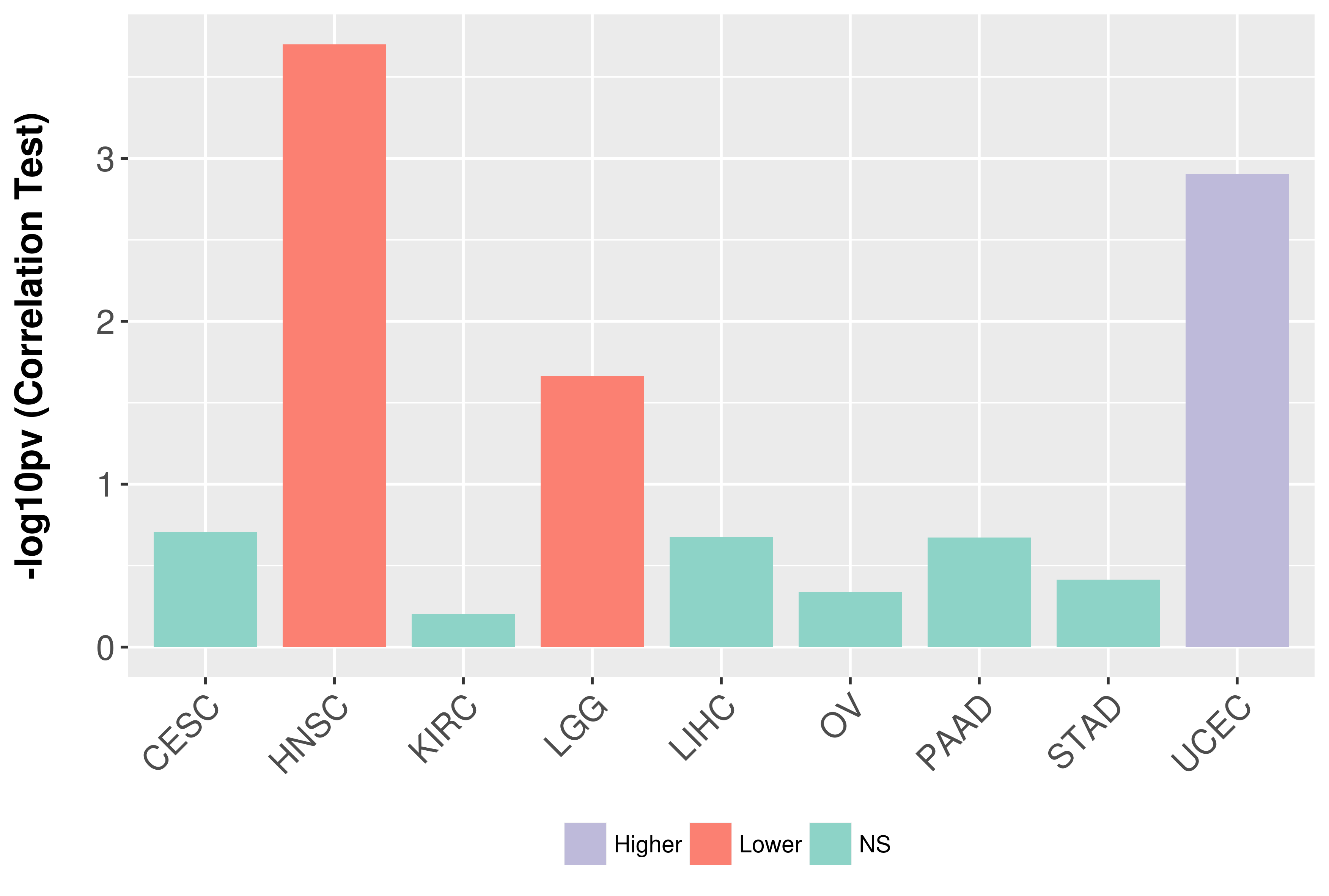
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SIRT2. |
| Summary | |
|---|---|
| Symbol | SIRT2 |
| Name | sirtuin 2 |
| Aliases | SIR2L; sirtuin (silent mating type information regulation 2, S.cerevisiae, homolog) 2; sirtuin (silent matin ...... |
| Location | 19q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|