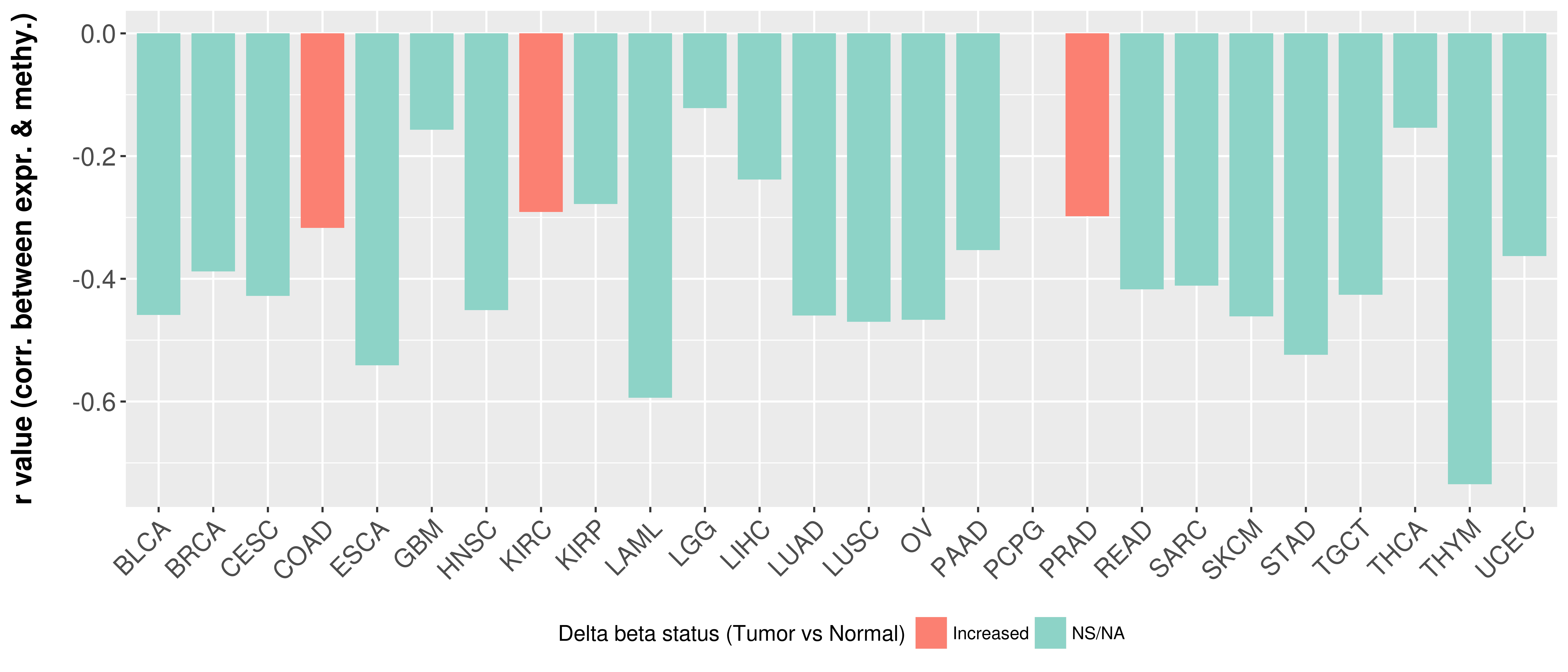
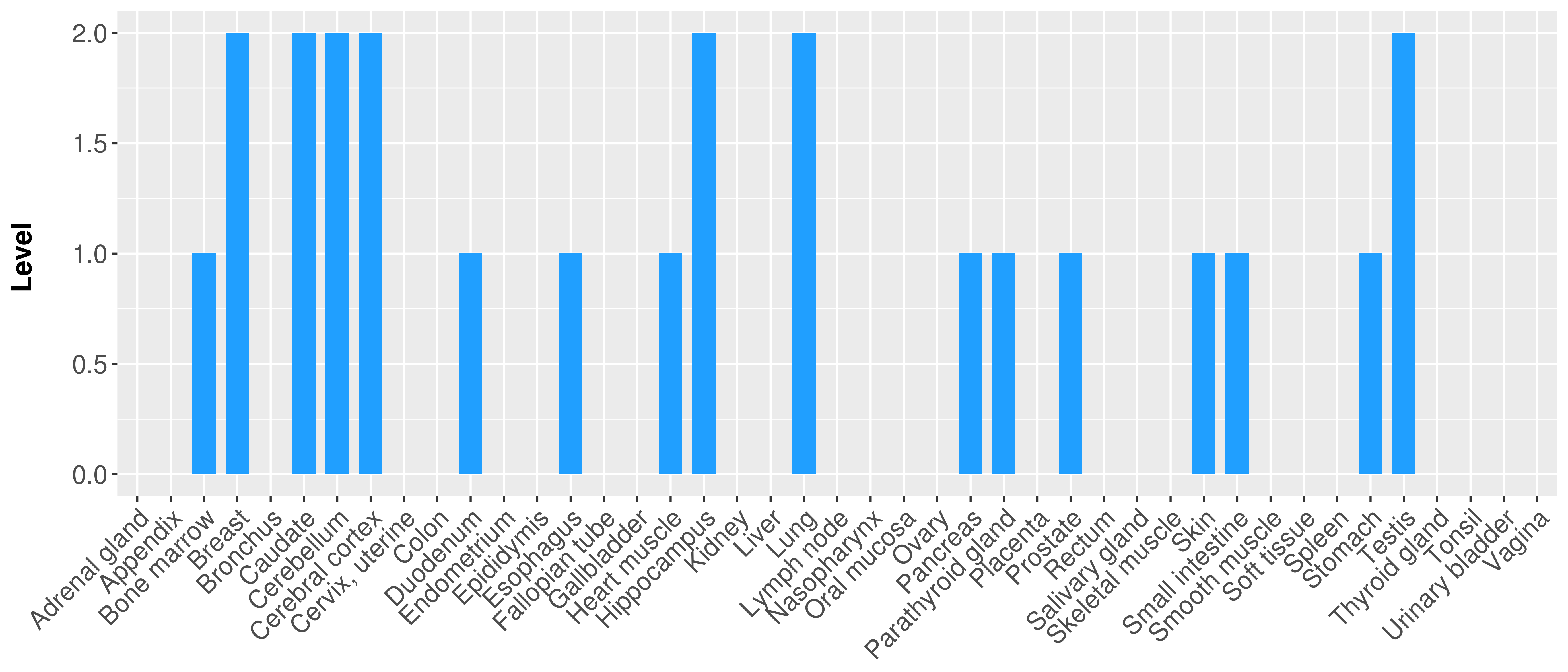
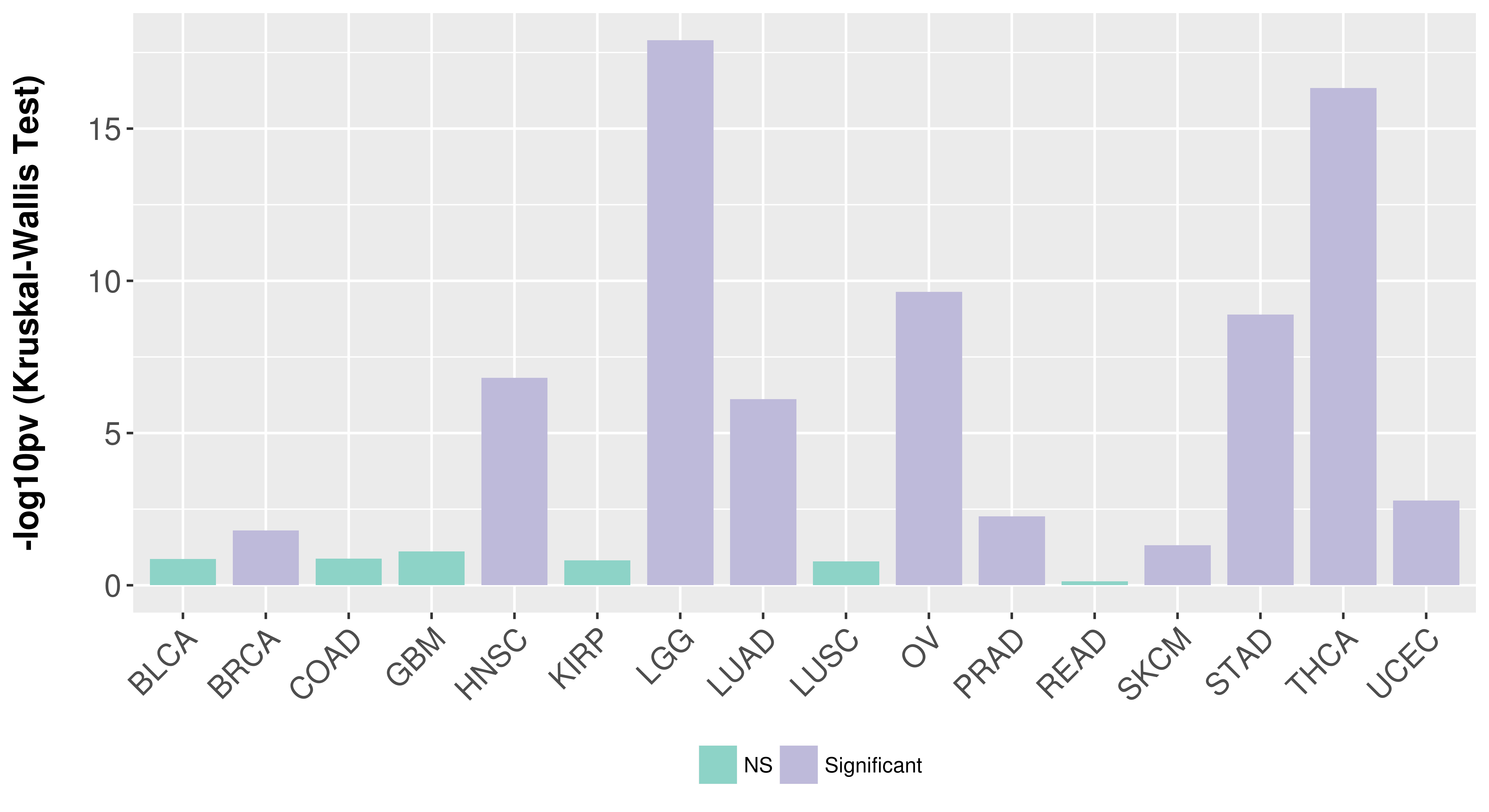
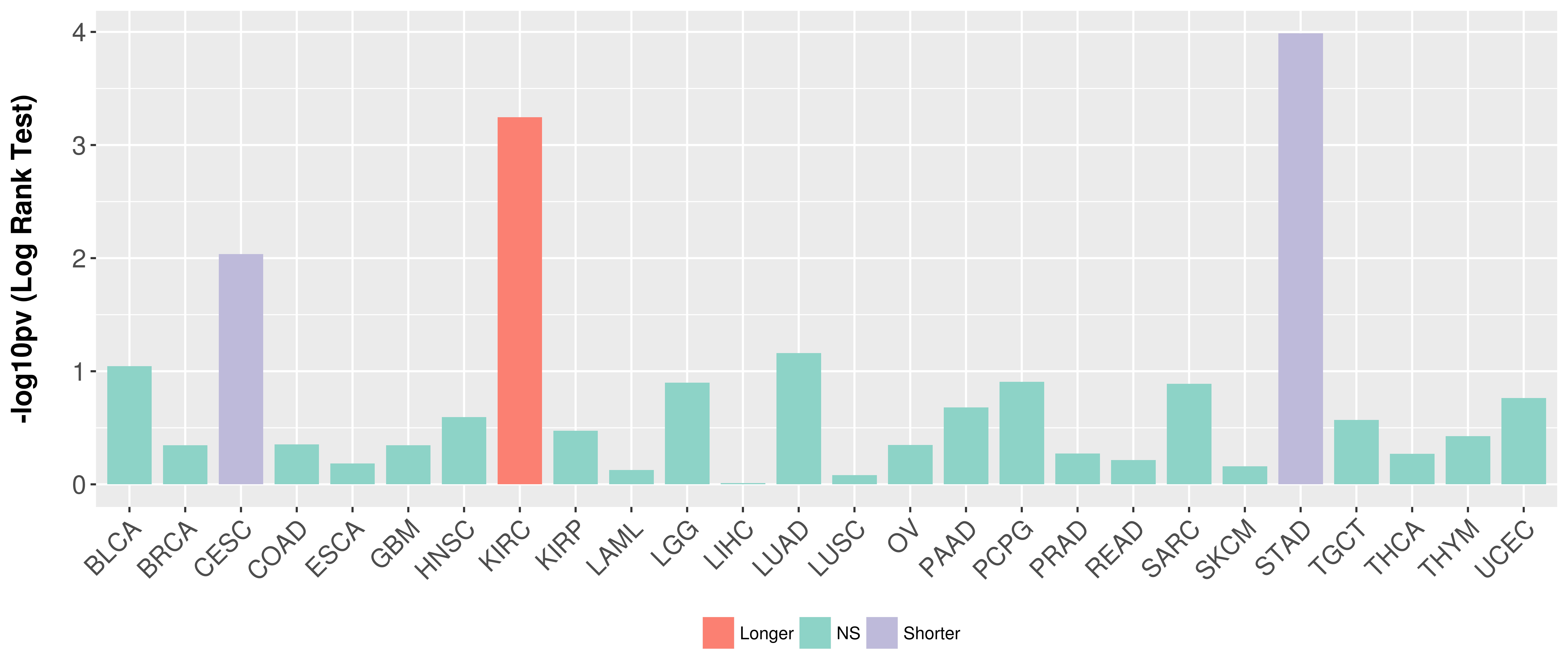
| COSM4728264 | c.1702T>A | p.F568I | Substitution - Missense | Large_intestine
|
| COSM3390403 | c.2565-4C>T | p.? | Unknown | Pancreas
|
| COSM3396412 | c.1505-3T>G | p.? | Unknown | Prostate
|
| COSM5367013 | c.399T>C | p.D133D | Substitution - coding silent | Large_intestine
|
| COSM4392338 | c.3030+1G>A | p.? | Unknown | Prostate
|
| COSM4436949 | c.3055C>G | p.L1019V | Substitution - Missense | Oesophagus
|
| COSM1331261 | c.1860C>G | p.F620L | Substitution - Missense | Ovary
|
| COSM1114785 | c.2147G>T | p.R716I | Substitution - Missense | Endometrium
|
| COSM5177205 | c.1127delA | p.N376fs*16 | Deletion - Frameshift | Large_intestine
|
| COSM2725347 | c.3043C>T | p.R1015C | Substitution - Missense | Cervix
|
| COSM118080 | c.2412T>C | p.I804I | Substitution - coding silent | Ovary
|
| COSM1114787 | c.1852C>T | p.R618C | Substitution - Missense | Endometrium
|
| COSM308155 | c.1594C>A | p.L532M | Substitution - Missense | Biliary_tract
|
| COSM4728262 | c.3105_3106delAG | p.R1035fs*9 | Deletion - Frameshift | Large_intestine
|
| COSM258058 | c.443G>A | p.R148H | Substitution - Missense | Large_intestine
|
| COSM1114781 | c.2557C>G | p.L853V | Substitution - Missense | Endometrium
|
| COSM1756379 | c.837G>T | p.M279I | Substitution - Missense | Urinary_tract
|
| COSM5435199 | c.2246C>T | p.P749L | Substitution - Missense | Oesophagus
|
| COSM4106534 | c.2743A>C | p.I915L | Substitution - Missense | Stomach
|
| COSM1132410 | c.2829C>A | p.Y943* | Substitution - Nonsense | Prostate
|
| COSM456720 | c.1384C>G | p.H462D | Substitution - Missense | Skin
|
| COSM3973224 | c.1300G>C | p.D434H | Substitution - Missense | Central_nervous_system
|
| COSM1664265 | c.2185T>G | p.F729V | Substitution - Missense | Kidney
|
| COSM755440 | c.1530G>T | p.Q510H | Substitution - Missense | Lung
|
| COSM1114824 | c.149C>T | p.T50M | Substitution - Missense | Endometrium
|
| COSM3713854 | c.824G>T | p.R275L | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM4386755 | c.548T>G | p.L183R | Substitution - Missense | Lung
|
| COSM3557677 | c.2587C>T | p.R863* | Substitution - Nonsense | Skin
|
| COSM456720 | c.1384C>G | p.H462D | Substitution - Missense | Breast
|
| COSM5750531 | c.929T>C | p.I310T | Substitution - Missense | Stomach
|
| COSM1114801 | c.1299A>C | p.K433N | Substitution - Missense | Endometrium
|
| COSM3396412 | c.1505-3T>G | p.? | Unknown | Prostate
|
| COSM5959316 | c.139A>G | p.T47A | Substitution - Missense | Thyroid
|
| COSM5005415 | c.1190G>A | p.R397H | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM755442 | c.2818-2A>C | p.? | Unknown | Lung
|
| COSM1114817 | c.468G>A | p.E156E | Substitution - coding silent | Endometrium
|
| COSM1490467 | c.1455T>A | p.S485R | Substitution - Missense | Breast
|
| COSM2725355 | c.2618A>T | p.E873V | Substitution - Missense | Large_intestine
|
| COSM251142 | c.478G>A | p.E160K | Substitution - Missense | Liver
|
| COSM3723639 | c.2769A>C | p.E923D | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM369386 | c.2226G>T | p.M742I | Substitution - Missense | Lung
|
| COSM5552876 | c.459A>T | p.E153D | Substitution - Missense | Prostate
|
| COSM1114797 | c.1497_1498insA | p.E500fs*29 | Insertion - Frameshift | Endometrium
|
| COSM1556051 | c.1029G>A | p.L343L | Substitution - coding silent | Haematopoietic_and_lymphoid_tissue
|
| COSM5577753 | c.538G>A | p.G180R | Substitution - Missense | Skin
|
| COSM3725137 | c.912C>G | p.H304Q | Substitution - Missense | Lung
|
| COSM3557681 | c.1816-1G>A | p.? | Unknown | Skin
|
| COSM1637190 | c.530-1G>A | p.? | Unknown | Bone
|
| COSM3424447 | c.1531A>G | p.M511V | Substitution - Missense | Large_intestine
|
| COSM1114824 | c.149C>T | p.T50M | Substitution - Missense | Large_intestine
|
| COSM4156575 | c.174G>A | p.E58E | Substitution - coding silent | Thyroid
|
| COSM1114826 | c.78C>T | p.D26D | Substitution - coding silent | Endometrium
|
| COSM1114803 | c.1242G>T | p.K414N | Substitution - Missense | Endometrium
|
| COSM82374 | c.585C>A | p.I195I | Substitution - coding silent | Ovary
|
| COSM755437 | c.446C>T | p.T149I | Substitution - Missense | Lung
|
| COSM253100 | c.2128delA | p.M710fs*6 | Deletion - Frameshift | Large_intestine
|
| COSM4393506 | c.2978G>C | p.R993P | Substitution - Missense | Prostate
|
| COSM5749641 | c.1109C>T | p.S370F | Substitution - Missense | Genital_tract
|
| COSM4792252 | c.319C>G | p.H107D | Substitution - Missense | Liver
|
| COSM1756379 | c.837G>T | p.M279I | Substitution - Missense | Urinary_tract
|
| COSM3800393 | c.2752C>G | p.Q918E | Substitution - Missense | Urinary_tract
|
| COSM755436 | c.417T>G | p.I139M | Substitution - Missense | Lung
|
| COSM5000569 | c.2314C>G | p.P772A | Substitution - Missense | Pancreas
|
| COSM253100 | c.2128delA | p.M710fs*6 | Deletion - Frameshift | Ovary
|
| COSM1114791 | c.1639G>T | p.E547* | Substitution - Nonsense | Endometrium
|
| COSM1114773 | c.2685C>A | p.V895V | Substitution - coding silent | Endometrium
|
| COSM79057 | c.757C>T | p.R253* | Substitution - Nonsense | Ovary
|
| COSM487922 | c.272G>A | p.R91Q | Substitution - Missense | Kidney
|
| COSM1643943 | c.2896G>A | p.E966K | Substitution - Missense | Stomach
|
| COSM1665759 | c.1462delA | p.M488fs*2 | Deletion - Frameshift | Kidney
|
| COSM255810 | c.1714C>T | p.L572L | Substitution - coding silent | Central_nervous_system
|
| COSM1114795 | c.1498G>T | p.E500* | Substitution - Nonsense | Endometrium
|
| COSM3372054 | c.534G>C | p.V178V | Substitution - coding silent | Thyroid
|
| COSM5064355 | c.917G>A | p.R306Q | Substitution - Missense | Stomach
|
| COSM1569706 | c.2351C>G | p.P784R | Substitution - Missense | Large_intestine
|
| COSM1465512 | c.794G>T | p.G265V | Substitution - Missense | Large_intestine
|
| COSM1114763 | c.3044G>A | p.R1015H | Substitution - Missense | Endometrium
|
| COSM1625488 | c.2954A>G | p.Y985C | Substitution - Missense | Liver
|
| COSM1114805 | c.1109C>A | p.S370Y | Substitution - Missense | Endometrium
|
| COSM5450622 | c.1551T>G | p.I517M | Substitution - Missense | Large_intestine
|
| COSM3843511 | c.1319C>T | p.S440F | Substitution - Missense | Breast
|
| COSM5467813 | c.2127A>C | p.K709N | Substitution - Missense | Large_intestine
|
| COSM1114799 | c.1458T>A | p.G486G | Substitution - coding silent | Endometrium
|
| COSM5812439 | c.546A>T | p.P182P | Substitution - coding silent | Liver
|
| COSM3800394 | c.2359C>T | p.Q787* | Substitution - Nonsense | Urinary_tract
|
| COSM1114803 | c.1242G>T | p.K414N | Substitution - Missense | Large_intestine
|
| COSM1490468 | c.962C>A | p.S321Y | Substitution - Missense | Breast
|
| COSM5016708 | c.2236_2242delATTGAAC | p.I746fs*64 | Deletion - Frameshift | Kidney
|
| COSM3770673 | c.1254delG | p.L419fs*1 | Deletion - Frameshift | Soft_tissue
|
| COSM1490471 | c.126G>A | p.A42A | Substitution - coding silent | Thyroid
|
| COSM1465508 | c.2257C>T | p.R753C | Substitution - Missense | Large_intestine
|
| COSM1114819 | c.460G>T | p.D154Y | Substitution - Missense | Endometrium
|
| COSM392049 | c.175-1delG | p.? | Unknown | Lung
|
| COSM3713854 | c.824G>T | p.R275L | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM3964684 | c.2851C>A | p.R951S | Substitution - Missense | Lung
|
| COSM4728263 | c.2958A>G | p.E986E | Substitution - coding silent | Large_intestine
|
| COSM1625488 | c.2954A>G | p.Y985C | Substitution - Missense | Liver
|
| COSM4938389 | c.889A>T | p.K297* | Substitution - Nonsense | Liver
|
| COSM367812 | c.619G>T | p.A207S | Substitution - Missense | Lung
|
| COSM362721 | c.2506G>A | p.D836N | Substitution - Missense | Lung
|
| COSM755435 | c.103C>G | p.Q35E | Substitution - Missense | Lung
|
| COSM5347923 | c.1986delA | p.E663fs*6 | Deletion - Frameshift | Liver
|
| COSM4106536 | c.2116C>T | p.R706C | Substitution - Missense | Stomach
|
| COSM487921 | c.1084T>C | p.F362L | Substitution - Missense | Kidney
|
| COSM3557680 | c.1819C>T | p.R607* | Substitution - Nonsense | Skin
|
| COSM5775127 | c.103C>T | p.Q35* | Substitution - Nonsense | Breast
|
| COSM4728265 | c.1611G>A | p.P537P | Substitution - coding silent | Large_intestine
|
| COSM4535459 | c.2194G>A | p.E732K | Substitution - Missense | Skin
|
| COSM5655920 | c.1922G>C | p.R641T | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM2725387 | c.858C>T | p.C286C | Substitution - coding silent | Biliary_tract
|
| COSM1490471 | c.126G>A | p.A42A | Substitution - coding silent | Breast
|
| COSM3424448 | c.705C>A | p.H235Q | Substitution - Missense | Large_intestine
|
| COSM3557679 | c.2424G>A | p.R808R | Substitution - coding silent | Skin
|
| COSM5647979 | c.2251C>T | p.R751* | Substitution - Nonsense | Oesophagus
|
| COSM1114765 | c.2996G>T | p.R999I | Substitution - Missense | Endometrium
|
| COSM5433242 | c.1384C>T | p.H462Y | Substitution - Missense | Oesophagus
|
| COSM1465521 | c.169_171delAAG | p.K57delK | Deletion - In frame | Large_intestine
|
| COSM5005567 | c.811-2A>G | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1114785 | c.2147G>T | p.R716I | Substitution - Missense | Endometrium
|
| COSM78542 | c.1574G>A | p.R525H | Substitution - Missense | Ovary
|
| COSM144478 | c.1004G>A | p.R335H | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4792252 | c.319C>G | p.H107D | Substitution - Missense | Liver
|
| COSM203615 | c.386G>A | p.R129Q | Substitution - Missense | Endometrium
|
| COSM5411960 | c.2863G>A | p.G955R | Substitution - Missense | Skin
|
| COSM2152834 | c.123C>T | p.A41A | Substitution - coding silent | Central_nervous_system
|
| COSM1465508 | c.2257C>T | p.R753C | Substitution - Missense | Stomach
|
| COSM1569707 | c.2761C>T | p.R921C | Substitution - Missense | Large_intestine
|
| COSM3405946 | c.2172A>C | p.Q724H | Substitution - Missense | Central_nervous_system
|
| COSM5002573 | c.1093G>T | p.A365S | Substitution - Missense | Pancreas
|
| COSM5665456 | c.421G>C | p.A141P | Substitution - Missense | Soft_tissue
|
| COSM2152834 | c.123C>T | p.A41A | Substitution - coding silent | Central_nervous_system
|
| COSM4728267 | c.393G>T | p.K131N | Substitution - Missense | Large_intestine
|
| COSM1114783 | c.2475T>C | p.A825A | Substitution - coding silent | Endometrium
|
| COSM1266206 | c.2213A>T | p.Q738L | Substitution - Missense | Oesophagus
|
| COSM5438788 | c.844G>T | p.E282* | Substitution - Nonsense | Oesophagus
|
| COSM1715512 | c.1675G>A | p.A559T | Substitution - Missense | Skin
|
| COSM1465507 | c.2716T>C | p.C906R | Substitution - Missense | Large_intestine
|
| COSM1114767 | c.2978G>A | p.R993Q | Substitution - Missense | Endometrium
|
| COSM1114815 | c.524C>A | p.P175H | Substitution - Missense | Endometrium
|
| COSM5029010 | c.962C>G | p.S321C | Substitution - Missense | Breast
|
| COSM1114789 | c.1794A>T | p.P598P | Substitution - coding silent | Endometrium
|
| COSM1114787 | c.1852C>T | p.R618C | Substitution - Missense | Large_intestine
|
| COSM298514 | c.2713C>T | p.R905C | Substitution - Missense | Large_intestine
|
| COSM1114809 | c.886G>T | p.E296* | Substitution - Nonsense | Endometrium
|
| COSM1114828 | c.2T>C | p.M1T | Substitution - Missense | Endometrium
|
| COSM3708428 | c.530-4C>T | p.? | Unknown | Liver
|
| COSM4487072 | c.3137C>T | p.P1046L | Substitution - Missense | Skin
|
| COSM277375 | c.2535G>A | p.E845E | Substitution - coding silent | Large_intestine
|
| COSM1114807 | c.1050G>A | p.L350L | Substitution - coding silent | Endometrium
|
| COSM5366854 | c.2825G>T | p.R942I | Substitution - Missense | Large_intestine
|
| COSM5435200 | c.793G>A | p.G265R | Substitution - Missense | Oesophagus
|
| COSM3843512 | c.1149C>T | p.L383L | Substitution - coding silent | Breast
|
| COSM4568086 | c.2781_2782AC>TT | p.Q927_R928>H* | Complex - compound substitution | Skin
|
| COSM1465521 | c.169_171delAAG | p.K57delK | Deletion - In frame | Oesophagus
|
| COSM5588982 | c.1904G>A | p.R635K | Substitution - Missense | Skin
|
| COSM2725355 | c.2618A>T | p.E873V | Substitution - Missense | Large_intestine
|
| COSM5064356 | c.859G>A | p.V287I | Substitution - Missense | Stomach
|
| COSM1114813 | c.668G>A | p.G223D | Substitution - Missense | Stomach
|
| COSM1331262 | c.2580G>C | p.W860C | Substitution - Missense | Ovary
|
| COSM21740 | c.1815+1G>A | p.? | Unknown | Skin
|
| COSM1114793 | c.1497_1500delAGAA | p.K499fs*12 | Deletion - Frameshift | Endometrium
|
| COSM5903299 | c.823C>T | p.R275C | Substitution - Missense | Skin
|
| COSM1490469 | c.433C>T | p.R145C | Substitution - Missense | Breast
|
| COSM4449547 | c.262-1G>A | p.? | Unknown | Skin
|
| COSM4635777 | c.2921T>C | p.L974S | Substitution - Missense | Large_intestine
|
| COSM2725393 | c.172G>A | p.E58K | Substitution - Missense | Thyroid
|
| COSM456719 | c.2359delC | p.Q787fs*25 | Deletion - Frameshift | Breast
|
| COSM4807444 | c.1793C>T | p.P598L | Substitution - Missense | Pancreas
|
| COSM1465508 | c.2257C>T | p.R753C | Substitution - Missense | Large_intestine
|
| COSM1465509 | c.983G>A | p.R328H | Substitution - Missense | Large_intestine
|
| COSM3557683 | c.514G>T | p.E172* | Substitution - Nonsense | Skin
|
| COSM1465521 | c.169_171delAAG | p.K57delK | Deletion - In frame | Large_intestine
|
| COSM4728266 | c.1134T>C | p.L378L | Substitution - coding silent | Large_intestine
|
| COSM2725382 | c.1370G>A | p.R457Q | Substitution - Missense | Large_intestine
|
| COSM405234 | c.3030_3030+1insTG | p.E1011fs*10 | Unknown | Lung
|
| COSM3939781 | c.271C>T | p.R91* | Substitution - Nonsense | Oesophagus
|
| COSM5910094 | c.2722G>A | p.E908K | Substitution - Missense | Skin
|
| COSM1114822 | c.177A>C | p.K59N | Substitution - Missense | Large_intestine
|
| COSM5007579 | c.3109G>T | p.E1037* | Substitution - Nonsense | Large_intestine
|
| COSM1114811 | c.829G>T | p.E277* | Substitution - Nonsense | Endometrium
|
| COSM1114769 | c.2875G>T | p.G959* | Substitution - Nonsense | Endometrium
|
| COSM3708428 | c.530-4C>T | p.? | Unknown | Liver
|
| COSM3557682 | c.719C>A | p.P240Q | Substitution - Missense | Skin
|
| COSM1465510 | c.891_892insA | p.S298fs*14 | Insertion - Frameshift | Large_intestine
|
| COSM1114822 | c.177A>C | p.K59N | Substitution - Missense | Endometrium
|
| COSM255810 | c.1714C>T | p.L572L | Substitution - coding silent | Central_nervous_system
|
| COSM1569705 | c.608A>G | p.N203S | Substitution - Missense | Large_intestine
|
| COSM4589172 | c.2013T>G | p.H671Q | Substitution - Missense | Bone
|
| COSM5681062 | c.2710G>A | p.E904K | Substitution - Missense | Soft_tissue
|
| COSM363027 | c.2754A>T | p.Q918H | Substitution - Missense | Lung
|
| COSM5421036 | c.1997_1998insA | p.Q667fs*14 | Insertion - Frameshift | Prostate
|
| COSM1114785 | c.2147G>T | p.R716I | Substitution - Missense | Kidney
|
| COSM1114787 | c.1852C>T | p.R618C | Substitution - Missense | Endometrium
|
| COSM4770921 | c.705C>T | p.H235H | Substitution - coding silent | Pleura
|
| COSM3738180 | c.1449C>A | p.S483R | Substitution - Missense | Soft_tissue
|
| COSM1465521 | c.169_171delAAG | p.K57delK | Deletion - In frame | Biliary_tract
|
| COSM367813 | c.149C>A | p.T50K | Substitution - Missense | Lung
|
| COSM3973225 | c.1270C>T | p.R424* | Substitution - Nonsense | Central_nervous_system
|
| COSM4892745 | c.955G>A | p.E319K | Substitution - Missense | Skin
|
| COSM1114767 | c.2978G>A | p.R993Q | Substitution - Missense | Skin
|
| COSM1465506 | c.2783G>A | p.R928Q | Substitution - Missense | Large_intestine
|
| COSM5951052 | c.193C>T | p.Q65* | Substitution - Nonsense | Prostate
|
| COSM2725350 | c.2852G>A | p.R951H | Substitution - Missense | Large_intestine
|
| COSM3701800 | c.2443-6T>G | p.? | Unknown | Liver
|
| COSM4901049 | c.3076G>A | p.E1026K | Substitution - Missense | Skin
|
| COSM3843510 | c.2542G>C | p.E848Q | Substitution - Missense | Breast
|
| COSM1756380 | c.92C>G | p.P31R | Substitution - Missense | Urinary_tract
|
| COSM1114767 | c.2978G>A | p.R993Q | Substitution - Missense | Stomach
|
| COSM4954000 | c.95C>A | p.S32Y | Substitution - Missense | Liver
|
| COSM4106535 | c.2384G>A | p.R795H | Substitution - Missense | Stomach
|
| COSM21740 | c.1815+1G>A | p.? | Unknown | Skin
|
| COSM1114775 | c.2673C>A | p.S891S | Substitution - coding silent | Endometrium
|
| COSM5617513 | c.3129T>G | p.T1043T | Substitution - coding silent | Lung
|
| COSM1490466 | c.2665G>T | p.G889C | Substitution - Missense | Breast
|
| COSM1756380 | c.92C>G | p.P31R | Substitution - Missense | Urinary_tract
|
| COSM3557678 | c.2490A>G | p.E830E | Substitution - coding silent | Skin
|
| COSM4106532 | c.2774G>A | p.R925K | Substitution - Missense | Stomach
|
| COSM3800395 | c.1099-2A>G | p.? | Unknown | Urinary_tract
|
| COSM755438 | c.513T>A | p.F171L | Substitution - Missense | Lung
|
| COSM1569705 | c.608A>G | p.N203S | Substitution - Missense | Liver
|
| COSM4106537 | c.2038A>T | p.K680* | Substitution - Nonsense | Stomach
|
| COSM2725377 | c.1690A>G | p.N564D | Substitution - Missense | Large_intestine
|
| COSM1114779 | c.2590G>A | p.D864N | Substitution - Missense | Endometrium
|
| COSM1114777 | c.2654G>A | p.R885Q | Substitution - Missense | Endometrium
|
| COSM4106533 | c.2773A>G | p.R925G | Substitution - Missense | Stomach
|
| COSM1114771 | c.2867C>A | p.T956N | Substitution - Missense | Endometrium
|
| COSM1569705 | c.608A>G | p.N203S | Substitution - Missense | Liver
|
| COSM251142 | c.478G>A | p.E160K | Substitution - Missense | Liver
|
| COSM4954000 | c.95C>A | p.S32Y | Substitution - Missense | Liver
|
| COSM755441 | c.1957A>T | p.I653F | Substitution - Missense | Lung
|
| COSM1114787 | c.1852C>T | p.R618C | Substitution - Missense | Endometrium
|
| COSM5411961 | c.886G>A | p.E296K | Substitution - Missense | Skin
|
| COSM4411470 | c.1971_1972insTT | p.N658fs*12 | Insertion - Frameshift | Kidney
|
| COSM21740 | c.1815+1G>A | p.? | Unknown | Skin
|
| COSM755439 | c.639G>T | p.G213G | Substitution - coding silent | Lung
|
| COSM4486317 | c.3036C>T | p.F1012F | Substitution - coding silent | Skin
|
| COSM1727831 | c.2427G>T | p.K809N | Substitution - Missense | Liver
|
| COSM1465509 | c.983G>A | p.R328H | Substitution - Missense | Large_intestine
|
| COSM1490470 | c.262-1G>C | p.? | Unknown | Breast
|
| COSM5367013 | c.399T>C | p.D133D | Substitution - coding silent | Large_intestine
|
| COSM5700284 | c.2010G>A | p.R670R | Substitution - coding silent | Soft_tissue
|
| COSM1114813 | c.668G>A | p.G223D | Substitution - Missense | Endometrium
|
| COSM1190630 | c.2312A>G | p.E771G | Substitution - Missense | Lung
|