Browse SMARCC1 in pancancer
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00249 Myb-like DNA-binding domain PF04433 SWIRM domain PF16495 SWIRM-associated region 1 PF16496 SWIRM-associated domain at the N-terminal PF16498 SWIRM-associated domain at the C-terminal |
||||||||||
| Function |
Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). May stimulate the ATPase activity of the catalytic subunit of the complex. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001655 urogenital system development GO:0006333 chromatin assembly or disassembly GO:0006337 nucleosome disassembly GO:0006338 chromatin remodeling GO:0008286 insulin receptor signaling pathway GO:0009894 regulation of catabolic process GO:0009895 negative regulation of catabolic process GO:0010498 proteasomal protein catabolic process GO:0030850 prostate gland development GO:0031329 regulation of cellular catabolic process GO:0031330 negative regulation of cellular catabolic process GO:0031498 chromatin disassembly GO:0032434 regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0032435 negative regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0032868 response to insulin GO:0032869 cellular response to insulin stimulus GO:0032984 macromolecular complex disassembly GO:0032986 protein-DNA complex disassembly GO:0034728 nucleosome organization GO:0042176 regulation of protein catabolic process GO:0042177 negative regulation of protein catabolic process GO:0043044 ATP-dependent chromatin remodeling GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043241 protein complex disassembly GO:0043434 response to peptide hormone GO:0045861 negative regulation of proteolysis GO:0048608 reproductive structure development GO:0048732 gland development GO:0061136 regulation of proteasomal protein catabolic process GO:0061458 reproductive system development GO:0071375 cellular response to peptide hormone stimulus GO:0071417 cellular response to organonitrogen compound GO:0071824 protein-DNA complex subunit organization GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1901799 negative regulation of proteasomal protein catabolic process GO:1903050 regulation of proteolysis involved in cellular protein catabolic process GO:1903051 negative regulation of proteolysis involved in cellular protein catabolic process GO:1903362 regulation of cellular protein catabolic process GO:1903363 negative regulation of cellular protein catabolic process |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001158 enhancer sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0031490 chromatin DNA binding GO:0031491 nucleosome binding GO:0031492 nucleosomal DNA binding GO:0035326 enhancer binding GO:0043566 structure-specific DNA binding GO:0047485 protein N-terminus binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000803 sex chromosome GO:0001741 XY body GO:0016514 SWI/SNF complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0071564 npBAF complex GO:0071565 nBAF complex GO:0090544 BAF-type complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214858: RMTs methylate histone arginines |
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
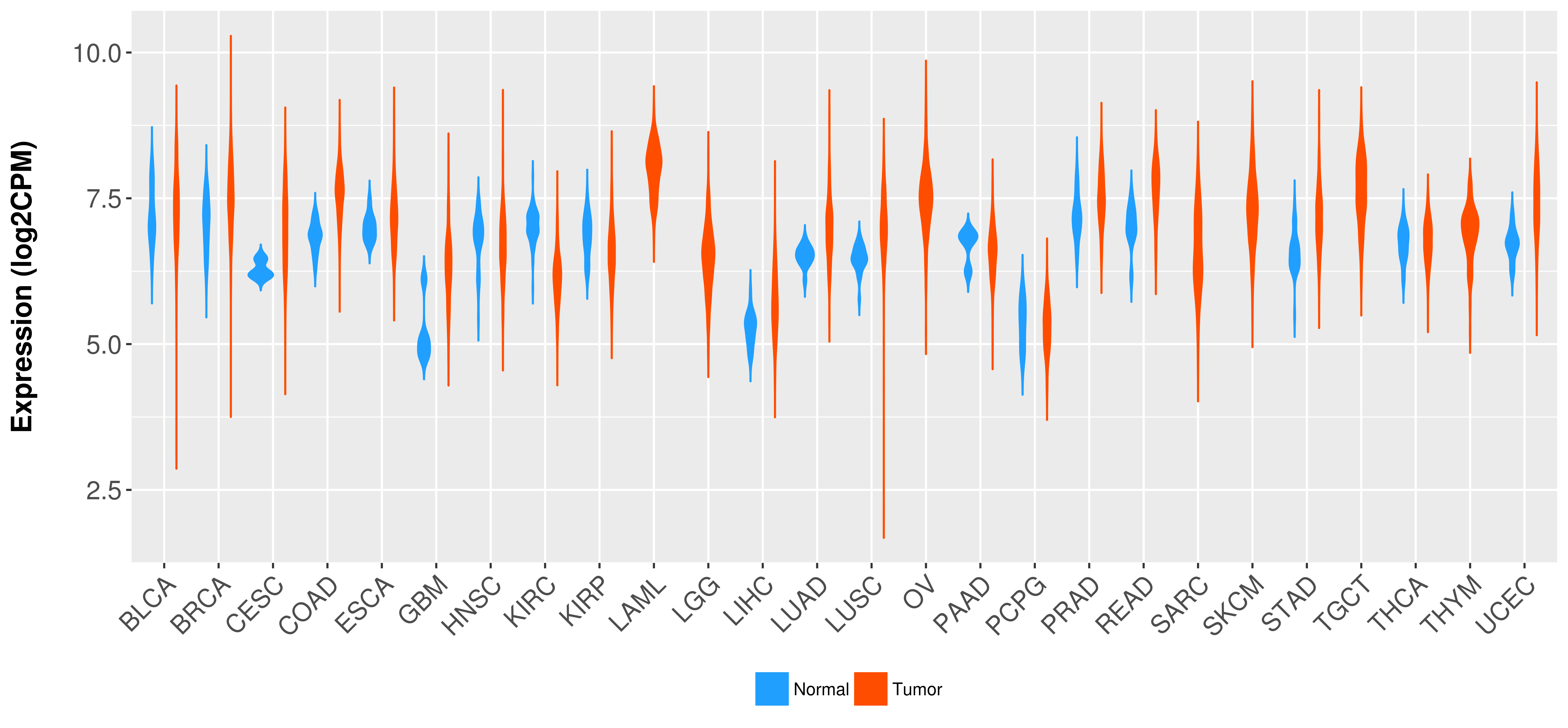
Differential expression analysis for cancers with more than 10 normal samples
|
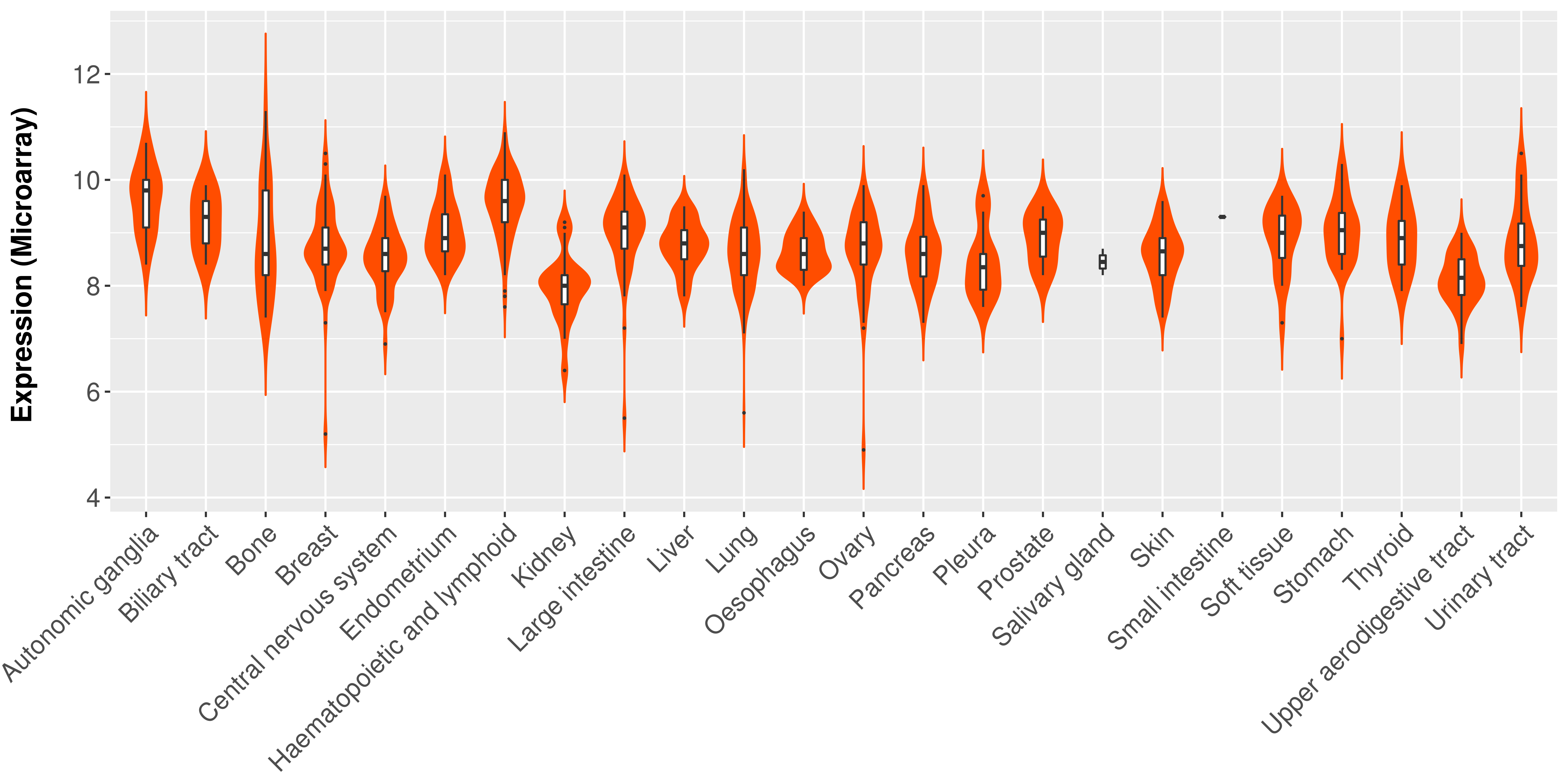
|

|
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
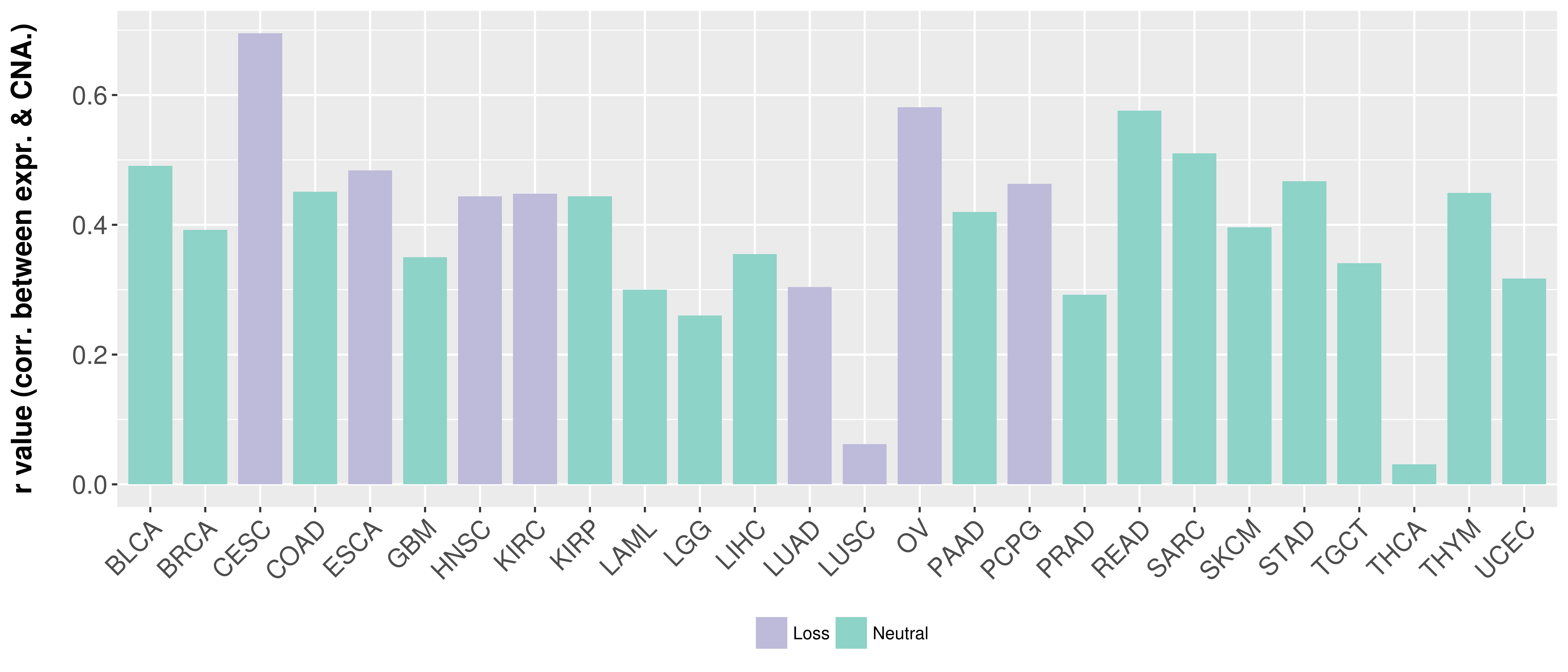
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
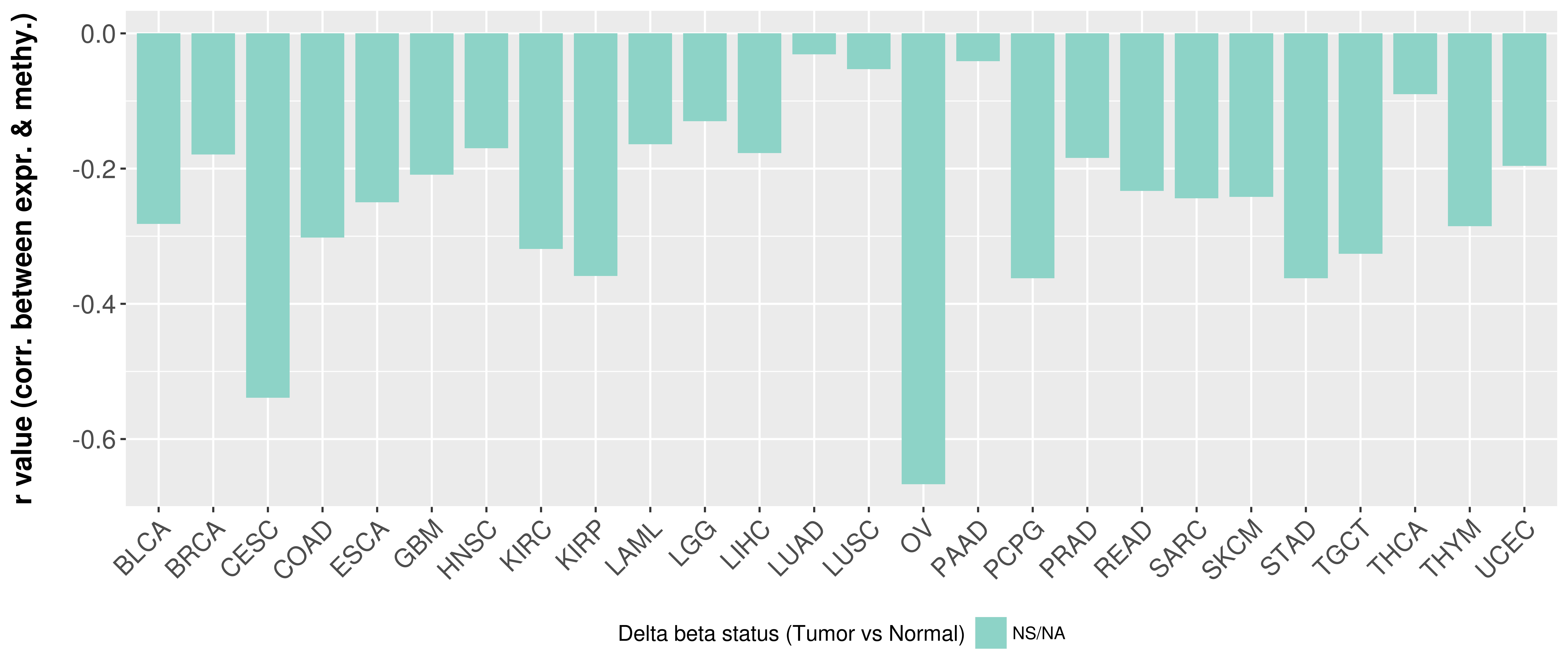
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
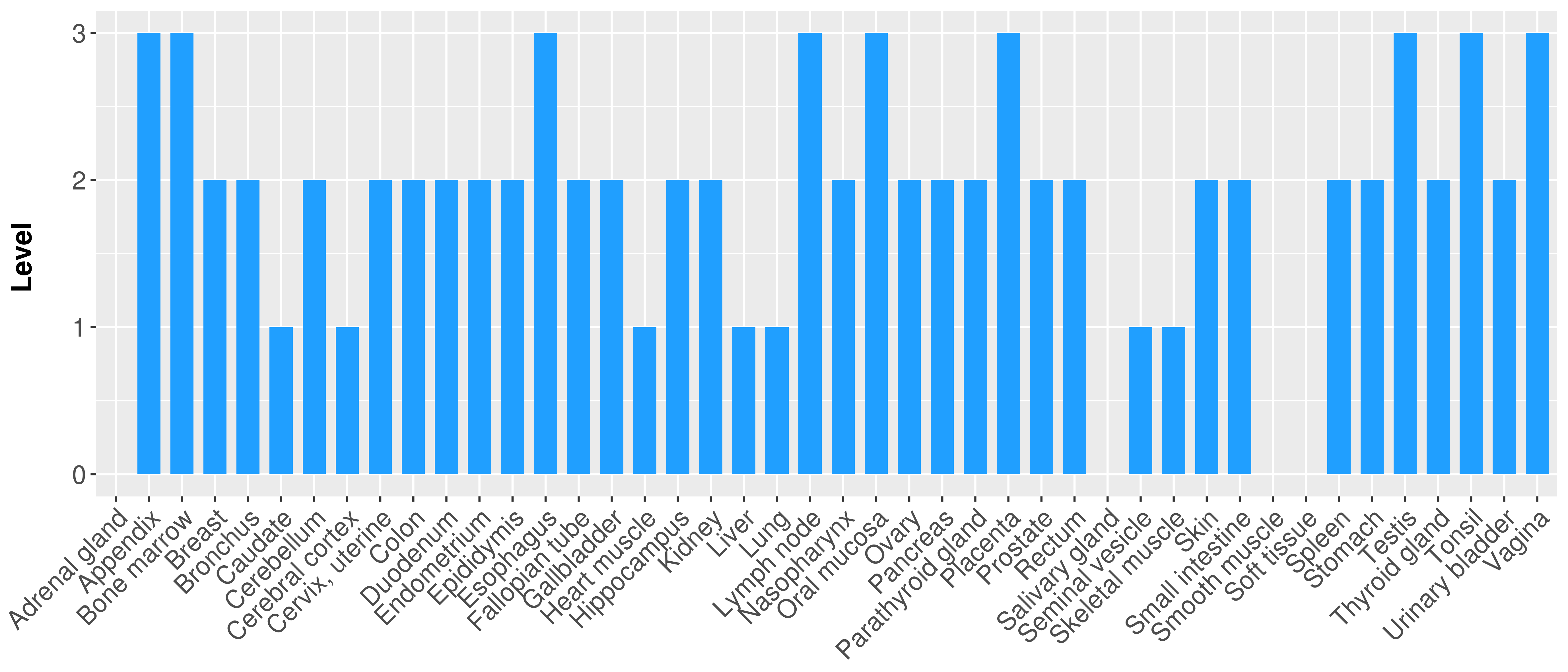
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
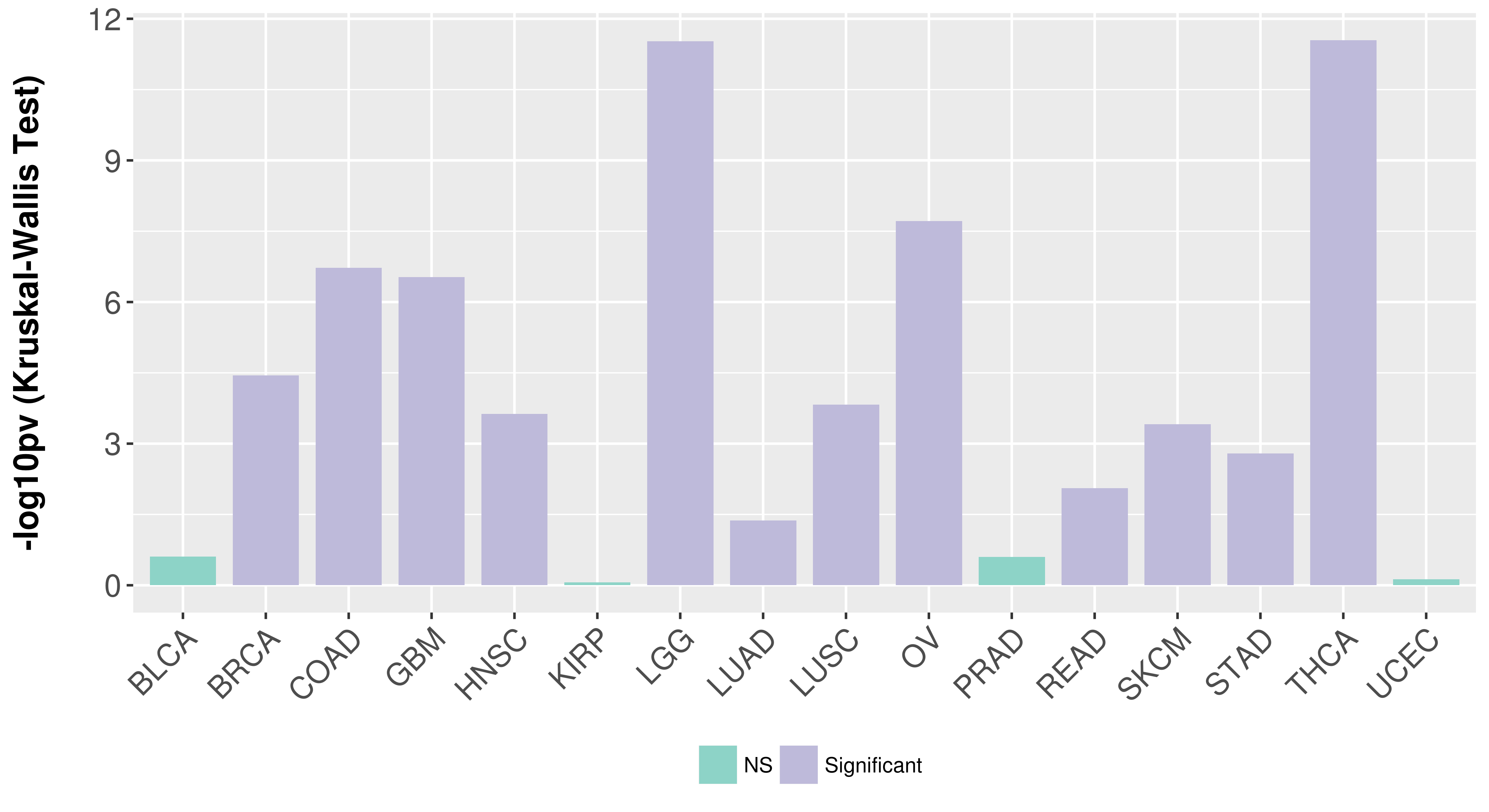
Association between expresson and subtype.
|
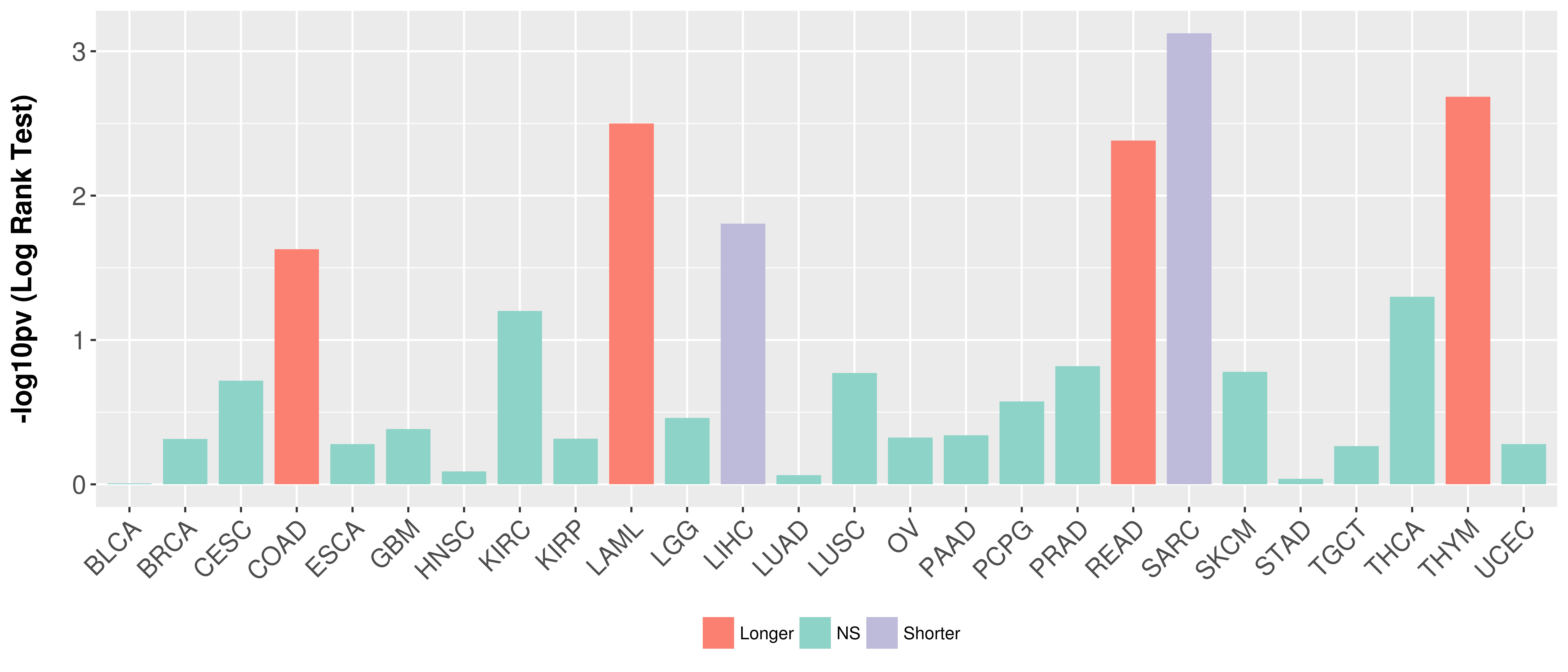
Overall survival analysis based on expression.
|
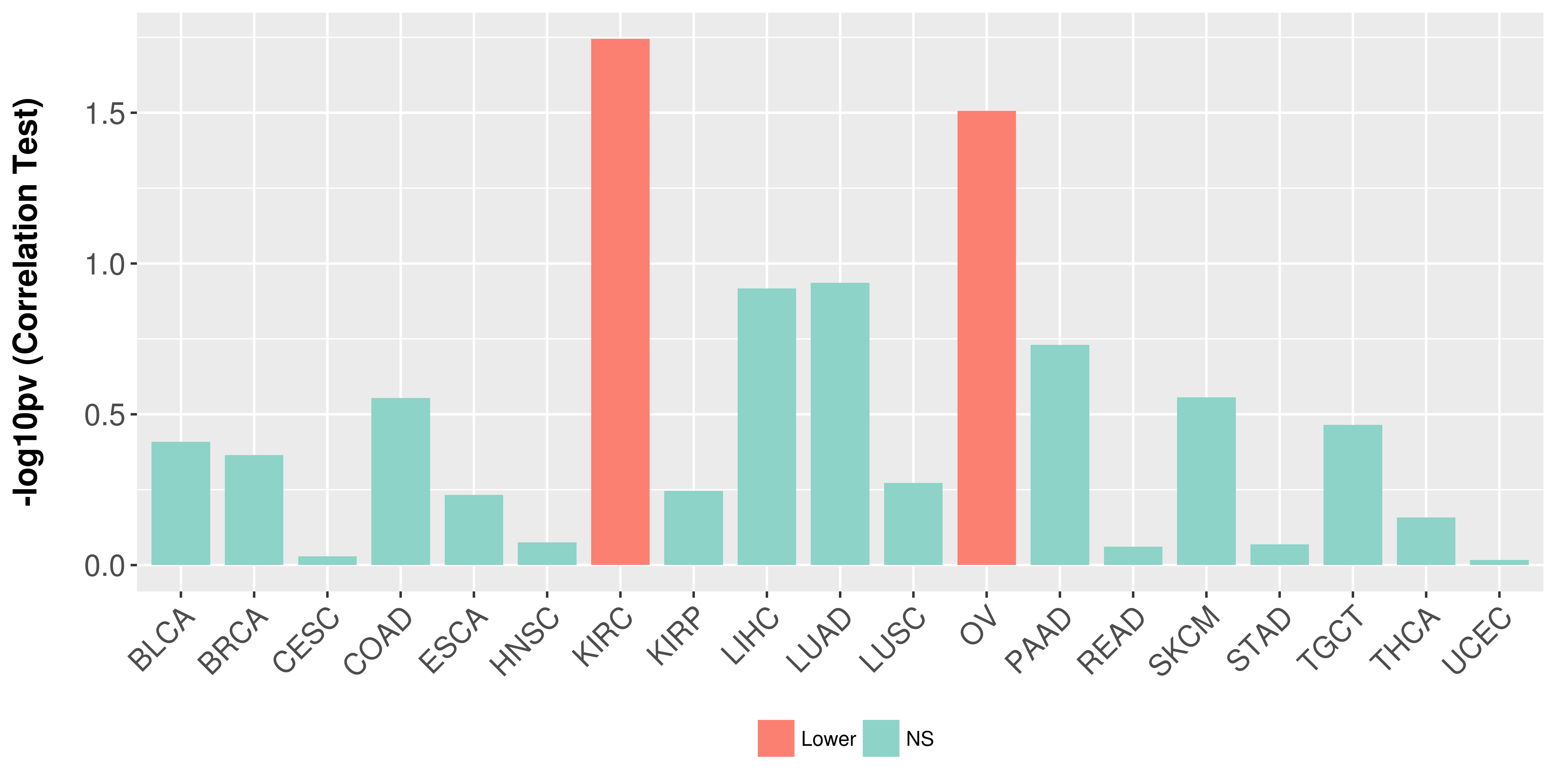
Association between expresson and stage.
|
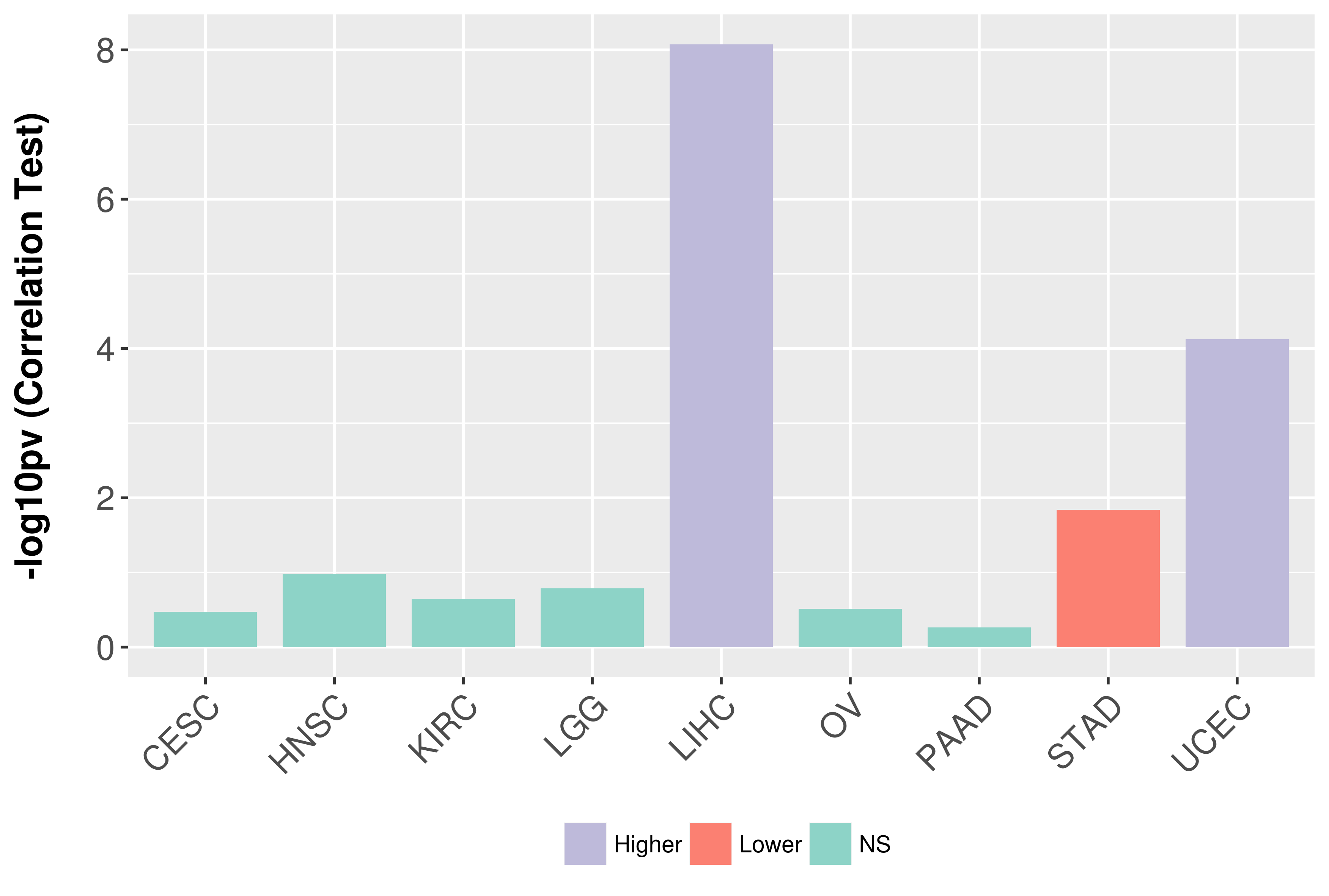
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SMARCC1. |
| Summary | |
|---|---|
| Symbol | SMARCC1 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| Aliases | BAF155; SRG3; CRACC1; SWI3; BRG1-associated factor 155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related mat ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SMARCC1. |