Browse SUV39H1 in pancancer
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00385 Chromo (CHRromatin Organisation MOdifier) domain PF05033 Pre-SET motif PF00856 SET domain |
||||||||||
| Function |
Histone methyltransferase that specifically trimethylates 'Lys-9' of histone H3 using monomethylated H3 'Lys-9' as substrate. Also weakly methylates histone H1 (in vitro). H3 'Lys-9' trimethylation represents a specific tag for epigenetic transcriptional repression by recruiting HP1 (CBX1, CBX3 and/or CBX5) proteins to methylated histones. Mainly functions in heterochromatin regions, thereby playing a central role in the establishment of constitutive heterochromatin at pericentric and telomere regions. H3 'Lys-9' trimethylation is also required to direct DNA methylation at pericentric repeats. SUV39H1 is targeted to histone H3 via its interaction with RB1 and is involved in many processes, such as repression of MYOD1-stimulated differentiation, regulation of the control switch for exiting the cell cycle and entering differentiation, repression by the PML-RARA fusion protein, BMP-induced repression, repression of switch recombination to IgA and regulation of telomere length. Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. The eNoSC complex is able to sense the energy status of cell: upon glucose starvation, elevation of NAD(+)/NADP(+) ratio activates SIRT1, leading to histone H3 deacetylation followed by dimethylation of H3 at 'Lys-9' (H3K9me2) by SUV39H1 and the formation of silent chromatin in the rDNA locus. Recruited by the large PER complex to the E-box elements of the circadian target genes such as PER2 itself or PER1, contributes to the conversion of local chromatin to a heterochromatin-like repressive state through H3 'Lys-9' trimethylation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000183 chromatin silencing at rDNA GO:0001666 response to hypoxia GO:0006342 chromatin silencing GO:0006364 rRNA processing GO:0006479 protein methylation GO:0007623 circadian rhythm GO:0008213 protein alkylation GO:0016072 rRNA metabolic process GO:0016458 gene silencing GO:0016570 histone modification GO:0016571 histone methylation GO:0018022 peptidyl-lysine methylation GO:0018023 peptidyl-lysine trimethylation GO:0018027 peptidyl-lysine dimethylation GO:0018205 peptidyl-lysine modification GO:0022613 ribonucleoprotein complex biogenesis GO:0032259 methylation GO:0034470 ncRNA processing GO:0034968 histone lysine methylation GO:0036123 histone H3-K9 dimethylation GO:0036124 histone H3-K9 trimethylation GO:0036293 response to decreased oxygen levels GO:0036294 cellular response to decreased oxygen levels GO:0040029 regulation of gene expression, epigenetic GO:0042254 ribosome biogenesis GO:0042752 regulation of circadian rhythm GO:0042754 negative regulation of circadian rhythm GO:0043414 macromolecule methylation GO:0045814 negative regulation of gene expression, epigenetic GO:0048511 rhythmic process GO:0051567 histone H3-K9 methylation GO:0061647 histone H3-K9 modification GO:0070482 response to oxygen levels GO:0071453 cellular response to oxygen levels GO:0071456 cellular response to hypoxia |
| Molecular Function |
GO:0003682 chromatin binding GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018024 histone-lysine N-methyltransferase activity GO:0042054 histone methyltransferase activity GO:0046974 histone methyltransferase activity (H3-K9 specific) GO:0047485 protein N-terminus binding |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0000785 chromatin GO:0000792 heterochromatin GO:0000793 condensed chromosome GO:0000794 condensed nuclear chromosome GO:0005652 nuclear lamina GO:0005677 chromatin silencing complex GO:0017053 transcriptional repressor complex GO:0033553 rDNA heterochromatin GO:0034399 nuclear periphery GO:0090568 nuclear transcriptional repressor complex GO:0098687 chromosomal region |
| KEGG |
hsa00310 Lysine degradation |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-5250941: Negative epigenetic regulation of rRNA expression R-HSA-3214841: PKMTs methylate histone lysines R-HSA-427359: SIRT1 negatively regulates rRNA Expression |
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SUV39H1. |
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
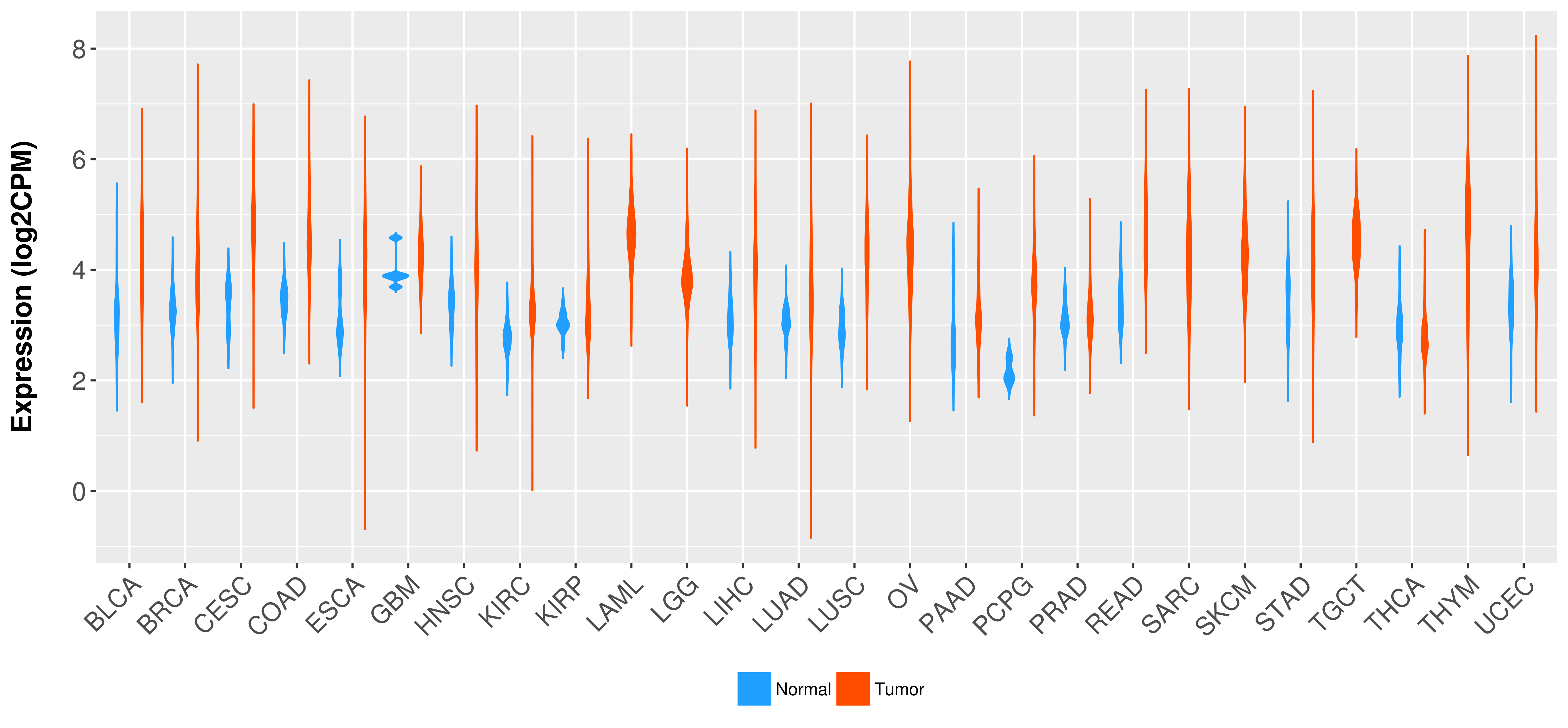
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
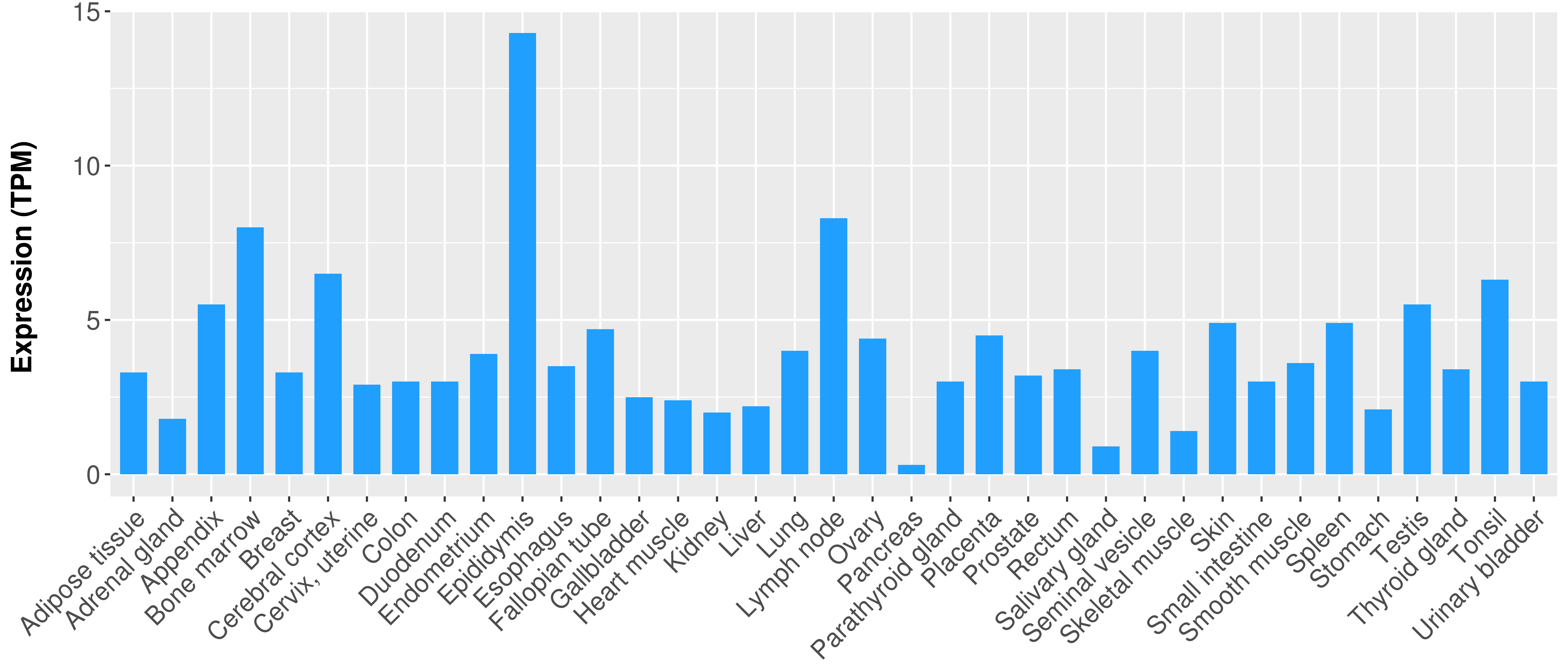
|
|
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
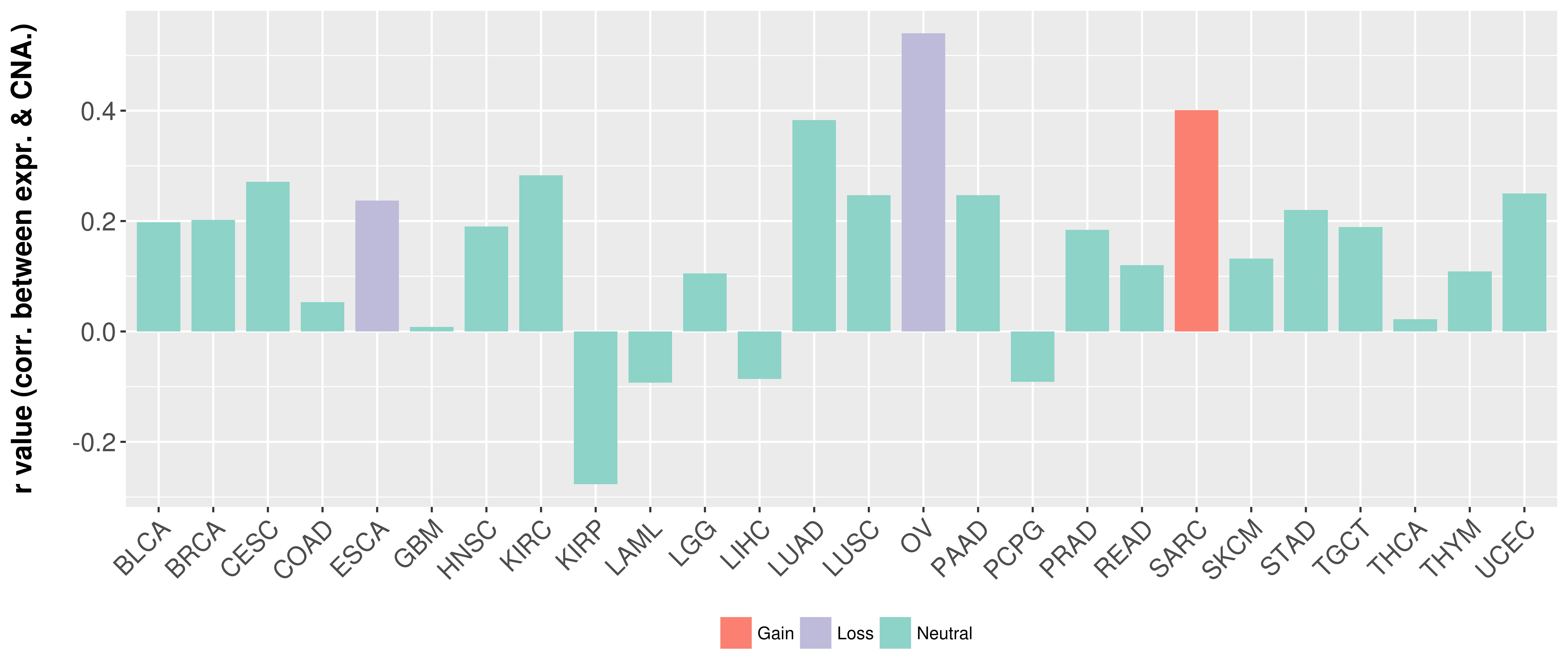
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
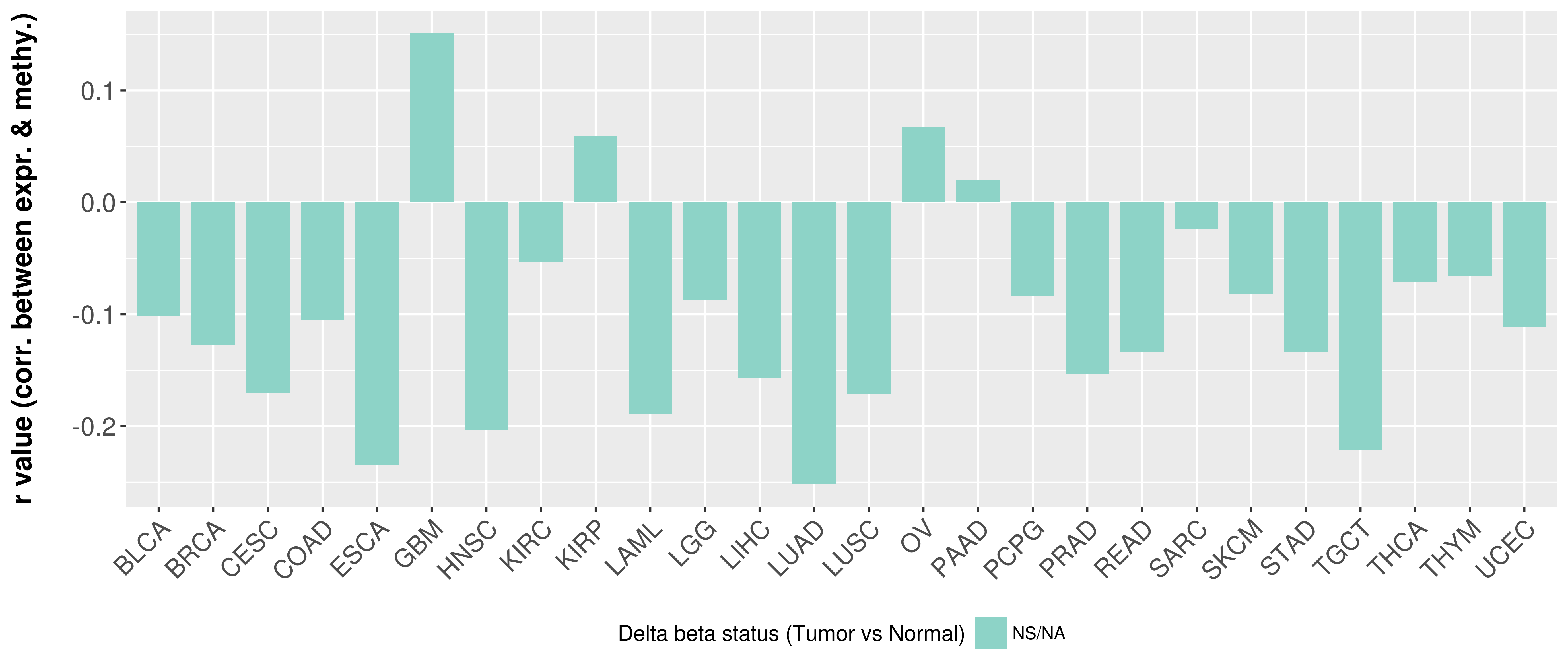
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
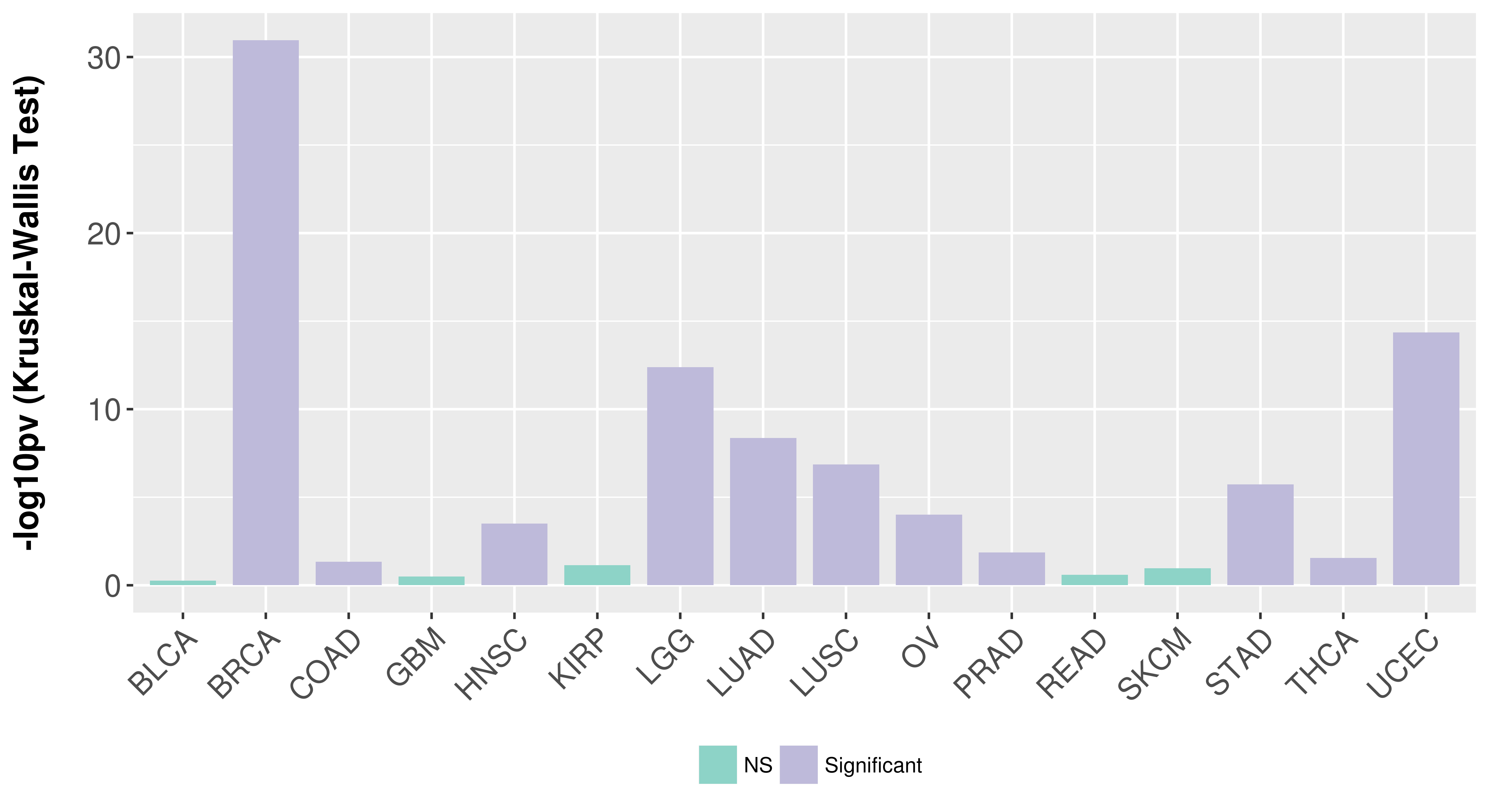
Association between expresson and subtype.
|
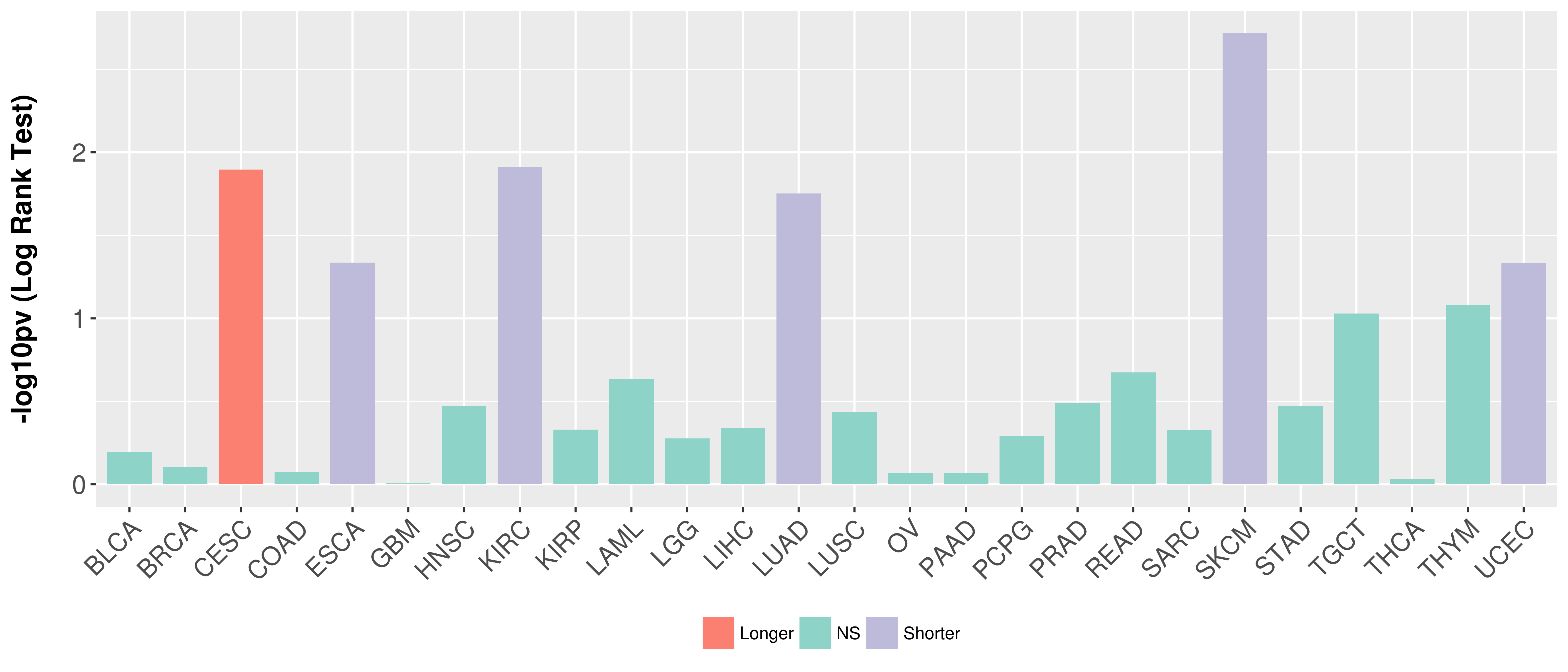
Overall survival analysis based on expression.
|
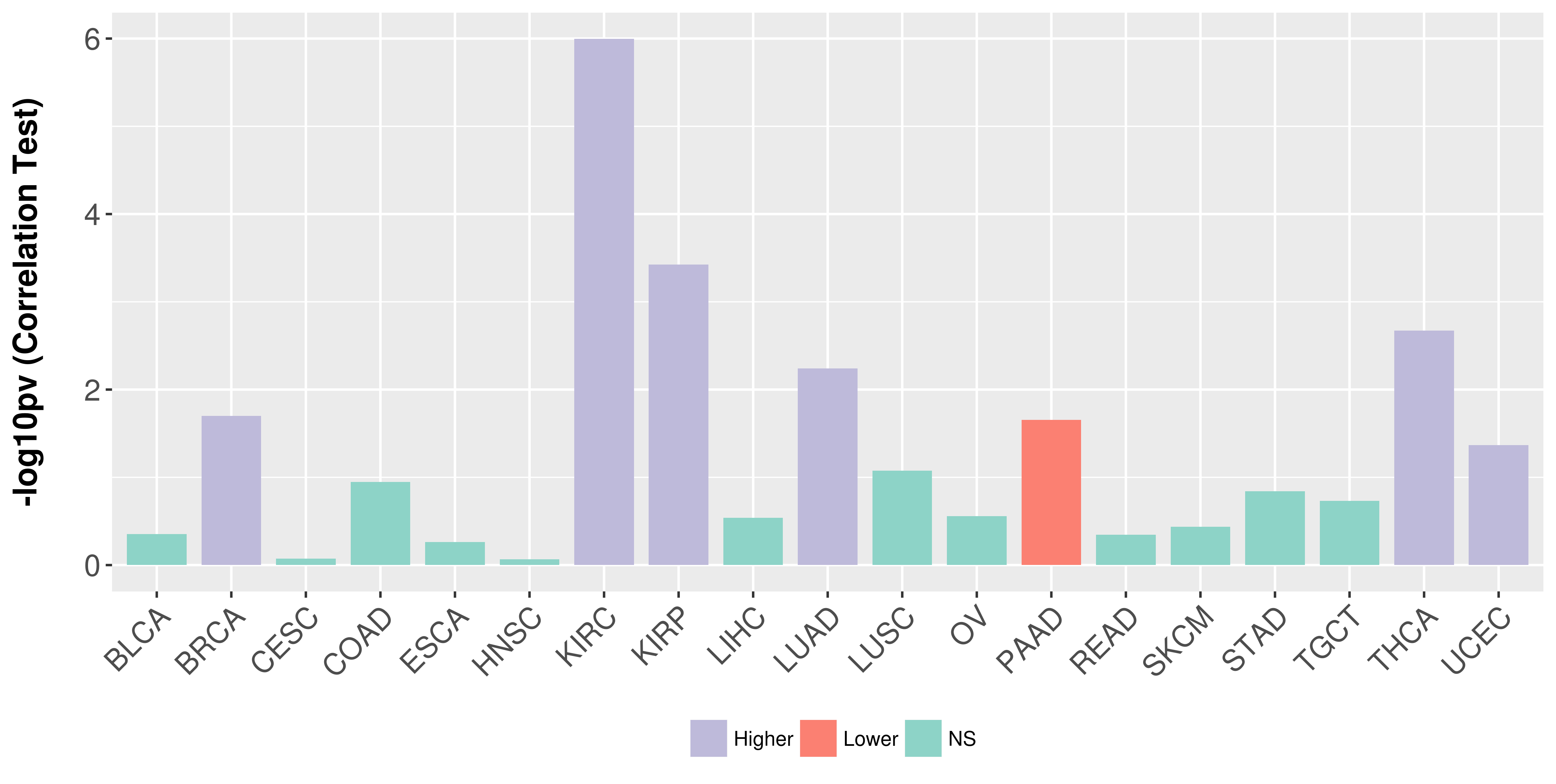
Association between expresson and stage.
|
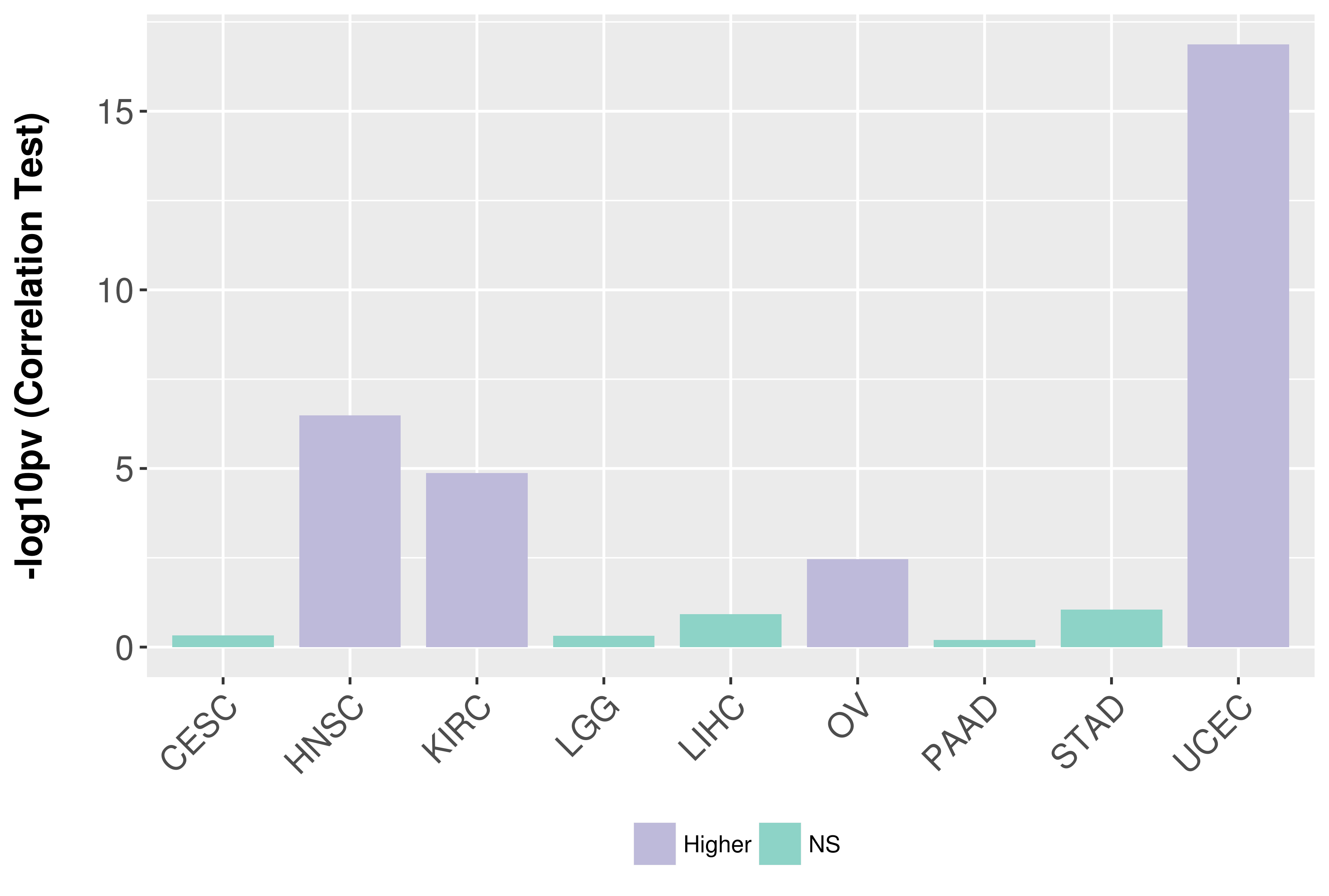
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SUV39H1. |
| Summary | |
|---|---|
| Symbol | SUV39H1 |
| Name | suppressor of variegation 3-9 homolog 1 |
| Aliases | KMT1A; SUV39H; suppressor of variegation 3-9 (Drosophila) homolog 1; H3-K9-HMTase 1; MG44; Su(var)3-9 homolo ...... |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|