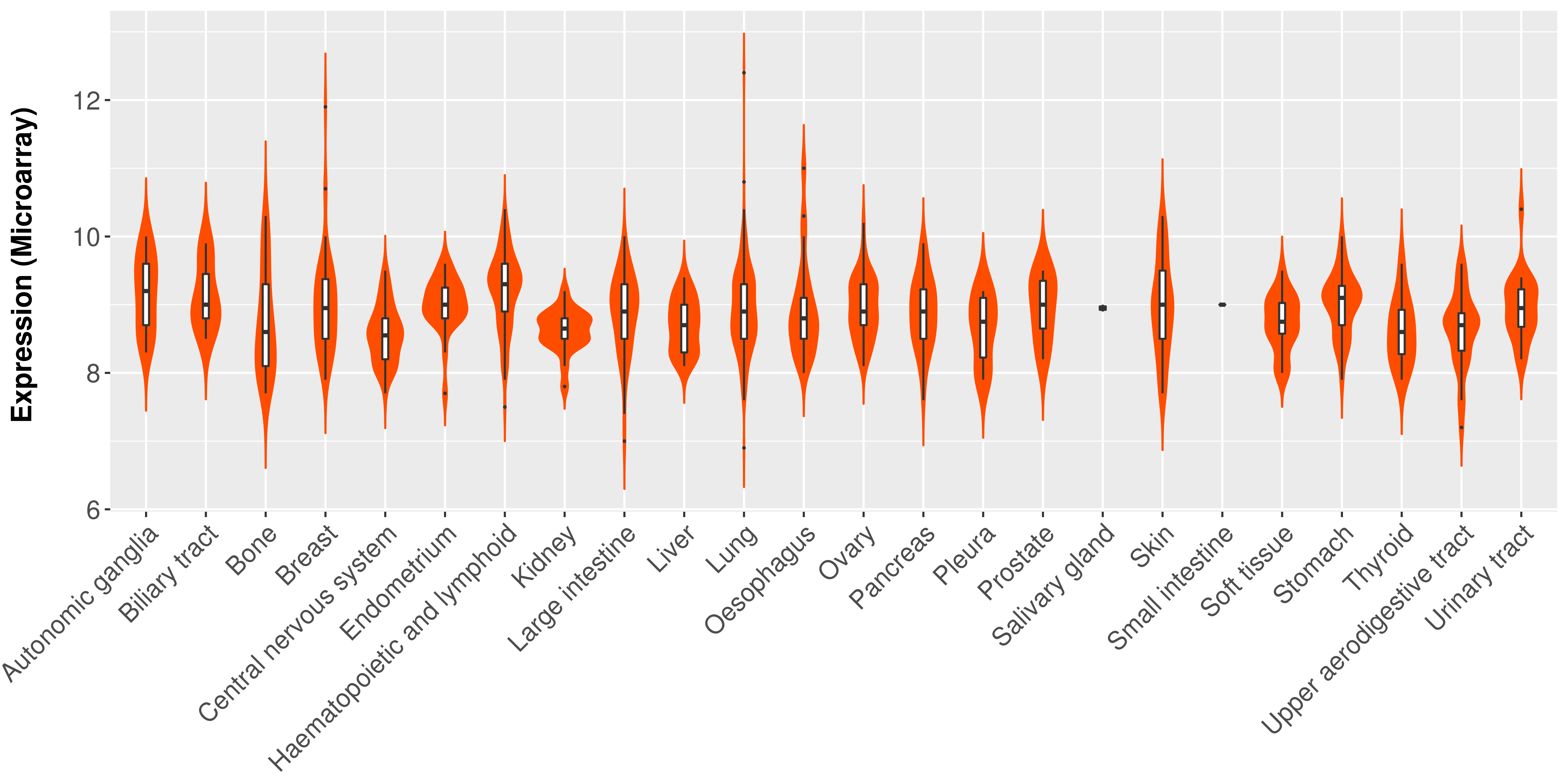
| Tissue |
Cell Line |
Expression Level (Microarray) |
| Autonomic ganglia |
CHP126 |
9.4
|
| Autonomic ganglia |
CHP212 |
8.7
|
| Autonomic ganglia |
IMR32 |
10
|
| Autonomic ganglia |
KELLY |
9.7
|
| Autonomic ganglia |
KPNRTBM1 |
9.6
|
| Autonomic ganglia |
KPNSI9S |
8.4
|
| Autonomic ganglia |
KPNYN |
9.5
|
| Autonomic ganglia |
MHHNB11 |
10
|
| Autonomic ganglia |
NB1 |
9.8
|
| Autonomic ganglia |
NH6 |
9
|
| Autonomic ganglia |
SHSY5Y |
9.1
|
| Autonomic ganglia |
SIMA |
9.4
|
| Autonomic ganglia |
SKNAS |
8.6
|
| Autonomic ganglia |
SKNBE2 |
8.5
|
| Autonomic ganglia |
SKNDZ |
9.2
|
| Autonomic ganglia |
SKNFI |
8.8
|
| Autonomic ganglia |
SKNSH |
8.3
|
| Biliary tract |
HUCCT1 |
8.8
|
| Biliary tract |
HUH28 |
9
|
| Biliary tract |
SNU1079 |
9.9
|
| Biliary tract |
SNU1196 |
8.5
|
| Biliary tract |
SNU245 |
8.8
|
| Biliary tract |
SNU308 |
9.7
|
| Biliary tract |
SNU478 |
9.2
|
| Bone |
143B |
10
|
| Bone |
A673 |
9.1
|
| Bone |
CADOES1 |
9.4
|
| Bone |
CAL78 |
8.3
|
| Bone |
G292CLONEA141B1 |
8.8
|
| Bone |
HOS |
8.7
|
| Bone |
HS706T |
8
|
| Bone |
HS737T |
8
|
| Bone |
HS819T |
7.7
|
| Bone |
HS821T |
8.2
|
| Bone |
HS822T |
7.8
|
| Bone |
HS863T |
8.1
|
| Bone |
HS870T |
8.1
|
| Bone |
HS888T |
8.6
|
| Bone |
MG63 |
8.8
|
| Bone |
MHHES1 |
9.8
|
| Bone |
OUMS27 |
8
|
| Bone |
RDES |
10.3
|
| Bone |
SJSA1 |
8.4
|
| Bone |
SKES1 |
10.1
|
| Bone |
SKNMC |
9.6
|
| Bone |
SW1353 |
8.5
|
| Bone |
T173 |
8
|
| Bone |
TC71 |
8.7
|
| Bone |
U2OS |
9.3
|
| Breast |
AU565 |
9.8
|
| Breast |
BT20 |
10.7
|
| Breast |
BT474 |
9.2
|
| Breast |
BT483 |
9.5
|
| Breast |
BT549 |
8.7
|
| Breast |
CAL120 |
8.9
|
| Breast |
CAL148 |
9.4
|
| Breast |
CAL51 |
9.6
|
| Breast |
CAL851 |
8.7
|
| Breast |
CAMA1 |
9.2
|
| Breast |
DU4475 |
9.3
|
| Breast |
EFM192A |
9.3
|
| Breast |
EFM19 |
9.2
|
| Breast |
EVSAT |
9
|
| Breast |
HCC1143 |
9.1
|
| Breast |
HCC1187 |
8.5
|
| Breast |
HCC1395 |
8.1
|
| Breast |
HCC1419 |
9.3
|
| Breast |
HCC1428 |
10
|
| Breast |
HCC1500 |
8.7
|
| Breast |
HCC1569 |
8.5
|
| Breast |
HCC1599 |
9.7
|
| Breast |
HCC1806 |
8.7
|
| Breast |
HCC1937 |
9.5
|
| Breast |
HCC1954 |
8.8
|
| Breast |
HCC202 |
9
|
| Breast |
HCC2157 |
9.1
|
| Breast |
HCC2218 |
8.8
|
| Breast |
HCC38 |
9
|
| Breast |
HCC70 |
9.5
|
| Breast |
HDQP1 |
9.1
|
| Breast |
HMC18 |
8.6
|
| Breast |
HS274T |
8.3
|
| Breast |
HS281T |
8.4
|
| Breast |
HS343T |
8.3
|
| Breast |
HS578T |
8.3
|
| Breast |
HS606T |
8
|
| Breast |
HS739T |
8.3
|
| Breast |
HS742T |
8
|
| Breast |
JIMT1 |
7.9
|
| Breast |
KPL1 |
11.9
|
| Breast |
MCF7 |
10.7
|
| Breast |
MDAMB134VI |
9.4
|
| Breast |
MDAMB157 |
8.4
|
| Breast |
MDAMB175VII |
8.3
|
| Breast |
MDAMB231 |
8.9
|
| Breast |
MDAMB361 |
8.7
|
| Breast |
MDAMB415 |
8.9
|
| Breast |
MDAMB436 |
8.6
|
| Breast |
MDAMB453 |
9.1
|
| Breast |
MDAMB468 |
9.5
|
| Breast |
SKBR3 |
9.6
|
| Breast |
T47D |
8.5
|
| Breast |
UACC812 |
9
|
| Breast |
UACC893 |
8
|
| Breast |
YMB1 |
8.5
|
| Breast |
ZR751 |
8.3
|
| Breast |
ZR7530 |
9.5
|
| Central nervous system |
1321N1 |
8.7
|
| Central nervous system |
42MGBA |
8.9
|
| Central nervous system |
8MGBA |
9.2
|
| Central nervous system |
A172 |
8.2
|
| Central nervous system |
AM38 |
8.5
|
| Central nervous system |
BECKER |
8
|
| Central nervous system |
CAS1 |
8.2
|
| Central nervous system |
CCFSTTG1 |
8.2
|
| Central nervous system |
D283MED |
9
|
| Central nervous system |
D341MED |
9.2
|
| Central nervous system |
DAOY |
9.4
|
| Central nervous system |
DBTRG05MG |
8
|
| Central nervous system |
DKMG |
8.3
|
| Central nervous system |
GAMG |
8.6
|
| Central nervous system |
GB1 |
9.5
|
| Central nervous system |
GI1 |
8.9
|
| Central nervous system |
GMS10 |
8.5
|
| Central nervous system |
GOS3 |
8.5
|
| Central nervous system |
H4 |
7.7
|
| Central nervous system |
HS683 |
8.6
|
| Central nervous system |
KALS1 |
9
|
| Central nervous system |
KG1C |
8
|
| Central nervous system |
KNS42 |
9
|
| Central nervous system |
KNS60 |
8.5
|
| Central nervous system |
KNS81 |
8.7
|
| Central nervous system |
KS1 |
8.4
|
| Central nervous system |
LN18 |
8.1
|
| Central nervous system |
LN229 |
8.1
|
| Central nervous system |
M059K |
8.4
|
| Central nervous system |
MOGGCCM |
8.6
|
| Central nervous system |
MOGGUVW |
8.5
|
| Central nervous system |
NMCG1 |
8.5
|
| Central nervous system |
ONS76 |
9.2
|
| Central nervous system |
SF126 |
8.1
|
| Central nervous system |
SF295 |
8.1
|
| Central nervous system |
SNB19 |
8.5
|
| Central nervous system |
SNU1105 |
8.7
|
| Central nervous system |
SNU201 |
8.4
|
| Central nervous system |
SNU466 |
8
|
| Central nervous system |
SNU489 |
8.3
|
| Central nervous system |
SNU626 |
8.7
|
| Central nervous system |
SNU738 |
9.4
|
| Central nervous system |
SW1088 |
8.6
|
| Central nervous system |
SW1783 |
8.2
|
| Central nervous system |
T98G |
8.7
|
| Central nervous system |
TM31 |
9.1
|
| Central nervous system |
U118MG |
8.8
|
| Central nervous system |
U138MG |
8
|
| Central nervous system |
U251MG |
8.7
|
| Central nervous system |
U87MG |
8.8
|
| Central nervous system |
YH13 |
8.6
|
| Central nervous system |
YKG1 |
8.8
|
| Endometrium |
AN3CA |
8.9
|
| Endometrium |
COLO684 |
9
|
| Endometrium |
EFE184 |
9.2
|
| Endometrium |
EN |
8.8
|
| Endometrium |
ESS1 |
8.8
|
| Endometrium |
HEC108 |
8.8
|
| Endometrium |
HEC151 |
8.7
|
| Endometrium |
HEC1A |
8.9
|
| Endometrium |
HEC1B |
9.4
|
| Endometrium |
HEC251 |
9.6
|
| Endometrium |
HEC265 |
8.3
|
| Endometrium |
HEC50B |
8.8
|
| Endometrium |
HEC59 |
9.6
|
| Endometrium |
HEC6 |
9.2
|
| Endometrium |
ISHIKAWAHERAKLIO02ER |
9.3
|
| Endometrium |
JHUEM1 |
9.1
|
| Endometrium |
JHUEM2 |
9.3
|
| Endometrium |
JHUEM3 |
9
|
| Endometrium |
KLE |
7.7
|
| Endometrium |
MFE280 |
9.5
|
| Endometrium |
MFE296 |
8.8
|
| Endometrium |
MFE319 |
9.2
|
| Endometrium |
RL952 |
9
|
| Endometrium |
SNGM |
9
|
| Endometrium |
SNU1077 |
9.2
|
| Endometrium |
SNU685 |
9.3
|
| Endometrium |
TEN |
8.9
|
| Haematopoietic and lymphoid |
697 |
9.9
|
| Haematopoietic and lymphoid |
A3KAW |
8.9
|
| Haematopoietic and lymphoid |
A4FUK |
9
|
| Haematopoietic and lymphoid |
ALLSIL |
9.6
|
| Haematopoietic and lymphoid |
AML193 |
9
|
| Haematopoietic and lymphoid |
AMO1 |
7.5
|
| Haematopoietic and lymphoid |
BCP1 |
8.2
|
| Haematopoietic and lymphoid |
BDCM |
8.7
|
| Haematopoietic and lymphoid |
BL41 |
9.3
|
| Haematopoietic and lymphoid |
BL70 |
9.3
|
| Haematopoietic and lymphoid |
BV173 |
9.9
|
| Haematopoietic and lymphoid |
CA46 |
9.1
|
| Haematopoietic and lymphoid |
CI1 |
9.4
|
| Haematopoietic and lymphoid |
CMK115 |
9.3
|
| Haematopoietic and lymphoid |
CMK86 |
9.9
|
| Haematopoietic and lymphoid |
CMK |
9.5
|
| Haematopoietic and lymphoid |
CMLT1 |
9.8
|
| Haematopoietic and lymphoid |
COLO775 |
8.6
|
| Haematopoietic and lymphoid |
DAUDI |
9.6
|
| Haematopoietic and lymphoid |
DB |
9.6
|
| Haematopoietic and lymphoid |
DEL |
8.5
|
| Haematopoietic and lymphoid |
DND41 |
9.4
|
| Haematopoietic and lymphoid |
DOHH2 |
9.5
|
| Haematopoietic and lymphoid |
EB1 |
9.4
|
| Haematopoietic and lymphoid |
EB2 |
9.6
|
| Haematopoietic and lymphoid |
EHEB |
9.4
|
| Haematopoietic and lymphoid |
EJM |
9.2
|
| Haematopoietic and lymphoid |
EM2 |
9.1
|
| Haematopoietic and lymphoid |
EOL1 |
9.4
|
| Haematopoietic and lymphoid |
F36P |
9.7
|
| Haematopoietic and lymphoid |
GA10 |
9.4
|
| Haematopoietic and lymphoid |
GDM1 |
9.4
|
| Haematopoietic and lymphoid |
GRANTA519 |
8.2
|
| Haematopoietic and lymphoid |
HDLM2 |
9.3
|
| Haematopoietic and lymphoid |
HDMYZ |
8.5
|
| Haematopoietic and lymphoid |
HEL9217 |
9.3
|
| Haematopoietic and lymphoid |
HEL |
9.2
|
| Haematopoietic and lymphoid |
HH |
10
|
| Haematopoietic and lymphoid |
HL60 |
9.6
|
| Haematopoietic and lymphoid |
HPBALL |
9.5
|
| Haematopoietic and lymphoid |
HS604T |
8.4
|
| Haematopoietic and lymphoid |
HS611T |
8.8
|
| Haematopoietic and lymphoid |
HS616T |
8.1
|
| Haematopoietic and lymphoid |
HS751T |
8.1
|
| Haematopoietic and lymphoid |
HT |
9.4
|
| Haematopoietic and lymphoid |
HTK |
9.7
|
| Haematopoietic and lymphoid |
HUNS1 |
8.7
|
| Haematopoietic and lymphoid |
HUT102 |
8.7
|
| Haematopoietic and lymphoid |
HUT78 |
9.1
|
| Haematopoietic and lymphoid |
JEKO1 |
8.9
|
| Haematopoietic and lymphoid |
JK1 |
10.1
|
| Haematopoietic and lymphoid |
JM1 |
9.4
|
| Haematopoietic and lymphoid |
JURKAT |
9.6
|
| Haematopoietic and lymphoid |
JURLMK1 |
9.2
|
| Haematopoietic and lymphoid |
JVM2 |
9.1
|
| Haematopoietic and lymphoid |
JVM3 |
9
|
| Haematopoietic and lymphoid |
K562 |
9.3
|
| Haematopoietic and lymphoid |
KARPAS299 |
8.8
|
| Haematopoietic and lymphoid |
KARPAS422 |
9.6
|
| Haematopoietic and lymphoid |
KARPAS620 |
8.8
|
| Haematopoietic and lymphoid |
KASUMI1 |
9.2
|
| Haematopoietic and lymphoid |
KASUMI2 |
9.6
|
| Haematopoietic and lymphoid |
KASUMI6 |
9.3
|
| Haematopoietic and lymphoid |
KCL22 |
9.1
|
| Haematopoietic and lymphoid |
KE37 |
10
|
| Haematopoietic and lymphoid |
KE97 |
9
|
| Haematopoietic and lymphoid |
KG1 |
9.1
|
| Haematopoietic and lymphoid |
KHM1B |
8.5
|
| Haematopoietic and lymphoid |
KIJK |
8.5
|
| Haematopoietic and lymphoid |
KMH2 |
9.3
|
| Haematopoietic and lymphoid |
KMM1 |
8.1
|
| Haematopoietic and lymphoid |
KMS11 |
8.5
|
| Haematopoietic and lymphoid |
KMS12BM |
8.9
|
| Haematopoietic and lymphoid |
KMS18 |
9.4
|
| Haematopoietic and lymphoid |
KMS20 |
8.7
|
| Haematopoietic and lymphoid |
KMS21BM |
8.6
|
| Haematopoietic and lymphoid |
KMS26 |
8.2
|
| Haematopoietic and lymphoid |
KMS27 |
8.7
|
| Haematopoietic and lymphoid |
KMS28BM |
8.1
|
| Haematopoietic and lymphoid |
KMS34 |
8.8
|
| Haematopoietic and lymphoid |
KO52 |
9.9
|
| Haematopoietic and lymphoid |
KOPN8 |
9.2
|
| Haematopoietic and lymphoid |
KU812 |
10.3
|
| Haematopoietic and lymphoid |
KYO1 |
9.3
|
| Haematopoietic and lymphoid |
L1236 |
9.3
|
| Haematopoietic and lymphoid |
L363 |
9.8
|
| Haematopoietic and lymphoid |
L428 |
10.4
|
| Haematopoietic and lymphoid |
L540 |
8.7
|
| Haematopoietic and lymphoid |
LAMA84 |
9.5
|
| Haematopoietic and lymphoid |
LOUCY |
9.5
|
| Haematopoietic and lymphoid |
LP1 |
8.5
|
| Haematopoietic and lymphoid |
M07E |
10.3
|
| Haematopoietic and lymphoid |
MC116 |
9.4
|
| Haematopoietic and lymphoid |
ME1 |
8.3
|
| Haematopoietic and lymphoid |
MEC1 |
9.5
|
| Haematopoietic and lymphoid |
MEC2 |
9.2
|
| Haematopoietic and lymphoid |
MEG01 |
9.5
|
| Haematopoietic and lymphoid |
MHHCALL2 |
9.7
|
| Haematopoietic and lymphoid |
MHHCALL3 |
9.8
|
| Haematopoietic and lymphoid |
MHHCALL4 |
9.7
|
| Haematopoietic and lymphoid |
MINO |
9.6
|
| Haematopoietic and lymphoid |
MJ |
9.4
|
| Haematopoietic and lymphoid |
MM1S |
7.9
|
| Haematopoietic and lymphoid |
MOLM13 |
9.2
|
| Haematopoietic and lymphoid |
MOLM16 |
10.1
|
| Haematopoietic and lymphoid |
MOLM6 |
9.8
|
| Haematopoietic and lymphoid |
MOLP2 |
8
|
| Haematopoietic and lymphoid |
MOLP8 |
9.3
|
| Haematopoietic and lymphoid |
MOLT13 |
10.1
|
| Haematopoietic and lymphoid |
MOLT16 |
9.9
|
| Haematopoietic and lymphoid |
MOLT4 |
9.5
|
| Haematopoietic and lymphoid |
MONOMAC1 |
9.1
|
| Haematopoietic and lymphoid |
MONOMAC6 |
9.3
|
| Haematopoietic and lymphoid |
MOTN1 |
9.1
|
| Haematopoietic and lymphoid |
MUTZ5 |
9.4
|
| Haematopoietic and lymphoid |
MV411 |
9.2
|
| Haematopoietic and lymphoid |
NALM19 |
9.3
|
| Haematopoietic and lymphoid |
NALM1 |
9.9
|
| Haematopoietic and lymphoid |
NALM6 |
9.4
|
| Haematopoietic and lymphoid |
NAMALWA |
9.6
|
| Haematopoietic and lymphoid |
NB4 |
9.3
|
| Haematopoietic and lymphoid |
NCIH929 |
8.2
|
| Haematopoietic and lymphoid |
NCO2 |
9.7
|
| Haematopoietic and lymphoid |
NOMO1 |
9
|
| Haematopoietic and lymphoid |
NUDHL1 |
8.8
|
| Haematopoietic and lymphoid |
NUDUL1 |
9.9
|
| Haematopoietic and lymphoid |
OCIAML2 |
9.4
|
| Haematopoietic and lymphoid |
OCIAML3 |
8.9
|
| Haematopoietic and lymphoid |
OCIAML5 |
9.3
|
| Haematopoietic and lymphoid |
OCILY10 |
9.5
|
| Haematopoietic and lymphoid |
OCILY19 |
9.7
|
| Haematopoietic and lymphoid |
OCILY3 |
8.6
|
| Haematopoietic and lymphoid |
OCIM1 |
10.3
|
| Haematopoietic and lymphoid |
OPM2 |
7.5
|
| Haematopoietic and lymphoid |
P12ICHIKAWA |
10
|
| Haematopoietic and lymphoid |
P31FUJ |
8.7
|
| Haematopoietic and lymphoid |
P3HR1 |
9.2
|
| Haematopoietic and lymphoid |
PCM6 |
8.8
|
| Haematopoietic and lymphoid |
PEER |
9.6
|
| Haematopoietic and lymphoid |
PF382 |
10.1
|
| Haematopoietic and lymphoid |
PFEIFFER |
9.3
|
| Haematopoietic and lymphoid |
PL21 |
9.4
|
| Haematopoietic and lymphoid |
RAJI |
9.3
|
| Haematopoietic and lymphoid |
RCHACV |
9.7
|
| Haematopoietic and lymphoid |
REC1 |
9.5
|
| Haematopoietic and lymphoid |
REH |
9.5
|
| Haematopoietic and lymphoid |
RI1 |
8.7
|
| Haematopoietic and lymphoid |
RL |
9.3
|
| Haematopoietic and lymphoid |
RPMI8226 |
8
|
| Haematopoietic and lymphoid |
RPMI8402 |
10.1
|
| Haematopoietic and lymphoid |
RS411 |
9.3
|
| Haematopoietic and lymphoid |
SEM |
9.6
|
| Haematopoietic and lymphoid |
SET2 |
9.6
|
| Haematopoietic and lymphoid |
SIGM5 |
9.2
|
| Haematopoietic and lymphoid |
SKM1 |
8.7
|
| Haematopoietic and lymphoid |
SKMM2 |
9.4
|
| Haematopoietic and lymphoid |
SR786 |
8.9
|
| Haematopoietic and lymphoid |
ST486 |
9.2
|
| Haematopoietic and lymphoid |
SUDHL10 |
9.4
|
| Haematopoietic and lymphoid |
SUDHL1 |
8.5
|
| Haematopoietic and lymphoid |
SUDHL4 |
8.9
|
| Haematopoietic and lymphoid |
SUDHL5 |
9.5
|
| Haematopoietic and lymphoid |
SUDHL6 |
9.8
|
| Haematopoietic and lymphoid |
SUDHL8 |
9.1
|
| Haematopoietic and lymphoid |
SUPB15 |
8.7
|
| Haematopoietic and lymphoid |
SUPHD1 |
8.9
|
| Haematopoietic and lymphoid |
SUPM2 |
8.5
|
| Haematopoietic and lymphoid |
SUPT11 |
9.6
|
| Haematopoietic and lymphoid |
SUPT1 |
9.6
|
| Haematopoietic and lymphoid |
TALL1 |
10
|
| Haematopoietic and lymphoid |
TF1 |
9.4
|
| Haematopoietic and lymphoid |
THP1 |
9.1
|
| Haematopoietic and lymphoid |
TO175T |
8.2
|
| Haematopoietic and lymphoid |
TOLEDO |
9.1
|
| Haematopoietic and lymphoid |
U266B1 |
8.7
|
| Haematopoietic and lymphoid |
U937 |
9.3
|
| Haematopoietic and lymphoid |
UT7 |
10.2
|
| Haematopoietic and lymphoid |
WSUDLCL2 |
9.4
|
| Kidney |
769P |
8.9
|
| Kidney |
786O |
8.7
|
| Kidney |
A498 |
8.5
|
| Kidney |
A704 |
8.7
|
| Kidney |
ACHN |
8.5
|
| Kidney |
BFTC909 |
8.1
|
| Kidney |
CAKI1 |
8.5
|
| Kidney |
CAKI2 |
8.4
|
| Kidney |
CAL54 |
8.6
|
| Kidney |
KMRC1 |
8.8
|
| Kidney |
KMRC20 |
8.8
|
| Kidney |
KMRC2 |
8.9
|
| Kidney |
KMRC3 |
8.6
|
| Kidney |
OSRC2 |
8.4
|
| Kidney |
RCC10RGB |
8.5
|
| Kidney |
SNU1272 |
8.8
|
| Kidney |
SNU349 |
7.8
|
| Kidney |
TUHR10TKB |
8.8
|
| Kidney |
TUHR14TKB |
9.2
|
| Kidney |
TUHR4TKB |
8.8
|
| Kidney |
VMRCRCW |
8.4
|
| Kidney |
VMRCRCZ |
9
|
| Large intestine |
C2BBE1 |
8.1
|
| Large intestine |
CCK81 |
8.3
|
| Large intestine |
CL11 |
9.4
|
| Large intestine |
CL14 |
9.3
|
| Large intestine |
CL34 |
9.4
|
| Large intestine |
CL40 |
8.9
|
| Large intestine |
COLO205 |
8.3
|
| Large intestine |
COLO320 |
9.5
|
| Large intestine |
COLO678 |
9.4
|
| Large intestine |
CW2 |
9.1
|
| Large intestine |
DLD1 |
9.1
|
| Large intestine |
GP2D |
9.7
|
| Large intestine |
HCC56 |
8.3
|
| Large intestine |
HCT116 |
9.1
|
| Large intestine |
HCT15 |
9.2
|
| Large intestine |
HS675T |
7.9
|
| Large intestine |
HS698T |
8.5
|
| Large intestine |
HT115 |
8.9
|
| Large intestine |
HT29 |
9.1
|
| Large intestine |
HT55 |
9.6
|
| Large intestine |
KM12 |
9.2
|
| Large intestine |
LOVO |
8.8
|
| Large intestine |
LS1034 |
8.9
|
| Large intestine |
LS123 |
8.9
|
| Large intestine |
LS180 |
8.4
|
| Large intestine |
LS411N |
8.9
|
| Large intestine |
LS513 |
8.7
|
| Large intestine |
MDST8 |
7.4
|
| Large intestine |
NCIH508 |
8.6
|
| Large intestine |
NCIH716 |
8.8
|
| Large intestine |
NCIH747 |
8.3
|
| Large intestine |
OUMS23 |
10
|
| Large intestine |
RCM1 |
8.9
|
| Large intestine |
RKO |
8.9
|
| Large intestine |
SKCO1 |
9.7
|
| Large intestine |
SNU1040 |
9.5
|
| Large intestine |
SNU1197 |
8.8
|
| Large intestine |
SNU175 |
9.3
|
| Large intestine |
SNU283 |
9.1
|
| Large intestine |
SNU407 |
9.4
|
| Large intestine |
SNU503 |
7.9
|
| Large intestine |
SNU61 |
8.6
|
| Large intestine |
SNU81 |
9.5
|
| Large intestine |
SNUC1 |
9.1
|
| Large intestine |
SNUC2A |
8.1
|
| Large intestine |
SNUC4 |
7
|
| Large intestine |
SNUC5 |
9.3
|
| Large intestine |
SW1116 |
8.6
|
| Large intestine |
SW1417 |
8.3
|
| Large intestine |
SW1463 |
9.1
|
| Large intestine |
SW403 |
9.2
|
| Large intestine |
SW480 |
8.5
|
| Large intestine |
SW48 |
9.8
|
| Large intestine |
SW620 |
9.2
|
| Large intestine |
SW837 |
8.5
|
| Large intestine |
SW948 |
8.9
|
| Large intestine |
T84 |
9.1
|
| Liver |
ALEXANDERCELLS |
8.1
|
| Liver |
C3A |
8.8
|
| Liver |
HEP3B217 |
9.1
|
| Liver |
HEPG2 |
9.4
|
| Liver |
HLE |
8.7
|
| Liver |
HLF |
8.3
|
| Liver |
HUH1 |
8.3
|
| Liver |
HUH6 |
9.3
|
| Liver |
HUH7 |
8.7
|
| Liver |
JHH1 |
8.2
|
| Liver |
JHH2 |
8.4
|
| Liver |
JHH4 |
8.8
|
| Liver |
JHH5 |
9
|
| Liver |
JHH6 |
8.8
|
| Liver |
JHH7 |
9
|
| Liver |
LI7 |
8.3
|
| Liver |
PLCPRF5 |
8.2
|
| Liver |
SKHEP1 |
8.6
|
| Liver |
SNU182 |
8.5
|
| Liver |
SNU387 |
9.1
|
| Liver |
SNU398 |
9.2
|
| Liver |
SNU423 |
8.6
|
| Liver |
SNU449 |
8.2
|
| Liver |
SNU475 |
8.8
|
| Liver |
SNU761 |
9
|
| Liver |
SNU878 |
8.3
|
| Liver |
SNU886 |
9.2
|
| Lung |
A549 |
7.8
|
| Lung |
ABC1 |
8.7
|
| Lung |
BEN |
8.3
|
| Lung |
CAL12T |
8.4
|
| Lung |
CALU1 |
8.6
|
| Lung |
CALU3 |
9.1
|
| Lung |
CALU6 |
8.5
|
| Lung |
CHAGOK1 |
7.7
|
| Lung |
COLO668 |
9.4
|
| Lung |
COLO699 |
9.5
|
| Lung |
CORL105 |
9.2
|
| Lung |
CORL23 |
9
|
| Lung |
CORL24 |
9.7
|
| Lung |
CORL279 |
9.6
|
| Lung |
CORL311 |
9.6
|
| Lung |
CORL47 |
8.7
|
| Lung |
CORL51 |
9.3
|
| Lung |
CORL88 |
9.5
|
| Lung |
CORL95 |
9
|
| Lung |
CPCN |
8.9
|
| Lung |
DMS114 |
9.4
|
| Lung |
DMS153 |
9
|
| Lung |
DMS273 |
9.1
|
| Lung |
DMS454 |
8.4
|
| Lung |
DMS53 |
8.9
|
| Lung |
DMS79 |
9.9
|
| Lung |
DV90 |
9.2
|
| Lung |
EBC1 |
8.6
|
| Lung |
EPLC272H |
7.9
|
| Lung |
HARA |
8.7
|
| Lung |
HCC1171 |
8.6
|
| Lung |
HCC1195 |
10.8
|
| Lung |
HCC15 |
8.7
|
| Lung |
HCC2279 |
8.8
|
| Lung |
HCC2935 |
8.8
|
| Lung |
HCC33 |
9.8
|
| Lung |
HCC366 |
9.3
|
| Lung |
HCC4006 |
8.8
|
| Lung |
HCC44 |
8.9
|
| Lung |
HCC78 |
8.9
|
| Lung |
HCC827 |
9.4
|
| Lung |
HCC95 |
7.6
|
| Lung |
HLC1 |
9.1
|
| Lung |
HLFA |
8.1
|
| Lung |
HS229T |
8.3
|
| Lung |
HS618T |
8
|
| Lung |
IALM |
9.1
|
| Lung |
KNS62 |
8.3
|
| Lung |
LC1F |
9.6
|
| Lung |
LC1SQSF |
9
|
| Lung |
LCLC103H |
8.9
|
| Lung |
LCLC97TM1 |
9.3
|
| Lung |
LK2 |
9
|
| Lung |
LOUNH91 |
8.6
|
| Lung |
LU65 |
8.3
|
| Lung |
LU99 |
9.2
|
| Lung |
LUDLU1 |
8
|
| Lung |
LXF289 |
8.4
|
| Lung |
MORCPR |
8.9
|
| Lung |
NCIH1048 |
9.3
|
| Lung |
NCIH1092 |
9.5
|
| Lung |
NCIH1105 |
10.4
|
| Lung |
NCIH1155 |
9
|
| Lung |
NCIH1184 |
9
|
| Lung |
NCIH1299 |
8.6
|
| Lung |
NCIH1339 |
9.2
|
| Lung |
NCIH1341 |
9.3
|
| Lung |
NCIH1355 |
8.7
|
| Lung |
NCIH1373 |
8.5
|
| Lung |
NCIH1385 |
9.3
|
| Lung |
NCIH1395 |
8.1
|
| Lung |
NCIH1435 |
9.3
|
| Lung |
NCIH1436 |
9.4
|
| Lung |
NCIH1437 |
9.4
|
| Lung |
NCIH146 |
8.7
|
| Lung |
NCIH1563 |
8.3
|
| Lung |
NCIH1568 |
9.6
|
| Lung |
NCIH1573 |
8.7
|
| Lung |
NCIH1581 |
9.5
|
| Lung |
NCIH1618 |
9.7
|
| Lung |
NCIH1623 |
9.3
|
| Lung |
NCIH1648 |
8.6
|
| Lung |
NCIH1650 |
9.1
|
| Lung |
NCIH1651 |
8.2
|
| Lung |
NCIH1666 |
9
|
| Lung |
NCIH1693 |
6.9
|
| Lung |
NCIH1694 |
10
|
| Lung |
NCIH1703 |
8.4
|
| Lung |
NCIH1734 |
8
|
| Lung |
NCIH1755 |
8.2
|
| Lung |
NCIH1781 |
8.7
|
| Lung |
NCIH1792 |
8.4
|
| Lung |
NCIH1793 |
8.7
|
| Lung |
NCIH1836 |
9.3
|
| Lung |
NCIH1838 |
8.4
|
| Lung |
NCIH1869 |
9.8
|
| Lung |
NCIH1876 |
10.1
|
| Lung |
NCIH1915 |
9.3
|
| Lung |
NCIH1930 |
9
|
| Lung |
NCIH1944 |
8.7
|
| Lung |
NCIH1963 |
9.9
|
| Lung |
NCIH196 |
8.5
|
| Lung |
NCIH1975 |
8.6
|
| Lung |
NCIH2009 |
9
|
| Lung |
NCIH2023 |
8.5
|
| Lung |
NCIH2029 |
8.9
|
| Lung |
NCIH2030 |
7.8
|
| Lung |
NCIH2066 |
8.7
|
| Lung |
NCIH2081 |
8.5
|
| Lung |
NCIH2085 |
8.7
|
| Lung |
NCIH2087 |
9.3
|
| Lung |
NCIH209 |
9.4
|
| Lung |
NCIH2106 |
10
|
| Lung |
NCIH2110 |
8.7
|
| Lung |
NCIH211 |
9.3
|
| Lung |
NCIH2122 |
8.3
|
| Lung |
NCIH2126 |
8.2
|
| Lung |
NCIH2141 |
9.4
|
| Lung |
NCIH2170 |
12.4
|
| Lung |
NCIH2171 |
9.5
|
| Lung |
NCIH2172 |
8.6
|
| Lung |
NCIH2196 |
9.3
|
| Lung |
NCIH2227 |
9.9
|
| Lung |
NCIH2228 |
8.5
|
| Lung |
NCIH226 |
8.7
|
| Lung |
NCIH2286 |
9
|
| Lung |
NCIH2291 |
9
|
| Lung |
NCIH2342 |
9.2
|
| Lung |
NCIH2347 |
8.4
|
| Lung |
NCIH23 |
10
|
| Lung |
NCIH2405 |
9
|
| Lung |
NCIH2444 |
8.6
|
| Lung |
NCIH292 |
8.9
|
| Lung |
NCIH322 |
8.6
|
| Lung |
NCIH3255 |
8.7
|
| Lung |
NCIH358 |
9.5
|
| Lung |
NCIH441 |
8.5
|
| Lung |
NCIH446 |
9.3
|
| Lung |
NCIH460 |
8
|
| Lung |
NCIH510 |
9.4
|
| Lung |
NCIH520 |
8.7
|
| Lung |
NCIH522 |
9.5
|
| Lung |
NCIH524 |
8.9
|
| Lung |
NCIH526 |
10.1
|
| Lung |
NCIH596 |
8.4
|
| Lung |
NCIH647 |
9
|
| Lung |
NCIH650 |
8.5
|
| Lung |
NCIH661 |
8.9
|
| Lung |
NCIH69 |
8.2
|
| Lung |
NCIH727 |
7.7
|
| Lung |
NCIH810 |
9.4
|
| Lung |
NCIH82 |
9.3
|
| Lung |
NCIH838 |
9.8
|
| Lung |
NCIH841 |
9.4
|
| Lung |
NCIH854 |
7.8
|
| Lung |
NCIH889 |
9.4
|
| Lung |
PC14 |
8.7
|
| Lung |
RERFLCAD1 |
8.8
|
| Lung |
RERFLCAD2 |
8.7
|
| Lung |
RERFLCAI |
8.3
|
| Lung |
RERFLCKJ |
9.4
|
| Lung |
RERFLCMS |
8.7
|
| Lung |
RERFLCSQ1 |
8.1
|
| Lung |
SBC5 |
8.5
|
| Lung |
SCLC21H |
9.6
|
| Lung |
SHP77 |
9.1
|
| Lung |
SKLU1 |
8.5
|
| Lung |
SKMES1 |
8.4
|
| Lung |
SQ1 |
8.8
|
| Lung |
SW1271 |
8.5
|
| Lung |
SW1573 |
9.3
|
| Lung |
SW900 |
8.6
|
| Lung |
VMRCLCD |
8.9
|
| Lung |
VMRCLCP |
9.4
|
| Oesophagus |
COLO680N |
8.8
|
| Oesophagus |
ECGI10 |
8.2
|
| Oesophagus |
KYSE140 |
8.8
|
| Oesophagus |
KYSE150 |
9.1
|
| Oesophagus |
KYSE180 |
9.2
|
| Oesophagus |
KYSE270 |
9.5
|
| Oesophagus |
KYSE30 |
9.4
|
| Oesophagus |
KYSE410 |
8
|
| Oesophagus |
KYSE450 |
8.3
|
| Oesophagus |
KYSE510 |
8.6
|
| Oesophagus |
KYSE520 |
8.3
|
| Oesophagus |
KYSE70 |
8.7
|
| Oesophagus |
OE19 |
8.5
|
| Oesophagus |
OE33 |
10
|
| Oesophagus |
TE10 |
8.5
|
| Oesophagus |
TE11 |
8.8
|
| Oesophagus |
TE14 |
9
|
| Oesophagus |
TE15 |
8.9
|
| Oesophagus |
TE1 |
9.1
|
| Oesophagus |
TE4 |
8.6
|
| Oesophagus |
TE5 |
8.3
|
| Oesophagus |
TE6 |
10.3
|
| Oesophagus |
TE8 |
8.7
|
| Oesophagus |
TE9 |
8.2
|
| Oesophagus |
TT |
11
|
| Ovary |
59M |
9.4
|
| Ovary |
A2780 |
9.3
|
| Ovary |
CAOV3 |
8.7
|
| Ovary |
CAOV4 |
8.5
|
| Ovary |
COLO704 |
9.2
|
| Ovary |
COV318 |
9.3
|
| Ovary |
COV362 |
8.4
|
| Ovary |
COV434 |
8.8
|
| Ovary |
COV504 |
8.4
|
| Ovary |
COV644 |
9.1
|
| Ovary |
EFO21 |
9.3
|
| Ovary |
EFO27 |
8.9
|
| Ovary |
ES2 |
8.8
|
| Ovary |
FUOV1 |
9.1
|
| Ovary |
HEYA8 |
8.4
|
| Ovary |
HS571T |
8.1
|
| Ovary |
IGROV1 |
9
|
| Ovary |
JHOC5 |
9.4
|
| Ovary |
JHOM1 |
8.8
|
| Ovary |
JHOM2B |
9.4
|
| Ovary |
JHOS2 |
8.7
|
| Ovary |
JHOS4 |
8.8
|
| Ovary |
KURAMOCHI |
9.8
|
| Ovary |
MCAS |
9.3
|
| Ovary |
NIHOVCAR3 |
8.9
|
| Ovary |
OAW28 |
9.5
|
| Ovary |
OAW42 |
8.9
|
| Ovary |
OC314 |
8.7
|
| Ovary |
OC316 |
8.7
|
| Ovary |
ONCODG1 |
9.2
|
| Ovary |
OV56 |
8.4
|
| Ovary |
OV7 |
8.6
|
| Ovary |
OV90 |
9
|
| Ovary |
OVCAR4 |
8.3
|
| Ovary |
OVCAR8 |
9.4
|
| Ovary |
OVISE |
9.1
|
| Ovary |
OVK18 |
8.5
|
| Ovary |
OVKATE |
8.7
|
| Ovary |
OVMANA |
9.7
|
| Ovary |
OVSAHO |
9.4
|
| Ovary |
OVTOKO |
8.5
|
| Ovary |
RMGI |
8.2
|
| Ovary |
RMUGS |
8.2
|
| Ovary |
SKOV3 |
9.4
|
| Ovary |
SNU119 |
10.2
|
| Ovary |
SNU840 |
9.4
|
| Ovary |
SNU8 |
8.9
|
| Ovary |
TOV112D |
9.1
|
| Ovary |
TOV21G |
9.2
|
| Ovary |
TYKNU |
8.7
|
| Pancreas |
ASPC1 |
8.6
|
| Pancreas |
BXPC3 |
9.2
|
| Pancreas |
CAPAN1 |
9.3
|
| Pancreas |
CAPAN2 |
8.1
|
| Pancreas |
CFPAC1 |
8.2
|
| Pancreas |
DANG |
8.5
|
| Pancreas |
HPAC |
9.8
|
| Pancreas |
HPAFII |
9.1
|
| Pancreas |
HS766T |
9
|
| Pancreas |
HUPT3 |
9.1
|
| Pancreas |
HUPT4 |
9.5
|
| Pancreas |
KCIMOH1 |
9.3
|
| Pancreas |
KLM1 |
8.8
|
| Pancreas |
KP2 |
9.8
|
| Pancreas |
KP3 |
8.5
|
| Pancreas |
KP4 |
9
|
| Pancreas |
L33 |
8.5
|
| Pancreas |
MIAPACA2 |
9.2
|
| Pancreas |
PANC0203 |
9.3
|
| Pancreas |
PANC0213 |
9.4
|
| Pancreas |
PANC0327 |
8.9
|
| Pancreas |
PANC0403 |
8.8
|
| Pancreas |
PANC0504 |
8.8
|
| Pancreas |
PANC0813 |
8.6
|
| Pancreas |
PANC1005 |
9.9
|
| Pancreas |
PANC1 |
8.1
|
| Pancreas |
PATU8902 |
8.5
|
| Pancreas |
PATU8988S |
8.4
|
| Pancreas |
PATU8988T |
8.5
|
| Pancreas |
PK1 |
8.7
|
| Pancreas |
PK45H |
7.6
|
| Pancreas |
PK59 |
8.9
|
| Pancreas |
PL45 |
9.7
|
| Pancreas |
PSN1 |
9.4
|
| Pancreas |
QGP1 |
9.4
|
| Pancreas |
SNU213 |
9.2
|
| Pancreas |
SNU324 |
9.2
|
| Pancreas |
SNU410 |
8.4
|
| Pancreas |
SU8686 |
8.9
|
| Pancreas |
SUIT2 |
9
|
| Pancreas |
SW1990 |
8.3
|
| Pancreas |
T3M4 |
8.8
|
| Pancreas |
TCCPAN2 |
7.9
|
| Pancreas |
YAPC |
8.7
|
| Pleura |
ACCMESO1 |
8.6
|
| Pleura |
DM3 |
8
|
| Pleura |
ISTMES1 |
8.8
|
| Pleura |
ISTMES2 |
9.2
|
| Pleura |
JL1 |
8.7
|
| Pleura |
MPP89 |
9.2
|
| Pleura |
MSTO211H |
8.8
|
| Pleura |
NCIH2052 |
8.1
|
| Pleura |
NCIH2452 |
7.9
|
| Pleura |
NCIH28 |
9.2
|
| Prostate |
22RV1 |
8.8
|
| Prostate |
DU145 |
8.2
|
| Prostate |
LNCAPCLONEFGC |
9
|
| Prostate |
MDAPCA2B |
9.3
|
| Prostate |
NCIH660 |
9.4
|
| Prostate |
PC3 |
8.5
|
| Prostate |
VCAP |
9.5
|
| Salivary gland |
A253 |
8.9
|
| Salivary gland |
YD15 |
9
|
| Skin |
A101D |
9.5
|
| Skin |
A2058 |
9.5
|
| Skin |
A375 |
9
|
| Skin |
C32 |
8.9
|
| Skin |
CHL1 |
9
|
| Skin |
CJM |
9.5
|
| Skin |
COLO679 |
9.2
|
| Skin |
COLO741 |
9.2
|
| Skin |
COLO783 |
9.1
|
| Skin |
COLO792 |
8
|
| Skin |
COLO800 |
9.1
|
| Skin |
COLO818 |
8.4
|
| Skin |
COLO829 |
9.2
|
| Skin |
COLO849 |
10
|
| Skin |
G361 |
9.2
|
| Skin |
GRM |
9
|
| Skin |
HMCB |
8.8
|
| Skin |
HS294T |
8.6
|
| Skin |
HS600T |
7.9
|
| Skin |
HS688AT |
7.7
|
| Skin |
HS695T |
7.7
|
| Skin |
HS839T |
7.7
|
| Skin |
HS852T |
8.8
|
| Skin |
HS895T |
7.9
|
| Skin |
HS934T |
8.3
|
| Skin |
HS936T |
8.8
|
| Skin |
HS939T |
8.4
|
| Skin |
HS940T |
8.1
|
| Skin |
HS944T |
9.2
|
| Skin |
HT144 |
8.2
|
| Skin |
IGR1 |
9.1
|
| Skin |
IGR37 |
9.2
|
| Skin |
IGR39 |
8.6
|
| Skin |
IPC298 |
10
|
| Skin |
K029AX |
10.2
|
| Skin |
LOXIMVI |
8.5
|
| Skin |
MALME3M |
8.5
|
| Skin |
MDAMB435S |
8.4
|
| Skin |
MELHO |
10.1
|
| Skin |
MELJUSO |
9
|
| Skin |
MEWO |
10
|
| Skin |
RPMI7951 |
8.5
|
| Skin |
RVH421 |
8.5
|
| Skin |
SH4 |
9.7
|
| Skin |
SKMEL1 |
8.2
|
| Skin |
SKMEL24 |
7.7
|
| Skin |
SKMEL28 |
10.3
|
| Skin |
SKMEL2 |
10.3
|
| Skin |
SKMEL30 |
9.7
|
| Skin |
SKMEL31 |
8.8
|
| Skin |
SKMEL3 |
9
|
| Skin |
SKMEL5 |
9.2
|
| Skin |
UACC257 |
9
|
| Skin |
UACC62 |
9.6
|
| Skin |
WM115 |
9.9
|
| Skin |
WM1799 |
8.8
|
| Skin |
WM2664 |
9.8
|
| Skin |
WM793 |
8.6
|
| Skin |
WM88 |
9.7
|
| Skin |
WM983B |
8.9
|
| Small intestine |
HUTU80 |
9
|
| Soft tissue |
A204 |
8.6
|
| Soft tissue |
G401 |
9
|
| Soft tissue |
G402 |
8.5
|
| Soft tissue |
GCT |
8.9
|
| Soft tissue |
HS729 |
8.6
|
| Soft tissue |
HT1080 |
8.6
|
| Soft tissue |
KYM1 |
8.6
|
| Soft tissue |
MESSA |
9.1
|
| Soft tissue |
RD |
9.2
|
| Soft tissue |
RH30 |
9.5
|
| Soft tissue |
RH41 |
9.1
|
| Soft tissue |
RKN |
8
|
| Soft tissue |
S117 |
9.2
|
| Soft tissue |
SJRH30 |
8.9
|
| Soft tissue |
SKLMS1 |
8
|
| Soft tissue |
SKUT1 |
8.8
|
| Soft tissue |
TE125T |
8.3
|
| Soft tissue |
TE159T |
8.1
|
| Soft tissue |
TE441T |
9
|
| Soft tissue |
TE617T |
8.7
|
| Stomach |
2313287 |
8.4
|
| Stomach |
AGS |
9.1
|
| Stomach |
AZ521 |
9.1
|
| Stomach |
ECC10 |
9.5
|
| Stomach |
ECC12 |
8.9
|
| Stomach |
FU97 |
8.5
|
| Stomach |
GCIY |
9.5
|
| Stomach |
GSS |
8.9
|
| Stomach |
GSU |
9
|
| Stomach |
HGC27 |
8.7
|
| Stomach |
HS746T |
8.2
|
| Stomach |
HUG1N |
9
|
| Stomach |
IM95 |
9.1
|
| Stomach |
KATOIII |
8.4
|
| Stomach |
KE39 |
9.3
|
| Stomach |
LMSU |
9.1
|
| Stomach |
MKN1 |
8.6
|
| Stomach |
MKN45 |
8.6
|
| Stomach |
MKN74 |
9.2
|
| Stomach |
MKN7 |
8.4
|
| Stomach |
NCCSTCK140 |
9
|
| Stomach |
NCIN87 |
9.3
|
| Stomach |
NUGC2 |
7.9
|
| Stomach |
NUGC3 |
9
|
| Stomach |
NUGC4 |
9.3
|
| Stomach |
OCUM1 |
9.2
|
| Stomach |
RERFGC1B |
9.2
|
| Stomach |
SH10TC |
8.4
|
| Stomach |
SNU16 |
9.2
|
| Stomach |
SNU1 |
9.2
|
| Stomach |
SNU216 |
9.1
|
| Stomach |
SNU520 |
9.3
|
| Stomach |
SNU5 |
8.7
|
| Stomach |
SNU601 |
9.5
|
| Stomach |
SNU620 |
9.1
|
| Stomach |
SNU668 |
9.7
|
| Stomach |
SNU719 |
10
|
| Stomach |
TGBC11TKB |
9.8
|
| Thyroid |
8305C |
8.2
|
| Thyroid |
8505C |
9
|
| Thyroid |
BCPAP |
8.6
|
| Thyroid |
BHT101 |
9.6
|
| Thyroid |
CAL62 |
8.9
|
| Thyroid |
CGTHW1 |
7.9
|
| Thyroid |
FTC133 |
8.3
|
| Thyroid |
FTC238 |
8.6
|
| Thyroid |
ML1 |
9.1
|
| Thyroid |
SW579 |
8.3
|
| Thyroid |
TT2609C02 |
8.7
|
| Thyroid |
TT |
8.1
|
| Upper aerodigestive tract |
BHY |
8.3
|
| Upper aerodigestive tract |
BICR16 |
7.6
|
| Upper aerodigestive tract |
BICR18 |
8.1
|
| Upper aerodigestive tract |
BICR22 |
8.9
|
| Upper aerodigestive tract |
BICR31 |
7.2
|
| Upper aerodigestive tract |
BICR56 |
9.3
|
| Upper aerodigestive tract |
BICR6 |
9.1
|
| Upper aerodigestive tract |
CAL27 |
9
|
| Upper aerodigestive tract |
CAL33 |
8.8
|
| Upper aerodigestive tract |
DETROIT562 |
8.1
|
| Upper aerodigestive tract |
FADU |
9.6
|
| Upper aerodigestive tract |
HS840T |
8.2
|
| Upper aerodigestive tract |
HSC2 |
8.8
|
| Upper aerodigestive tract |
HSC3 |
8.8
|
| Upper aerodigestive tract |
HSC4 |
9.6
|
| Upper aerodigestive tract |
PECAPJ15 |
9.2
|
| Upper aerodigestive tract |
PECAPJ34CLONEC12 |
8.7
|
| Upper aerodigestive tract |
PECAPJ41CLONED2 |
8.8
|
| Upper aerodigestive tract |
PECAPJ49 |
7.6
|
| Upper aerodigestive tract |
SCC15 |
8.7
|
| Upper aerodigestive tract |
SCC25 |
8.7
|
| Upper aerodigestive tract |
SCC4 |
8.8
|
| Upper aerodigestive tract |
SCC9 |
8.6
|
| Upper aerodigestive tract |
SNU1076 |
8.6
|
| Upper aerodigestive tract |
SNU1214 |
8.2
|
| Upper aerodigestive tract |
SNU46 |
8.4
|
| Upper aerodigestive tract |
SNU899 |
8.4
|
| Upper aerodigestive tract |
YD10B |
8.7
|
| Upper aerodigestive tract |
YD38 |
8.8
|
| Upper aerodigestive tract |
YD8 |
9.4
|
| Urinary tract |
5637 |
8.3
|
| Urinary tract |
639V |
9.3
|
| Urinary tract |
647V |
9.4
|
| Urinary tract |
BC3C |
8.7
|
| Urinary tract |
BFTC905 |
9.3
|
| Urinary tract |
CAL29 |
8.9
|
| Urinary tract |
HS172T |
8.2
|
| Urinary tract |
HT1197 |
9
|
| Urinary tract |
HT1376 |
8.8
|
| Urinary tract |
J82 |
8.6
|
| Urinary tract |
JMSU1 |
9.2
|
| Urinary tract |
KMBC2 |
9
|
| Urinary tract |
KU1919 |
10.4
|
| Urinary tract |
RT11284 |
9.2
|
| Urinary tract |
RT112 |
8.8
|
| Urinary tract |
RT4 |
9.4
|
| Urinary tract |
SCABER |
8.8
|
| Urinary tract |
SW1710 |
9.2
|
| Urinary tract |
SW780 |
9.2
|
| Urinary tract |
T24 |
8.7
|
| Urinary tract |
TCCSUP |
8.6
|
| Urinary tract |
UMUC1 |
8.2
|
| Urinary tract |
UMUC3 |
8.6
|
| Urinary tract |
VMCUB1 |
9.3
|