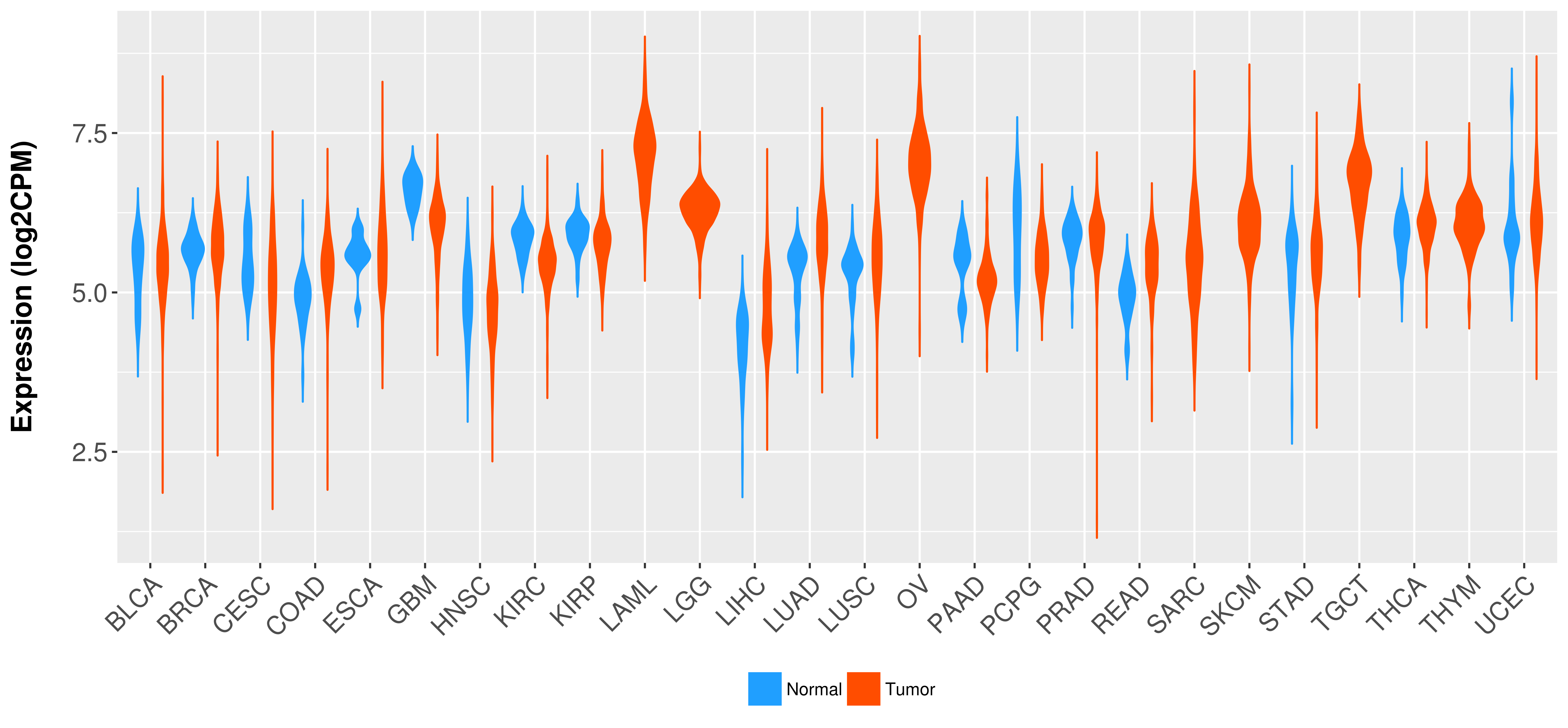
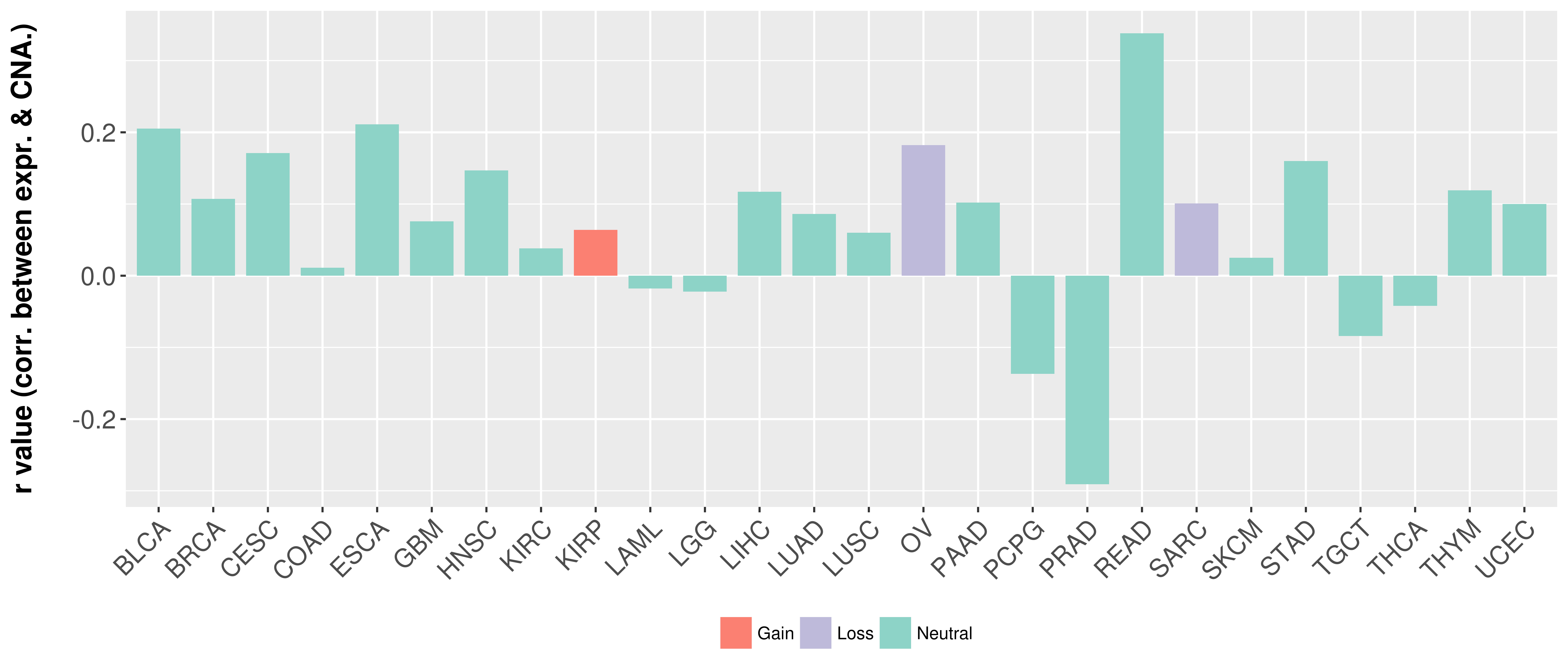
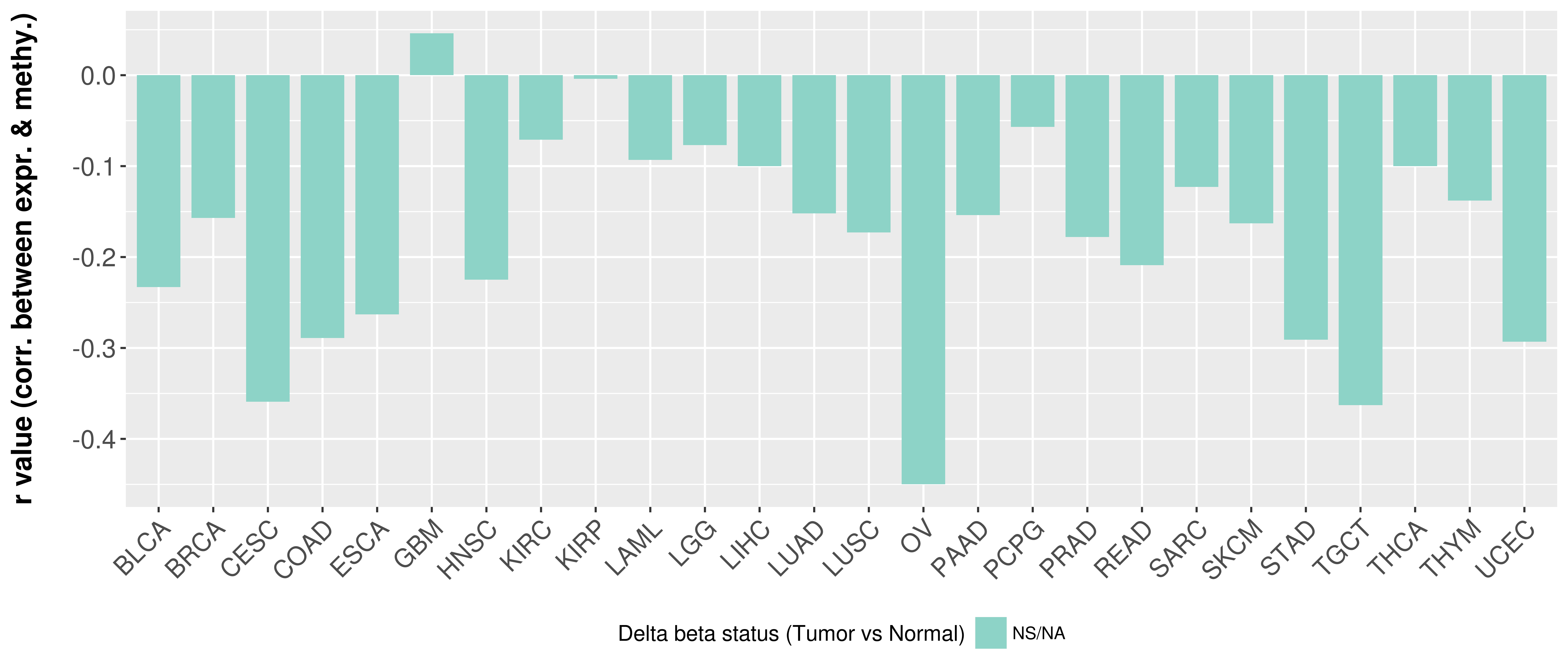
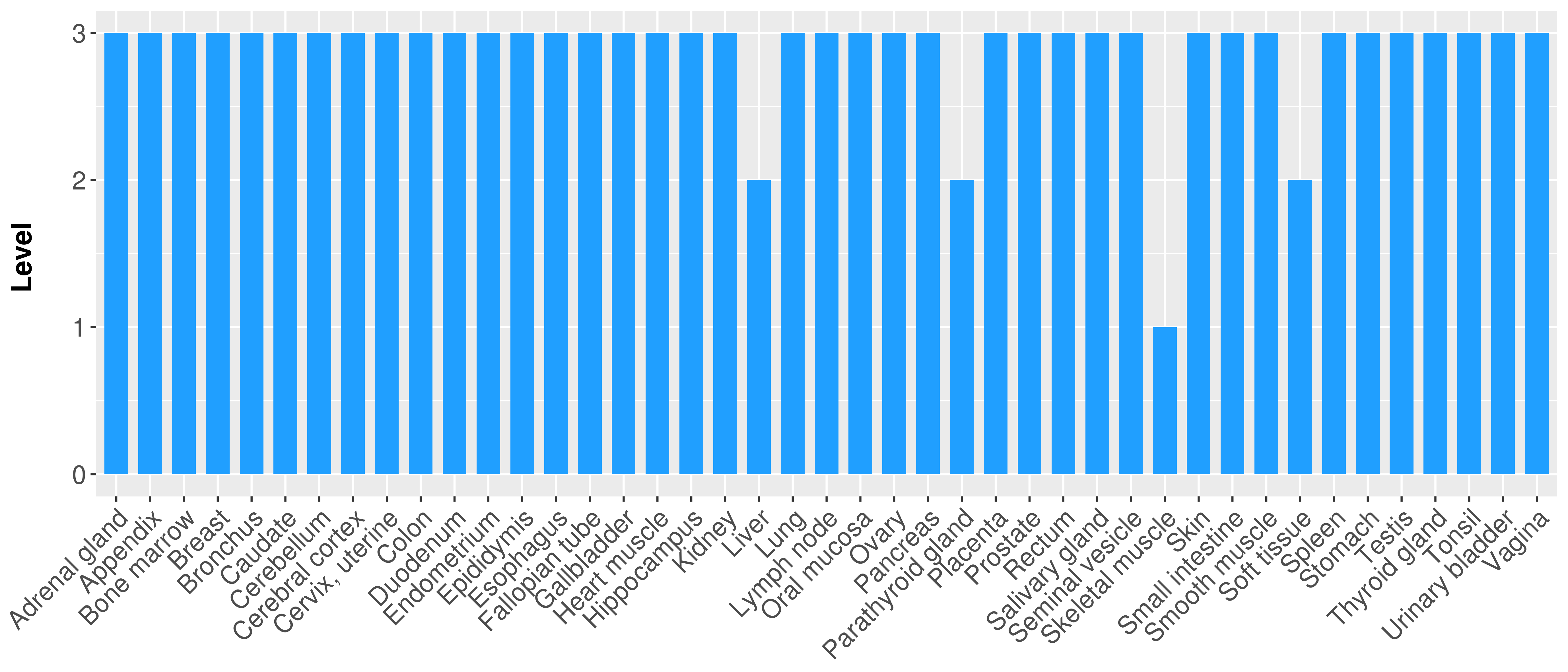
| COSM3724250 | c.2661G>A | p.P887P | Substitution - coding silent | Upper_aerodigestive_tract
|
| COSM1178926 | c.143_144insC | p.G49fs*13 | Insertion - Frameshift | Autonomic_ganglia
|
| COSM273498 | c.3064C>T | p.P1022S | Substitution - Missense | Large_intestine
|
| COSM3737488 | c.2229C>A | p.C743* | Substitution - Nonsense | Soft_tissue
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM457977 | c.3908A>G | p.Y1303C | Substitution - Missense | Breast
|
| COSM3095629 | c.193G>A | p.G65S | Substitution - Missense | Large_intestine
|
| COSM1735242 | c.143delC | p.P48fs*65 | Deletion - Frameshift | Large_intestine
|
| COSM1124807 | c.2685G>A | p.P895P | Substitution - coding silent | Endometrium
|
| COSM1293056 | c.3905T>C | p.F1302S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3719816 | c.3899A>G | p.Y1300C | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1124800 | c.3250A>C | p.T1084P | Substitution - Missense | Endometrium
|
| COSM2153280 | c.1409T>C | p.L470P | Substitution - Missense | Central_nervous_system
|
| COSM5413026 | c.1754G>A | p.G585E | Substitution - Missense | Skin
|
| COSM4435593 | c.772G>A | p.E258K | Substitution - Missense | Oesophagus
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Breast
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3563106 | c.2809G>A | p.E937K | Substitution - Missense | Skin
|
| COSM5030636 | c.245_246insG | p.L85fs*4 | Insertion - Frameshift | Bone
|
| COSM1315668 | c.881C>G | p.S294* | Substitution - Nonsense | Urinary_tract
|
| COSM1715114 | c.2719G>A | p.E907K | Substitution - Missense | Skin
|
| COSM1233530 | c.3205A>G | p.N1069D | Substitution - Missense | Large_intestine
|
| COSM5715829 | c.3450C>T | p.N1150N | Substitution - coding silent | Skin
|
| COSM3965483 | c.802G>C | p.D268H | Substitution - Missense | Lung
|
| COSM255663 | c.1939C>T | p.R647* | Substitution - Nonsense | Endometrium
|
| COSM1757283 | c.3602G>A | p.G1201E | Substitution - Missense | Urinary_tract
|
| COSM5030636 | c.245_246insG | p.L85fs*4 | Insertion - Frameshift | Bone
|
| COSM146125 | c.2542G>A | p.V848M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3379677 | c.3317G>C | p.C1106S | Substitution - Missense | Pancreas
|
| COSM248485 | c.4064G>A | p.R1355H | Substitution - Missense | Prostate
|
| COSM4381993 | c.1300G>C | p.D434H | Substitution - Missense | Large_intestine
|
| COSM5005363 | c.2843_2844insCT | p.S950fs*8 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM5701738 | c.1186_1187CC>AA | p.P396K | Substitution - Missense | Soft_tissue
|
| COSM1124815 | c.1487A>G | p.Y496C | Substitution - Missense | Endometrium
|
| COSM1124819 | c.683C>T | p.A228V | Substitution - Missense | Endometrium
|
| COSM3767043 | c.1068C>T | p.C356C | Substitution - coding silent | Liver
|
| COSM1132268 | c.199G>T | p.E67* | Substitution - Nonsense | Prostate
|
| COSM4457338 | c.1040C>T | p.P347L | Substitution - Missense | Skin
|
| COSM4742912 | c.3987G>A | p.E1329E | Substitution - coding silent | Large_intestine
|
| COSM146124 | c.3418C>G | p.L1140V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1124808 | c.2638C>T | p.H880Y | Substitution - Missense | Endometrium
|
| COSM255676 | c.3595C>G | p.Q1199E | Substitution - Missense | Central_nervous_system
|
| COSM1469179 | c.3271C>T | p.R1091C | Substitution - Missense | Large_intestine
|
| COSM1124810 | c.2242C>A | p.L748I | Substitution - Missense | Endometrium
|
| COSM4879339 | c.2923C>T | p.R975* | Substitution - Nonsense | Prostate
|
| COSM4742913 | c.3970C>T | p.R1324W | Substitution - Missense | Large_intestine
|
| COSM1315671 | c.515C>T | p.S172F | Substitution - Missense | Urinary_tract
|
| COSM248486 | c.3080delC | p.T1027fs*9 | Deletion - Frameshift | Prostate
|
| COSM3563109 | c.2052G>T | p.Q684H | Substitution - Missense | Skin
|
| COSM4742917 | c.647C>G | p.P216R | Substitution - Missense | Large_intestine
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Prostate
|
| COSM1124821 | c.443C>A | p.S148Y | Substitution - Missense | Endometrium
|
| COSM383655 | c.1649G>T | p.R550L | Substitution - Missense | Lung
|
| COSM3708718 | c.724C>T | p.R242C | Substitution - Missense | Liver
|
| COSM5702439 | c.1656_1657CC>AA | p.S552_L553>RI | Complex - compound substitution | Soft_tissue
|
| COSM1469183 | c.1416C>T | p.H472H | Substitution - coding silent | Large_intestine
|
| COSM4495125 | c.446C>T | p.P149L | Substitution - Missense | Skin
|
| COSM255320 | c.3332delT | p.L1111fs*9 | Deletion - Frameshift | Central_nervous_system
|
| COSM3765033 | c.3611C>T | p.S1204L | Substitution - Missense | Central_nervous_system
|
| COSM292119 | c.1489C>A | p.P497T | Substitution - Missense | Large_intestine
|
| COSM1124809 | c.2249G>A | p.R750H | Substitution - Missense | Endometrium
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Breast
|
| COSM5715829 | c.3450C>T | p.N1150N | Substitution - coding silent | Skin
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM4152566 | c.491C>T | p.S164F | Substitution - Missense | Kidney
|
| COSM757658 | c.1286A>C | p.H429P | Substitution - Missense | Lung
|
| COSM4742913 | c.3970C>T | p.R1324W | Substitution - Missense | Small_intestine
|
| COSM4436956 | c.3247G>C | p.E1083Q | Substitution - Missense | Oesophagus
|
| COSM4769937 | c.1254C>T | p.V418V | Substitution - coding silent | Biliary_tract
|
| COSM1715115 | c.1321C>T | p.R441W | Substitution - Missense | Skin
|
| COSM5715829 | c.3450C>T | p.N1150N | Substitution - coding silent | Skin
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM4390073 | c.721G>T | p.E241* | Substitution - Nonsense | Urinary_tract
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM4950261 | c.634_635insG | p.S213fs*2 | Insertion - Frameshift | Liver
|
| COSM4110861 | c.750A>G | p.K250K | Substitution - coding silent | Stomach
|
| COSM1729897 | c.472G>T | p.E158* | Substitution - Nonsense | Liver
|
| COSM1178128 | c.974A>G | p.Y325C | Substitution - Missense | Central_nervous_system
|
| COSM4423478 | c.397A>G | p.N133D | Substitution - Missense | Central_nervous_system
|
| COSM1469184 | c.1364G>T | p.G455V | Substitution - Missense | Large_intestine
|
| COSM4837754 | c.1909C>T | p.R637W | Substitution - Missense | Cervix
|
| COSM316700 | c.162G>T | p.S54S | Substitution - coding silent | Soft_tissue
|
| COSM1124823 | c.24T>C | p.S8S | Substitution - coding silent | Endometrium
|
| COSM2150924 | c.2219G>A | p.R740H | Substitution - Missense | Central_nervous_system
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM457977 | c.3908A>G | p.Y1303C | Substitution - Missense | Prostate
|
| COSM3973835 | c.3222G>T | p.W1074C | Substitution - Missense | Central_nervous_system
|
| COSM385747 | c.1903C>G | p.H635D | Substitution - Missense | Lung
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3670287 | c.1648C>T | p.R550C | Substitution - Missense | Large_intestine
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Prostate
|
| COSM4742919 | c.252_253insG | p.L85fs*4 | Insertion - Frameshift | Breast
|
| COSM79112 | c.2649C>A | p.C883* | Substitution - Nonsense | Ovary
|
| COSM1270991 | c.2174G>A | p.R725H | Substitution - Missense | Stomach
|
| COSM5420997 | c.140_143delCCCC | p.P47fs*65 | Deletion - Frameshift | Prostate
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1682715 | c.3736C>A | p.P1246T | Substitution - Missense | Large_intestine
|
| COSM3845307 | c.2214G>C | p.Q738H | Substitution - Missense | Breast
|
| COSM255663 | c.1939C>T | p.R647* | Substitution - Nonsense | Central_nervous_system
|
| COSM5100802 | c.1757_1758insG | p.I587fs*17 | Insertion - Frameshift | Large_intestine
|
| COSM4765831 | c.253_254insG | p.L85fs*4 | Insertion - Frameshift | Stomach
|
| COSM4742919 | c.252_253insG | p.L85fs*4 | Insertion - Frameshift | Large_intestine
|
| COSM3845306 | c.3562C>T | p.R1188* | Substitution - Nonsense | Breast
|
| COSM757657 | c.1278C>T | p.S426S | Substitution - coding silent | Lung
|
| COSM1124796 | c.3827G>T | p.R1276I | Substitution - Missense | Endometrium
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1124813 | c.1831G>C | p.V611L | Substitution - Missense | Endometrium
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM757662 | c.3057G>T | p.L1019L | Substitution - coding silent | Lung
|
| COSM1178926 | c.143_144insC | p.G49fs*13 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1124803 | c.3154T>G | p.S1052A | Substitution - Missense | Endometrium
|
| COSM4005163 | c.1976G>T | p.G659V | Substitution - Missense | Urinary_tract
|
| COSM4110857 | c.3537C>T | p.L1179L | Substitution - coding silent | Stomach
|
| COSM146124 | c.3418C>G | p.L1140V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4950261 | c.634_635insG | p.S213fs*2 | Insertion - Frameshift | Liver
|
| COSM3800739 | c.2827G>A | p.E943K | Substitution - Missense | Urinary_tract
|
| COSM5366928 | c.3385C>T | p.R1129* | Substitution - Nonsense | Large_intestine
|
| COSM5693979 | c.762A>T | p.V254V | Substitution - coding silent | Soft_tissue
|
| COSM5508241 | c.1077G>C | p.E359D | Substitution - Missense | Biliary_tract
|
| COSM1757284 | c.1199C>G | p.S400C | Substitution - Missense | Urinary_tract
|
| COSM2153280 | c.1409T>C | p.L470P | Substitution - Missense | Central_nervous_system
|
| COSM3694792 | c.162G>A | p.S54S | Substitution - coding silent | Large_intestine
|
| COSM5546969 | c.591_592delGA | p.E197fs*17 | Deletion - Frameshift | Prostate
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1293058 | c.1195C>T | p.Q399* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM316701 | c.1178A>T | p.Q393L | Substitution - Missense | Lung
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3364098 | c.1472A>C | p.K491T | Substitution - Missense | Kidney
|
| COSM305337 | c.166G>T | p.A56S | Substitution - Missense | Central_nervous_system
|
| COSM5413025 | c.1879C>T | p.R627W | Substitution - Missense | Skin
|
| COSM372350 | c.2387C>A | p.P796Q | Substitution - Missense | Lung
|
| COSM5813029 | c.645C>G | p.P215P | Substitution - coding silent | Liver
|
| COSM307173 | c.702G>A | p.P234P | Substitution - coding silent | Central_nervous_system
|
| COSM303614 | c.3292delC | p.R1098fs*22 | Deletion - Frameshift | Central_nervous_system
|
| COSM1293057 | c.3337_3339delTAC | p.Y1113delY | Deletion - In frame | Haematopoietic_and_lymphoid_tissue
|
| COSM5681151 | c.1380C>A | p.T460T | Substitution - coding silent | Soft_tissue
|
| COSM488568 | c.2269C>A | p.Q757K | Substitution - Missense | Kidney
|
| COSM757660 | c.2384C>G | p.T795S | Substitution - Missense | Lung
|
| COSM316699 | c.1389G>T | p.G463G | Substitution - coding silent | Lung
|
| COSM4522292 | c.1154G>T | p.C385F | Substitution - Missense | Skin
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM5004851 | c.1073+4C>T | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4980593 | c.3382G>T | p.E1128* | Substitution - Nonsense | Oesophagus
|
| COSM4828446 | c.877C>T | p.R293C | Substitution - Missense | Small_intestine
|
| COSM1132269 | c.778+1G>T | p.? | Unknown | Prostate
|
| COSM3737488 | c.2229C>A | p.C743* | Substitution - Nonsense | Soft_tissue
|
| COSM1757284 | c.1199C>G | p.S400C | Substitution - Missense | Urinary_tract
|
| COSM3670287 | c.1648C>T | p.R550C | Substitution - Missense | Central_nervous_system
|
| COSM1472314 | c.3886_3887CC>AG | p.P1296S | Substitution - Missense | Prostate
|
| COSM5498463 | c.1470+6G>A | p.? | Unknown | Biliary_tract
|
| COSM1270990 | c.2350C>G | p.Q784E | Substitution - Missense | Oesophagus
|
| COSM248487 | c.2548A>G | p.M850V | Substitution - Missense | Prostate
|
| COSM5910116 | c.2412G>A | p.K804K | Substitution - coding silent | Skin
|
| COSM5413024 | c.3276T>C | p.F1092F | Substitution - coding silent | Skin
|
| COSM5731903 | c.2308delC | p.L770fs*1 | Deletion - Frameshift | Pancreas
|
| COSM5437781 | c.2454T>C | p.P818P | Substitution - coding silent | Oesophagus
|
| COSM255663 | c.1939C>T | p.R647* | Substitution - Nonsense | Stomach
|
| COSM4881556 | c.3750C>A | p.T1250T | Substitution - coding silent | Upper_aerodigestive_tract
|
| COSM5419315 | c.3631A>G | p.T1211A | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM5557979 | c.1038G>C | p.K346N | Substitution - Missense | Prostate
|
| COSM457978 | c.3121C>T | p.R1041W | Substitution - Missense | Breast
|
| COSM1124795 | c.4018G>A | p.D1340N | Substitution - Missense | Endometrium
|
| COSM1756657 | c.3297C>T | p.I1099I | Substitution - coding silent | Urinary_tract
|
| COSM5417986 | c.2686-13_2686-2delTTCGTGCCTCCA | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4742914 | c.3492delT | p.F1164fs*9 | Deletion - Frameshift | Large_intestine
|
| COSM5497345 | c.420_435del16 | p.L141fs*5 | Deletion - Frameshift | Biliary_tract
|
| COSM5413028 | c.908C>T | p.S303F | Substitution - Missense | Skin
|
| COSM3563108 | c.2400G>A | p.G800G | Substitution - coding silent | Skin
|
| COSM3406562 | c.1288C>A | p.R430R | Substitution - coding silent | Central_nervous_system
|
| COSM4839655 | c.46C>A | p.P16T | Substitution - Missense | Cervix
|
| COSM4340380 | c.619C>T | p.Q207* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4830111 | c.3912C>T | p.L1304L | Substitution - coding silent | Cervix
|
| COSM4110856 | c.3551C>T | p.T1184M | Substitution - Missense | Stomach
|
| COSM316701 | c.1178A>T | p.Q393L | Substitution - Missense | Soft_tissue
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3563105 | c.2910G>T | p.L970L | Substitution - coding silent | Skin
|
| COSM1124816 | c.1350T>C | p.C450C | Substitution - coding silent | Endometrium
|
| COSM3708718 | c.724C>T | p.R242C | Substitution - Missense | Small_intestine
|
| COSM5988913 | c.2575G>T | p.E859* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM757661 | c.2493G>C | p.K831N | Substitution - Missense | Lung
|
| COSM1756658 | c.1253T>G | p.V418G | Substitution - Missense | Urinary_tract
|
| COSM1124817 | c.1092G>T | p.K364N | Substitution - Missense | Endometrium
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Large_intestine
|
| COSM3682160 | c.258C>T | p.L86L | Substitution - coding silent | Haematopoietic_and_lymphoid_tissue
|
| COSM5894190 | c.1735C>T | p.Q579* | Substitution - Nonsense | Skin
|
| COSM3563110 | c.1938C>T | p.L646L | Substitution - coding silent | Skin
|
| COSM5646109 | c.1349G>C | p.C450S | Substitution - Missense | Oesophagus
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1124802 | c.3180C>A | p.T1060T | Substitution - coding silent | Endometrium
|
| COSM3765033 | c.3611C>T | p.S1204L | Substitution - Missense | Central_nervous_system
|
| COSM1178926 | c.143_144insC | p.G49fs*13 | Insertion - Frameshift | Prostate
|
| COSM422498 | c.945G>A | p.R315R | Substitution - coding silent | Urinary_tract
|
| COSM1124820 | c.558C>T | p.S186S | Substitution - coding silent | Endometrium
|
| COSM4791908 | c.2261A>C | p.Q754P | Substitution - Missense | Liver
|
| COSM3563114 | c.224G>A | p.G75E | Substitution - Missense | Skin
|
| COSM388318 | c.386G>T | p.G129V | Substitution - Missense | Lung
|
| COSM3563111 | c.1932G>A | p.E644E | Substitution - coding silent | Skin
|
| COSM3563107 | c.2437C>T | p.P813S | Substitution - Missense | Skin
|
| COSM1497337 | c.2960A>G | p.N987S | Substitution - Missense | Kidney
|
| COSM1124798 | c.3485T>C | p.L1162P | Substitution - Missense | Endometrium
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Large_intestine
|
| COSM2150924 | c.2219G>A | p.R740H | Substitution - Missense | Central_nervous_system
|
| COSM4167359 | c.1279G>A | p.V427M | Substitution - Missense | Breast
|
| COSM757654 | c.144T>A | p.P48P | Substitution - coding silent | Lung
|
| COSM3379678 | c.1097C>T | p.S366L | Substitution - Missense | Large_intestine
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Large_intestine
|
| COSM3095621 | c.405T>C | p.C135C | Substitution - coding silent | Large_intestine
|
| COSM4970096 | c.1258C>A | p.H420N | Substitution - Missense | Central_nervous_system
|
| COSM3973834 | c.3313G>A | p.A1105T | Substitution - Missense | Central_nervous_system
|
| COSM1240713 | c.1276A>G | p.S426G | Substitution - Missense | Oesophagus
|
| COSM1132271 | c.3887C>G | p.P1296R | Substitution - Missense | Prostate
|
| COSM1124799 | c.3280+1G>T | p.? | Unknown | Endometrium
|
| COSM1469181 | c.2791G>A | p.D931N | Substitution - Missense | Large_intestine
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM242345 | c.204_205delAG | p.D69fs*9 | Deletion - Frameshift | Prostate
|
| COSM3095603 | c.1613G>T | p.G538V | Substitution - Missense | Large_intestine
|
| COSM4110860 | c.1235G>A | p.C412Y | Substitution - Missense | Stomach
|
| COSM4889763 | c.1864G>T | p.D622Y | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM3563113 | c.410C>T | p.P137L | Substitution - Missense | Skin
|
| COSM4983369 | c.410C>G | p.P137R | Substitution - Missense | Oesophagus
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM5413027 | c.1207C>T | p.P403S | Substitution - Missense | Skin
|
| COSM3095566 | c.4071T>A | p.I1357I | Substitution - coding silent | Large_intestine
|
| COSM4110856 | c.3551C>T | p.T1184M | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM146125 | c.2542G>A | p.V848M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM255676 | c.3595C>G | p.Q1199E | Substitution - Missense | Central_nervous_system
|
| COSM757656 | c.481G>A | p.E161K | Substitution - Missense | Lung
|
| COSM2156506 | c.2532C>A | p.V844V | Substitution - coding silent | Central_nervous_system
|
| COSM4655147 | c.2865T>G | p.L955L | Substitution - coding silent | Large_intestine
|
| COSM5435217 | c.727G>A | p.V243I | Substitution - Missense | Oesophagus
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3379707 | c.3502C>G | p.L1168V | Substitution - Missense | Pancreas
|
| COSM1124794 | c.4049G>A | p.R1350H | Substitution - Missense | Endometrium
|
| COSM4773570 | c.3411T>G | p.Y1137* | Substitution - Nonsense | Stomach
|
| COSM4808384 | c.2863C>T | p.L955F | Substitution - Missense | Pancreas
|
| COSM2156506 | c.2532C>A | p.V844V | Substitution - coding silent | Central_nervous_system
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM1497338 | c.3619G>A | p.V1207I | Substitution - Missense | Kidney
|
| COSM4340433 | c.3965C>T | p.P1322L | Substitution - Missense | Bone
|
| COSM4921634 | c.2671T>C | p.S891P | Substitution - Missense | Liver
|
| COSM3095571 | c.3712C>T | p.R1238W | Substitution - Missense | Large_intestine
|
| COSM5043992 | c.3111+1G>C | p.? | Unknown | Liver
|
| COSM457981 | c.275C>T | p.P92L | Substitution - Missense | Breast
|
| COSM4742915 | c.3004A>G | p.K1002E | Substitution - Missense | Large_intestine
|
| COSM4167359 | c.1279G>A | p.V427M | Substitution - Missense | Meninges
|
| COSM5475355 | c.3946G>A | p.D1316N | Substitution - Missense | Large_intestine
|
| COSM5900441 | c.1160C>T | p.S387F | Substitution - Missense | Skin
|
| COSM1124818 | c.699G>A | p.K233K | Substitution - coding silent | Endometrium
|
| COSM255663 | c.1939C>T | p.R647* | Substitution - Nonsense | Central_nervous_system
|
| COSM1735242 | c.143delC | p.P48fs*65 | Deletion - Frameshift | Pancreas
|
| COSM3095571 | c.3712C>T | p.R1238W | Substitution - Missense | Large_intestine
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM4110858 | c.2383A>T | p.T795S | Substitution - Missense | Stomach
|
| COSM4171962 | c.2204G>A | p.W735* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM4806297 | c.3593A>T | p.K1198M | Substitution - Missense | Liver
|
| COSM3390769 | c.3331C>T | p.L1111F | Substitution - Missense | Pancreas
|
| COSM294974 | c.3068A>G | p.E1023G | Substitution - Missense | Large_intestine
|
| COSM1124804 | c.3130C>T | p.L1044F | Substitution - Missense | Endometrium
|
| COSM4962090 | c.4088G>A | p.R1363H | Substitution - Missense | Pancreas
|
| COSM4828446 | c.877C>T | p.R293C | Substitution - Missense | Cervix
|
| COSM4928146 | c.1989G>A | p.L663L | Substitution - coding silent | Liver
|
| COSM3095605 | c.1575C>T | p.S525S | Substitution - coding silent | Large_intestine
|
| COSM4492840 | c.4048C>T | p.R1350C | Substitution - Missense | Skin
|
| COSM1315670 | c.754G>A | p.E252K | Substitution - Missense | Urinary_tract
|
| COSM1682715 | c.3736C>A | p.P1246T | Substitution - Missense | Large_intestine
|
| COSM1124806 | c.2794A>G | p.I932V | Substitution - Missense | Endometrium
|
| COSM303613 | c.120C>A | p.A40A | Substitution - coding silent | Central_nervous_system
|
| COSM1293059 | c.159delC | p.S54fs*59 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4791908 | c.2261A>C | p.Q754P | Substitution - Missense | Liver
|
| COSM146125 | c.2542G>A | p.V848M | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3390770 | c.1739G>A | p.R580H | Substitution - Missense | Pancreas
|
| COSM5513819 | c.1433G>A | p.R478Q | Substitution - Missense | Biliary_tract
|
| COSM1316858 | c.1477A>T | p.T493S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1756657 | c.3297C>T | p.I1099I | Substitution - coding silent | Urinary_tract
|
| COSM1124808 | c.2638C>T | p.H880Y | Substitution - Missense | Oesophagus
|
| COSM1124805 | c.3011C>A | p.P1004H | Substitution - Missense | Endometrium
|
| COSM1469182 | c.2530G>A | p.V844I | Substitution - Missense | Large_intestine
|
| COSM5789573 | c.3871A>C | p.N1291H | Substitution - Missense | Breast
|
| COSM4806297 | c.3593A>T | p.K1198M | Substitution - Missense | Liver
|
| COSM757659 | c.1509C>G | p.T503T | Substitution - coding silent | Lung
|
| COSM3095590 | c.2391G>T | p.E797D | Substitution - Missense | Large_intestine
|
| COSM1735242 | c.143delC | p.P48fs*65 | Deletion - Frameshift | Biliary_tract
|
| COSM5419561 | c.2579_2580GG>AA | p.W860* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM5894190 | c.1735C>T | p.Q579* | Substitution - Nonsense | Skin
|
| COSM5064853 | c.583G>A | p.V195M | Substitution - Missense | Stomach
|
| COSM3845308 | c.1172C>T | p.A391V | Substitution - Missense | Breast
|
| COSM457979 | c.2041_2044delTACT | p.Y681fs*89 | Deletion - Frameshift | Breast
|
| COSM3379678 | c.1097C>T | p.S366L | Substitution - Missense | Pancreas
|
| COSM3694704 | c.2555C>G | p.S852C | Substitution - Missense | Large_intestine
|
| COSM4835800 | c.1338G>C | p.L446L | Substitution - coding silent | Cervix
|
| COSM3965482 | c.3312C>T | p.L1104L | Substitution - coding silent | Lung
|
| COSM1178926 | c.143_144insC | p.G49fs*13 | Insertion - Frameshift | Breast
|
| COSM757655 | c.364G>A | p.E122K | Substitution - Missense | Lung
|
| COSM1491237 | c.3634C>G | p.L1212V | Substitution - Missense | Breast
|
| COSM4381993 | c.1300G>C | p.D434H | Substitution - Missense | Large_intestine
|
| COSM1124822 | c.136G>C | p.A46P | Substitution - Missense | Endometrium
|
| COSM1491239 | c.3111G>A | p.K1037K | Substitution - coding silent | Breast
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3973836 | c.3080C>G | p.T1027S | Substitution - Missense | Central_nervous_system
|
| COSM5631384 | c.2637G>A | p.M879I | Substitution - Missense | Oesophagus
|
| COSM4831808 | c.186C>A | p.T62T | Substitution - coding silent | Cervix
|
| COSM1124801 | c.3239C>T | p.A1080V | Substitution - Missense | Endometrium
|
| COSM5589420 | c.110C>T | p.S37F | Substitution - Missense | Skin
|
| COSM248484 | c.3407A>C | p.Y1136S | Substitution - Missense | Prostate
|
| COSM5941155 | c.1737+7C>T | p.? | Unknown | Skin
|
| COSM5616360 | c.667G>T | p.G223* | Substitution - Nonsense | Lung
|
| COSM1124797 | c.3821G>A | p.R1274Q | Substitution - Missense | Endometrium
|
| COSM1315669 | c.763G>C | p.D255H | Substitution - Missense | Urinary_tract
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM4110859 | c.1695A>C | p.T565T | Substitution - coding silent | Stomach
|
| COSM4888117 | c.1445C>A | p.T482K | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM5023333 | c.94G>A | p.E32K | Substitution - Missense | Bone
|
| COSM5715829 | c.3450C>T | p.N1150N | Substitution - coding silent | Skin
|
| COSM4608193 | c.962G>A | p.G321E | Substitution - Missense | Adrenal_gland
|
| COSM1735242 | c.143delC | p.P48fs*65 | Deletion - Frameshift | Large_intestine
|
| COSM1637772 | c.1287C>T | p.H429H | Substitution - coding silent | Bone
|
| COSM4742916 | c.2173C>T | p.R725C | Substitution - Missense | Large_intestine
|
| COSM1178128 | c.974A>G | p.Y325C | Substitution - Missense | Prostate
|
| COSM4886952 | c.1143C>T | p.C381C | Substitution - coding silent | Upper_aerodigestive_tract
|
| COSM1316859 | c.1475A>T | p.N492I | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM488569 | c.150T>C | p.P50P | Substitution - coding silent | Kidney
|
| COSM1124812 | c.1849C>T | p.R617C | Substitution - Missense | Endometrium
|
| COSM5511337 | c.3433-1G>A | p.? | Unknown | Biliary_tract
|
| COSM1331711 | c.277C>G | p.P93A | Substitution - Missense | Ovary
|
| COSM3563112 | c.1385C>T | p.T462I | Substitution - Missense | Skin
|
| COSM5748960 | c.2206C>T | p.R736C | Substitution - Missense | Pancreas
|
| COSM3563104 | c.3039C>T | p.F1013F | Substitution - coding silent | Skin
|
| COSM5366928 | c.3385C>T | p.R1129* | Substitution - Nonsense | Large_intestine
|
| COSM1124814 | c.1786A>G | p.S596G | Substitution - Missense | Endometrium
|
| COSM130349 | c.2342C>G | p.T781R | Substitution - Missense | Upper_aerodigestive_tract
|
| COSM1491238 | c.3483C>T | p.Y1161Y | Substitution - coding silent | Breast
|
| COSM316700 | c.162G>T | p.S54S | Substitution - coding silent | Lung
|
| COSM4742918 | c.451A>C | p.T151P | Substitution - Missense | Large_intestine
|
| COSM1757283 | c.3602G>A | p.G1201E | Substitution - Missense | Urinary_tract
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Breast
|
| COSM1469185 | c.252delG | p.L85fs*28 | Deletion - Frameshift | Large_intestine
|
| COSM1233531 | c.1243A>G | p.T415A | Substitution - Missense | Large_intestine
|
| COSM3670287 | c.1648C>T | p.R550C | Substitution - Missense | Central_nervous_system
|
| COSM1124811 | c.1986G>A | p.L662L | Substitution - coding silent | Endometrium
|
| COSM1132270 | c.3886C>A | p.P1296T | Substitution - Missense | Prostate
|