This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-3p | ABCA1 | -0.07 | 0.92099 | -0.11 | 0.8728 | mirMAP; miRNATAP | -0.39 | 0 | NA | |
| 2 | hsa-let-7a-3p | ABCC9 | -0.07 | 0.92099 | 0.17 | 0.63712 | mirMAP | -0.8 | 0 | NA | |
| 3 | hsa-let-7a-3p | ABCD2 | -0.07 | 0.92099 | -0.13 | 0.76186 | mirMAP | -0.51 | 5.0E-5 | NA | |
| 4 | hsa-miR-125a-5p | ABHD3 | 0.06 | 0.95785 | -0.09 | 0.90386 | PITA; miRanda | -0.19 | 0.00818 | NA | |
| 5 | hsa-let-7a-3p | ABI3BP | -0.07 | 0.92099 | -0.37 | 0.41633 | miRNATAP | -0.62 | 2.0E-5 | NA | |
| 6 | hsa-miR-15a-5p | ABL2 | 0.11 | 0.90237 | 0.04 | 0.95748 | MirTarget; miRNATAP | -0.16 | 5.0E-5 | NA | |
| 7 | hsa-miR-125a-5p | ACPP | 0.06 | 0.95785 | -0.43 | 0.34039 | MirTarget; miRanda | -0.36 | 0.02701 | NA | |
| 8 | hsa-miR-125a-5p | ACTL6A | 0.06 | 0.95785 | 0.05 | 0.95251 | miRanda | -0.12 | 0.00186 | NA | |
| 9 | hsa-let-7a-3p | ACVR1 | -0.07 | 0.92099 | -0.02 | 0.98204 | MirTarget; miRNATAP | -0.12 | 0.00024 | NA | |
| 10 | hsa-miR-125a-5p | ADAM10 | 0.06 | 0.95785 | -0.21 | 0.80651 | miRanda | -0.11 | 0.04343 | NA | |
| 11 | hsa-let-7a-3p | ADAM23 | -0.07 | 0.92099 | -0.36 | 0.37035 | mirMAP | -0.96 | 0 | NA | |
| 12 | hsa-miR-15a-5p | ADAMTS18 | 0.11 | 0.90237 | -0.6 | 0.24444 | miRNATAP | -1 | 0 | NA | |
| 13 | hsa-miR-15a-5p | ADAMTS3 | 0.11 | 0.90237 | -0.15 | 0.57772 | miRNATAP | -0.59 | 0 | NA | |
| 14 | hsa-let-7a-3p | ADAMTS5 | -0.07 | 0.92099 | 0.35 | 0.38267 | mirMAP | -0.36 | 0 | NA | |
| 15 | hsa-miR-15a-5p | ADAMTS5 | 0.11 | 0.90237 | 0.35 | 0.38267 | miRNATAP | -0.39 | 0 | NA | |
| 16 | hsa-let-7a-3p | ADAMTSL3 | -0.07 | 0.92099 | -0.2 | 0.64161 | MirTarget | -1.14 | 0 | NA | |
| 17 | hsa-miR-15a-5p | ADAMTSL3 | 0.11 | 0.90237 | -0.2 | 0.64161 | MirTarget | -1.46 | 0 | NA | |
| 18 | hsa-miR-15a-5p | ADCY1 | 0.11 | 0.90237 | 0.38 | 0.25371 | mirMAP | -0.32 | 0.01019 | NA | |
| 19 | hsa-miR-15a-5p | ADCY5 | 0.11 | 0.90237 | -0.55 | 0.2334 | MirTarget; miRNATAP | -1.61 | 0 | NA | |
| 20 | hsa-let-7a-3p | ADCYAP1 | -0.07 | 0.92099 | 0.13 | 0.79662 | miRNATAP | -1.12 | 0 | NA | |
| 21 | hsa-let-7a-3p | ADD2 | -0.07 | 0.92099 | -0.16 | 0.76694 | mirMAP | -0.74 | 0 | NA | |
| 22 | hsa-miR-125a-5p | ADH5 | 0.06 | 0.95785 | 0.01 | 0.99261 | miRanda | -0.12 | 0.0022 | NA | |
| 23 | hsa-miR-15a-5p | ADRB2 | 0.11 | 0.90237 | -0.01 | 0.98489 | MirTarget | -0.79 | 0 | NA | |
| 24 | hsa-let-7a-3p | AFF2 | -0.07 | 0.92099 | -0.3 | 0.56096 | mirMAP | -0.35 | 0.012 | NA | |
| 25 | hsa-let-7a-3p | AFF3 | -0.07 | 0.92099 | -0.5 | 0.15768 | MirTarget | -0.46 | 0.00048 | NA | |
| 26 | hsa-miR-15a-5p | AFF4 | 0.11 | 0.90237 | -0.1 | 0.9096 | MirTarget; miRNATAP | -0.11 | 0.00251 | NA | |
| 27 | hsa-miR-15a-5p | AGPAT4 | 0.11 | 0.90237 | 0.33 | 0.44742 | mirMAP | -0.46 | 2.0E-5 | NA | |
| 28 | hsa-let-7a-3p | AKAP12 | -0.07 | 0.92099 | 0.17 | 0.82172 | miRNATAP | -0.88 | 0 | NA | |
| 29 | hsa-miR-15a-5p | AKT3 | 0.11 | 0.90237 | 0.06 | 0.90908 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.72 | 0 | NA | |
| 30 | hsa-let-7a-3p | ALDH1L2 | -0.07 | 0.92099 | 0.28 | 0.55075 | MirTarget | -0.5 | 0 | NA | |
| 31 | hsa-miR-125a-5p | ALDH6A1 | 0.06 | 0.95785 | -0.27 | 0.59927 | miRanda | -0.22 | 0.00306 | NA | |
| 32 | hsa-let-7a-3p | AMIGO2 | -0.07 | 0.92099 | 0.21 | 0.75965 | MirTarget; mirMAP | -0.39 | 5.0E-5 | NA | |
| 33 | hsa-let-7a-3p | AMOTL1 | -0.07 | 0.92099 | -0.11 | 0.85689 | mirMAP | -0.64 | 0 | NA | |
| 34 | hsa-miR-15a-5p | AMOTL1 | 0.11 | 0.90237 | -0.11 | 0.85689 | MirTarget; miRNATAP | -0.94 | 0 | NA | |
| 35 | hsa-miR-125a-5p | ANGPT2 | 0.06 | 0.95785 | 0.03 | 0.95669 | miRanda | -0.19 | 0.01088 | NA | |
| 36 | hsa-miR-15a-5p | ANK2 | 0.11 | 0.90237 | 0.04 | 0.93166 | MirTarget; miRNATAP | -1.21 | 0 | NA | |
| 37 | hsa-miR-15a-5p | ANKRD29 | 0.11 | 0.90237 | 0.28 | 0.38764 | MirTarget | -0.39 | 0.00308 | NA | |
| 38 | hsa-let-7a-3p | ANKRD50 | -0.07 | 0.92099 | -0.1 | 0.88159 | mirMAP | -0.19 | 4.0E-5 | NA | |
| 39 | hsa-miR-125a-5p | ANKS1A | 0.06 | 0.95785 | -0.12 | 0.88556 | miRanda | -0.11 | 0.00232 | NA | |
| 40 | hsa-let-7a-3p | ANO5 | -0.07 | 0.92099 | 0.58 | 0.27012 | mirMAP | -0.59 | 0.00256 | NA | |
| 41 | hsa-miR-125a-5p | ANP32A | 0.06 | 0.95785 | -0.06 | 0.95269 | miRanda | -0.11 | 0.003 | NA | |
| 42 | hsa-let-7a-3p | ANTXR1 | -0.07 | 0.92099 | -0.05 | 0.96265 | mirMAP; miRNATAP | -0.79 | 0 | NA | |
| 43 | hsa-miR-125a-5p | API5 | 0.06 | 0.95785 | -0.01 | 0.99409 | miRanda | -0.13 | 3.0E-5 | NA | |
| 44 | hsa-miR-15a-5p | APLNR | 0.11 | 0.90237 | 0.01 | 0.99326 | MirTarget | -0.35 | 0.00048 | NA | |
| 45 | hsa-miR-15a-5p | APP | 0.11 | 0.90237 | -0.18 | 0.89833 | miRNAWalker2 validate; miRNATAP | -0.14 | 0.00073 | NA | |
| 46 | hsa-let-7a-3p | AQP9 | -0.07 | 0.92099 | 0.18 | 0.73115 | MirTarget | -0.89 | 0 | NA | |
| 47 | hsa-miR-15a-5p | AR | 0.11 | 0.90237 | -0.54 | 0.32591 | mirMAP; miRNATAP | -1.06 | 0 | NA | |
| 48 | hsa-miR-125a-5p | ARHGAP11B | 0.06 | 0.95785 | -0.31 | 0.23724 | miRanda | -0.19 | 0.01801 | NA | |
| 49 | hsa-let-7a-3p | ARHGAP20 | -0.07 | 0.92099 | 0.1 | 0.77766 | MirTarget; miRNATAP | -0.61 | 0 | NA | |
| 50 | hsa-miR-15a-5p | ARHGAP20 | 0.11 | 0.90237 | 0.1 | 0.77766 | MirTarget; miRNATAP | -0.85 | 0 | NA | |
| 51 | hsa-let-7a-3p | ARHGAP28 | -0.07 | 0.92099 | -0.23 | 0.43033 | mirMAP | -0.39 | 0.00025 | NA | |
| 52 | hsa-let-7a-3p | ARHGAP29 | -0.07 | 0.92099 | 0.16 | 0.78525 | mirMAP | -0.27 | 0.00025 | NA | |
| 53 | hsa-miR-125a-5p | ARID4A | 0.06 | 0.95785 | -0.06 | 0.92409 | miRanda | -0.12 | 0.00166 | NA | |
| 54 | hsa-let-7a-3p | ARL10 | -0.07 | 0.92099 | -0.24 | 0.42733 | mirMAP | -0.55 | 0 | NA | |
| 55 | hsa-miR-15a-5p | ARL2 | 0.11 | 0.90237 | 0.02 | 0.97901 | MirTarget; miRNATAP | -0.11 | 0.01332 | NA | |
| 56 | hsa-miR-15a-5p | ARMCX2 | 0.11 | 0.90237 | 0.29 | 0.5542 | MirTarget; miRNATAP | -0.57 | 0 | NA | |
| 57 | hsa-miR-15a-5p | ARNT2 | 0.11 | 0.90237 | -0.47 | 0.3134 | mirMAP | -0.78 | 0 | NA | |
| 58 | hsa-let-7a-3p | ARSB | -0.07 | 0.92099 | -0.08 | 0.88514 | mirMAP | -0.17 | 0.00258 | NA | |
| 59 | hsa-let-7a-3p | ASAH1 | -0.07 | 0.92099 | -0.05 | 0.96217 | MirTarget | -0.14 | 0.00225 | NA | |
| 60 | hsa-let-7a-3p | ASAP1 | -0.07 | 0.92099 | -0.09 | 0.91083 | mirMAP | -0.19 | 0.0006 | NA | |
| 61 | hsa-miR-15a-5p | ASH1L | 0.11 | 0.90237 | -0.14 | 0.87278 | MirTarget; miRNATAP | -0.12 | 0.00706 | NA | |
| 62 | hsa-miR-15a-5p | ASTN1 | 0.11 | 0.90237 | 0.05 | 0.94222 | MirTarget | -1.25 | 0 | NA | |
| 63 | hsa-let-7a-3p | ASXL2 | -0.07 | 0.92099 | -0.3 | 0.6827 | mirMAP; miRNATAP | -0.18 | 0.00081 | NA | |
| 64 | hsa-miR-15a-5p | ASXL2 | 0.11 | 0.90237 | -0.3 | 0.6827 | miRNAWalker2 validate | -0.13 | 0.03086 | NA | |
| 65 | hsa-let-7a-3p | ATE1 | -0.07 | 0.92099 | -0.24 | 0.65828 | mirMAP | -0.13 | 0.00265 | NA | |
| 66 | hsa-let-7a-3p | ATF2 | -0.07 | 0.92099 | -0.24 | 0.68896 | MirTarget; mirMAP; miRNATAP | -0.15 | 4.0E-5 | NA | |
| 67 | hsa-miR-15a-5p | ATF2 | 0.11 | 0.90237 | -0.24 | 0.68896 | miRNAWalker2 validate | -0.12 | 0.00218 | NA | |
| 68 | hsa-miR-125a-5p | ATL2 | 0.06 | 0.95785 | -0.06 | 0.94297 | miRNAWalker2 validate; miRanda | -0.12 | 0.00034 | NA | |
| 69 | hsa-let-7a-3p | ATP6AP2 | -0.07 | 0.92099 | -0.05 | 0.96232 | mirMAP | -0.13 | 0.00027 | NA | |
| 70 | hsa-let-7a-3p | ATRN | -0.07 | 0.92099 | -0.46 | 0.56279 | miRNATAP | -0.26 | 8.0E-5 | NA | |
| 71 | hsa-let-7a-3p | ATRX | -0.07 | 0.92099 | -0.24 | 0.77958 | MirTarget; miRNATAP | -0.13 | 0.00051 | NA | |
| 72 | hsa-miR-15a-5p | ATXN1L | 0.11 | 0.90237 | -0.14 | 0.85841 | miRNATAP | -0.11 | 0.00148 | NA | |
| 73 | hsa-let-7a-3p | B3GALNT1 | -0.07 | 0.92099 | 0.4 | 0.30968 | mirMAP | -0.37 | 0.00031 | NA | |
| 74 | hsa-miR-15a-5p | BACE1 | 0.11 | 0.90237 | -0.16 | 0.82465 | miRNAWalker2 validate; miRNATAP | -0.33 | 0 | NA | |
| 75 | hsa-miR-15a-5p | BACH2 | 0.11 | 0.90237 | -0.35 | 0.26727 | MirTarget; miRNATAP | -0.78 | 0 | NA | |
| 76 | hsa-let-7a-3p | BAG4 | -0.07 | 0.92099 | -0.21 | 0.6077 | mirMAP | -0.2 | 0.00021 | NA | |
| 77 | hsa-miR-125a-5p | BAG4 | 0.06 | 0.95785 | -0.21 | 0.6077 | MirTarget | -0.17 | 0.00683 | NA | |
| 78 | hsa-miR-125a-5p | BAK1 | 0.06 | 0.95785 | 0.22 | 0.78301 | miRNAWalker2 validate; PITA; miRanda; miRNATAP | -0.18 | 0.0026 | NA | |
| 79 | hsa-let-7a-3p | BASP1 | -0.07 | 0.92099 | 0.06 | 0.91132 | miRNATAP | -0.46 | 0 | NA | |
| 80 | hsa-miR-125a-5p | BATF2 | 0.06 | 0.95785 | -0.01 | 0.99363 | miRanda | -0.18 | 0.04948 | NA | |
| 81 | hsa-miR-125a-5p | BAZ1A | 0.06 | 0.95785 | -0.03 | 0.97529 | miRanda | -0.15 | 0.00012 | NA | |
| 82 | hsa-miR-15a-5p | BCL2 | 0.11 | 0.90237 | -0.44 | 0.34621 | miRNAWalker2 validate; miRTarBase | -0.22 | 0.02911 | 25623762; 25594541; 26915294; 18931683; 22335947 | miR 15a and miR 16 modulate drug resistance by targeting bcl 2 in human colon cancer cells; To investigate the reversal effect of targeted modulation of bcl-2 expression by miR-15a and miR-16 on drug resistance of human colon cancer cells;MicroRNAs miRNAs encoded by the miR-15 cluster are known to induce G1 arrest and apoptosis by targeting G1 checkpoints and the anti-apoptotic B cell lymphoma 2 BCL-2 gene;As a result transcript levels of the tumor-suppressive miR-15 and let-7 families increased which targeted and decreased the expression of the crucial prosurvival genes BCL-2 and BCL-XL respectively;MicroRNAs miRNAs are noncoding small RNAs that repress protein translation by targeting specific messenger RNAs miR-15a and miR-16-1 act as putative tumor suppressors by targeting the oncogene BCL2;The expression of MiR-15a was significantly inhibited by Bcl-2 P < 0.05 |
| 83 | hsa-miR-125a-5p | BCL2L14 | 0.06 | 0.95785 | -0.05 | 0.93549 | MirTarget; PITA; miRanda | -0.16 | 0.04212 | NA | |
| 84 | hsa-miR-15a-5p | BCL2L2 | 0.11 | 0.90237 | 0 | 0.99812 | MirTarget; miRNATAP | -0.14 | 0.00012 | 25874488 | MicroRNA 15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non small cell lung cancer; Furthermore we identified that BCL2L2 was a target of miR-15a and negatively regulated by miR-15a at the translational level; miR-15a acts as a tumor suppressor in NSCLC by directly targeting BCL2L2 and may serve as a potential diagnostic biomarker and therapeutic target for NSCLC |
| 85 | hsa-let-7a-3p | BCL6 | -0.07 | 0.92099 | -0.02 | 0.97924 | miRNATAP | -0.17 | 0.00802 | NA | |
| 86 | hsa-let-7a-3p | BDNF | -0.07 | 0.92099 | -0.01 | 0.97997 | MirTarget; miRNATAP | -0.24 | 0.00935 | NA | |
| 87 | hsa-miR-15a-5p | BDNF | 0.11 | 0.90237 | -0.01 | 0.97997 | MirTarget; miRNATAP | -0.36 | 0.00046 | 26581909 | MicroRNA 15a 5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF; BDNF was then overexpressed in HepG2 and SNU-182 cells to evaluate its selective effect on miR-15a-5p in HCC modulation; MiR-15a-5p selectively and negatively regulated BDNF at both gene and protein levels in HCC cells; Forced overexpression of BDNF effectively reversed the tumor suppressive functions of miR-15a-5p on HCC proliferation and cell division in vitro; Our study demonstrated that miR-15a-5p is a tumor suppressor in HCC and its regulation is through BDNF in HCC |
| 88 | hsa-let-7a-3p | BEND4 | -0.07 | 0.92099 | 0.03 | 0.96594 | mirMAP | -0.41 | 0.00473 | NA | |
| 89 | hsa-let-7a-3p | BEND6 | -0.07 | 0.92099 | 0.06 | 0.87652 | mirMAP | -0.77 | 0 | NA | |
| 90 | hsa-miR-15a-5p | BHLHE41 | 0.11 | 0.90237 | 0.27 | 0.66334 | miRNATAP | -0.38 | 0.00052 | NA | |
| 91 | hsa-let-7a-3p | BICD2 | -0.07 | 0.92099 | -0.05 | 0.94512 | mirMAP | -0.1 | 0.00472 | NA | |
| 92 | hsa-miR-125a-5p | BIK | 0.06 | 0.95785 | 0.15 | 0.80372 | miRanda | -0.21 | 0.04414 | NA | |
| 93 | hsa-let-7a-3p | BMP2K | -0.07 | 0.92099 | -0.19 | 0.72063 | mirMAP | -0.17 | 6.0E-5 | NA | |
| 94 | hsa-let-7a-3p | BMP3 | -0.07 | 0.92099 | -0.26 | 0.66775 | MirTarget; mirMAP | -0.82 | 0.00029 | NA | |
| 95 | hsa-let-7a-3p | BMPR1B | -0.07 | 0.92099 | -0.25 | 0.60455 | miRNATAP | -0.61 | 0.00028 | NA | |
| 96 | hsa-let-7a-3p | BMPR2 | -0.07 | 0.92099 | -0.17 | 0.84777 | MirTarget; miRNATAP | -0.18 | 0.00013 | NA | |
| 97 | hsa-miR-15a-5p | BMX | 0.11 | 0.90237 | -0.01 | 0.97533 | MirTarget; miRNATAP | -0.82 | 0 | NA | |
| 98 | hsa-let-7a-3p | BNC2 | -0.07 | 0.92099 | -0.03 | 0.94679 | mirMAP | -0.77 | 0 | NA | |
| 99 | hsa-miR-125a-5p | BRCC3 | 0.06 | 0.95785 | -0.17 | 0.83954 | miRanda | -0.13 | 0.01416 | NA | |
| 100 | hsa-miR-125a-5p | BRPF3 | 0.06 | 0.95785 | -0.19 | 0.82481 | MirTarget | -0.13 | 0.0056 | NA | |
| 101 | hsa-let-7a-3p | BST1 | -0.07 | 0.92099 | 0.23 | 0.52012 | MirTarget | -0.38 | 0.00024 | NA | |
| 102 | hsa-miR-15a-5p | BTBD19 | 0.11 | 0.90237 | 0.16 | 0.58395 | mirMAP | -0.42 | 7.0E-5 | NA | |
| 103 | hsa-let-7a-3p | BTC | -0.07 | 0.92099 | -0.19 | 0.42376 | mirMAP | -0.2 | 0.00283 | NA | |
| 104 | hsa-miR-125a-5p | BTC | 0.06 | 0.95785 | -0.19 | 0.42376 | miRanda | -0.16 | 0.04529 | NA | |
| 105 | hsa-miR-15a-5p | BTG2 | 0.11 | 0.90237 | 0.01 | 0.99517 | MirTarget; miRNATAP | -0.17 | 0.00744 | NA | |
| 106 | hsa-let-7a-3p | BVES | -0.07 | 0.92099 | 0.04 | 0.90028 | mirMAP | -0.64 | 0 | NA | |
| 107 | hsa-miR-15a-5p | BVES | 0.11 | 0.90237 | 0.04 | 0.90028 | MirTarget | -1.02 | 0 | NA | |
| 108 | hsa-let-7a-3p | CACNA1C | -0.07 | 0.92099 | -0.15 | 0.80484 | MirTarget | -0.23 | 0.03112 | NA | |
| 109 | hsa-miR-15a-5p | CACNA1E | 0.11 | 0.90237 | 0.16 | 0.74358 | MirTarget | -0.85 | 0 | NA | |
| 110 | hsa-let-7a-3p | CACNA2D1 | -0.07 | 0.92099 | -0.25 | 0.60439 | mirMAP | -1.24 | 0 | NA | |
| 111 | hsa-miR-15a-5p | CACNB4 | 0.11 | 0.90237 | 0.17 | 0.63328 | miRNATAP | -0.51 | 9.0E-5 | NA | |
| 112 | hsa-let-7a-3p | CACNG4 | -0.07 | 0.92099 | -0.11 | 0.86743 | MirTarget | -0.55 | 0.03405 | NA | |
| 113 | hsa-miR-15a-5p | CADM1 | 0.11 | 0.90237 | 0.14 | 0.76681 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.83 | 0 | NA | |
| 114 | hsa-miR-15a-5p | CALD1 | 0.11 | 0.90237 | 0.1 | 0.9298 | miRNAWalker2 validate | -0.9 | 0 | NA | |
| 115 | hsa-miR-15a-5p | CALU | 0.11 | 0.90237 | 0.06 | 0.9593 | MirTarget | -0.22 | 0 | NA | |
| 116 | hsa-miR-125a-5p | CANT1 | 0.06 | 0.95785 | 0.06 | 0.95485 | mirMAP | -0.12 | 0.04622 | NA | |
| 117 | hsa-miR-15a-5p | CAPN6 | 0.11 | 0.90237 | 0.87 | 0.21395 | miRNATAP | -1.14 | 1.0E-5 | NA | |
| 118 | hsa-miR-125a-5p | CAPZA2 | 0.06 | 0.95785 | -0.02 | 0.98174 | miRanda | -0.1 | 0.02005 | NA | |
| 119 | hsa-miR-15a-5p | CARD8 | 0.11 | 0.90237 | 0.12 | 0.83502 | miRNAWalker2 validate | -0.18 | 2.0E-5 | NA | |
| 120 | hsa-miR-125a-5p | CASP6 | 0.06 | 0.95785 | -0.09 | 0.9061 | miRanda | -0.13 | 0.02252 | NA | |
| 121 | hsa-let-7a-3p | CCDC102B | -0.07 | 0.92099 | 0.33 | 0.16371 | MirTarget | -0.29 | 0.00016 | NA | |
| 122 | hsa-let-7a-3p | CCDC50 | -0.07 | 0.92099 | 0.08 | 0.92311 | miRNATAP | -0.14 | 0 | NA | |
| 123 | hsa-miR-125a-5p | CCNC | 0.06 | 0.95785 | -0.05 | 0.95565 | miRanda | -0.16 | 0.00022 | NA | |
| 124 | hsa-miR-15a-5p | CCND1 | 0.11 | 0.90237 | -0.18 | 0.87202 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.13 | 0.01564 | 22922827 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR |
| 125 | hsa-miR-125a-5p | CCNJ | 0.06 | 0.95785 | -0.09 | 0.87726 | MirTarget; PITA; miRanda; miRNATAP | -0.1 | 0.02175 | NA | |
| 126 | hsa-miR-15a-5p | CCNJL | 0.11 | 0.90237 | -0.5 | 0.32399 | miRNATAP | -0.22 | 0.04258 | NA | |
| 127 | hsa-let-7a-3p | CCPG1 | -0.07 | 0.92099 | 0.07 | 0.93571 | mirMAP | -0.18 | 0.00016 | NA | |
| 128 | hsa-miR-125a-5p | CCT2 | 0.06 | 0.95785 | 0.01 | 0.99233 | miRanda | -0.11 | 0.01193 | NA | |
| 129 | hsa-let-7a-3p | CD109 | -0.07 | 0.92099 | 0.31 | 0.60219 | mirMAP | -0.58 | 7.0E-5 | NA | |
| 130 | hsa-miR-15a-5p | CD28 | 0.11 | 0.90237 | -0.11 | 0.71453 | MirTarget; miRNATAP | -0.26 | 0.03384 | NA | |
| 131 | hsa-let-7a-3p | CD302 | -0.07 | 0.92099 | -0.02 | 0.97427 | mirMAP | -0.17 | 0.00116 | NA | |
| 132 | hsa-let-7a-3p | CD38 | -0.07 | 0.92099 | 0.36 | 0.40643 | MirTarget | -0.66 | 4.0E-5 | NA | |
| 133 | hsa-let-7a-3p | CDC14A | -0.07 | 0.92099 | -0.08 | 0.8497 | mirMAP | -0.15 | 0.00458 | NA | |
| 134 | hsa-miR-125a-5p | CDC14C | 0.06 | 0.95785 | -0.06 | 0.82588 | miRanda | -0.24 | 0.00202 | NA | |
| 135 | hsa-miR-125a-5p | CDC23 | 0.06 | 0.95785 | -0.02 | 0.98133 | miRanda | -0.1 | 0.00117 | NA | |
| 136 | hsa-let-7a-3p | CDC42EP3 | -0.07 | 0.92099 | 0 | 0.99642 | MirTarget; mirMAP | -0.25 | 3.0E-5 | NA | |
| 137 | hsa-let-7a-3p | CDK17 | -0.07 | 0.92099 | -0.03 | 0.96173 | MirTarget; miRNATAP | -0.12 | 0.00022 | NA | |
| 138 | hsa-miR-15a-5p | CDK17 | 0.11 | 0.90237 | -0.03 | 0.96173 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 139 | hsa-let-7a-3p | CDS2 | -0.07 | 0.92099 | -0.27 | 0.71103 | mirMAP | -0.12 | 0.00961 | NA | |
| 140 | hsa-miR-15a-5p | CDS2 | 0.11 | 0.90237 | -0.27 | 0.71103 | mirMAP; miRNATAP | -0.25 | 0 | NA | |
| 141 | hsa-miR-15a-5p | CECR6 | 0.11 | 0.90237 | -0.51 | 0.04771 | MirTarget; miRNATAP | -0.2 | 0.03797 | NA | |
| 142 | hsa-miR-125a-5p | CENPO | 0.06 | 0.95785 | -0.07 | 0.87469 | mirMAP | -0.13 | 0.00452 | NA | |
| 143 | hsa-miR-125a-5p | CES3 | 0.06 | 0.95785 | 0.23 | 0.73891 | miRanda | -0.37 | 0.02945 | NA | |
| 144 | hsa-let-7a-3p | CFL2 | -0.07 | 0.92099 | -0 | 0.9933 | mirMAP | -0.42 | 0 | NA | |
| 145 | hsa-miR-15a-5p | CHD5 | 0.11 | 0.90237 | 0.26 | 0.55655 | miRNATAP | -0.46 | 0.0018 | NA | |
| 146 | hsa-miR-15a-5p | CHD9 | 0.11 | 0.90237 | -0.09 | 0.90367 | miRNATAP | -0.2 | 1.0E-5 | NA | |
| 147 | hsa-let-7a-3p | CHL1 | -0.07 | 0.92099 | -0.21 | 0.62739 | MirTarget; miRNATAP | -0.94 | 0 | NA | |
| 148 | hsa-let-7a-3p | CHM | -0.07 | 0.92099 | -0.21 | 0.75281 | MirTarget | -0.13 | 0.00246 | NA | |
| 149 | hsa-let-7a-3p | CHST2 | -0.07 | 0.92099 | 0.09 | 0.79889 | MirTarget | -0.3 | 0.0002 | NA | |
| 150 | hsa-miR-125a-5p | CKS1B | 0.06 | 0.95785 | 0.14 | 0.85897 | miRanda | -0.1 | 0.0406 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEUROGENESIS | 137 | 1402 | 1.608e-19 | 3.742e-16 |
| 2 | CELL DEVELOPMENT | 139 | 1426 | 1.037e-19 | 3.742e-16 |
| 3 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 94 | 788 | 4.632e-19 | 5.388e-16 |
| 4 | CIRCULATORY SYSTEM DEVELOPMENT | 94 | 788 | 4.632e-19 | 5.388e-16 |
| 5 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 152 | 1672 | 1.088e-18 | 1.013e-15 |
| 6 | REGULATION OF CELL DEVELOPMENT | 94 | 836 | 2.391e-17 | 1.854e-14 |
| 7 | REGULATION OF CELL DIFFERENTIATION | 137 | 1492 | 3.389e-17 | 2.253e-14 |
| 8 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 122 | 1275 | 1.091e-16 | 6.346e-14 |
| 9 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 103 | 1021 | 1.119e-15 | 5.207e-13 |
| 10 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 111 | 1142 | 1.064e-15 | 5.207e-13 |
| 11 | CELLULAR COMPONENT MORPHOGENESIS | 94 | 900 | 2.67e-15 | 1.13e-12 |
| 12 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 82 | 750 | 1.493e-14 | 5.789e-12 |
| 13 | NEURON DIFFERENTIATION | 90 | 874 | 2.559e-14 | 9.158e-12 |
| 14 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 64 | 513 | 3.473e-14 | 1.154e-11 |
| 15 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 89 | 872 | 6.05e-14 | 1.877e-11 |
| 16 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 82 | 771 | 6.886e-14 | 2.002e-11 |
| 17 | VASCULATURE DEVELOPMENT | 60 | 469 | 7.427e-14 | 2.033e-11 |
| 18 | TISSUE DEVELOPMENT | 128 | 1518 | 2.677e-13 | 6.921e-11 |
| 19 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 120 | 1395 | 4.337e-13 | 1.062e-10 |
| 20 | RESPONSE TO ENDOGENOUS STIMULUS | 123 | 1450 | 5.597e-13 | 1.302e-10 |
| 21 | BLOOD VESSEL MORPHOGENESIS | 50 | 364 | 6.396e-13 | 1.353e-10 |
| 22 | HEART DEVELOPMENT | 58 | 466 | 6.367e-13 | 1.353e-10 |
| 23 | LOCOMOTION | 102 | 1114 | 7.556e-13 | 1.529e-10 |
| 24 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 91 | 957 | 1.832e-12 | 3.279e-10 |
| 25 | REGULATION OF DEVELOPMENTAL GROWTH | 43 | 289 | 1.826e-12 | 3.279e-10 |
| 26 | NEURON PROJECTION DEVELOPMENT | 63 | 545 | 1.75e-12 | 3.279e-10 |
| 27 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 82 | 823 | 2.275e-12 | 3.92e-10 |
| 28 | ORGAN MORPHOGENESIS | 83 | 841 | 2.731e-12 | 4.538e-10 |
| 29 | HEAD DEVELOPMENT | 74 | 709 | 3.121e-12 | 5.007e-10 |
| 30 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 72 | 689 | 5.892e-12 | 9.138e-10 |
| 31 | TUBE DEVELOPMENT | 62 | 552 | 9.147e-12 | 1.373e-09 |
| 32 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 79 | 801 | 9.855e-12 | 1.433e-09 |
| 33 | MESENCHYME DEVELOPMENT | 33 | 190 | 1.028e-11 | 1.45e-09 |
| 34 | MUSCLE STRUCTURE DEVELOPMENT | 53 | 432 | 1.13e-11 | 1.546e-09 |
| 35 | POSITIVE REGULATION OF LOCOMOTION | 52 | 420 | 1.258e-11 | 1.672e-09 |
| 36 | NEURON DEVELOPMENT | 71 | 687 | 1.396e-11 | 1.804e-09 |
| 37 | REGULATION OF NEURON DIFFERENTIATION | 61 | 554 | 3.107e-11 | 3.907e-09 |
| 38 | CELL PROJECTION ORGANIZATION | 83 | 902 | 9.811e-11 | 1.201e-08 |
| 39 | POSITIVE REGULATION OF CELL DEVELOPMENT | 54 | 472 | 1.051e-10 | 1.254e-08 |
| 40 | MESENCHYMAL CELL DIFFERENTIATION | 26 | 134 | 1.295e-10 | 1.506e-08 |
| 41 | RESPONSE TO GROWTH FACTOR | 54 | 475 | 1.333e-10 | 1.513e-08 |
| 42 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 89 | 1008 | 1.695e-10 | 1.877e-08 |
| 43 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 63 | 609 | 2e-10 | 2.165e-08 |
| 44 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 127 | 1656 | 2.135e-10 | 2.18e-08 |
| 45 | HEART MORPHOGENESIS | 33 | 212 | 2.109e-10 | 2.18e-08 |
| 46 | REGULATION OF CELL MORPHOGENESIS | 59 | 552 | 2.156e-10 | 2.18e-08 |
| 47 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 134 | 1784 | 2.499e-10 | 2.474e-08 |
| 48 | REGULATION OF CELL DEATH | 116 | 1472 | 2.914e-10 | 2.825e-08 |
| 49 | PROTEIN PHOSPHORYLATION | 84 | 944 | 3.951e-10 | 3.751e-08 |
| 50 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 50 | 437 | 5.231e-10 | 4.868e-08 |
| 51 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 143 | 1977 | 7.564e-10 | 6.901e-08 |
| 52 | STEM CELL DIFFERENTIATION | 30 | 190 | 9.841e-10 | 8.806e-08 |
| 53 | TISSUE MORPHOGENESIS | 56 | 533 | 1.189e-09 | 1.044e-07 |
| 54 | REGULATION OF MEMBRANE POTENTIAL | 42 | 343 | 1.701e-09 | 1.466e-07 |
| 55 | INTRACELLULAR SIGNAL TRANSDUCTION | 119 | 1572 | 1.89e-09 | 1.599e-07 |
| 56 | REGULATION OF GROWTH | 62 | 633 | 2.443e-09 | 2.016e-07 |
| 57 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 134 | 1848 | 2.47e-09 | 2.016e-07 |
| 58 | BEHAVIOR | 54 | 516 | 2.726e-09 | 2.15e-07 |
| 59 | BIOLOGICAL ADHESION | 87 | 1032 | 2.692e-09 | 2.15e-07 |
| 60 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 121 | 1618 | 2.795e-09 | 2.168e-07 |
| 61 | TUBE MORPHOGENESIS | 40 | 323 | 2.998e-09 | 2.287e-07 |
| 62 | NEGATIVE REGULATION OF CELL DEATH | 77 | 872 | 3.149e-09 | 2.363e-07 |
| 63 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 41 | 337 | 3.221e-09 | 2.379e-07 |
| 64 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 131 | 1805 | 3.648e-09 | 2.652e-07 |
| 65 | REGULATION OF SYSTEM PROCESS | 53 | 507 | 3.985e-09 | 2.809e-07 |
| 66 | MUSCLE CELL DIFFERENTIATION | 33 | 237 | 3.935e-09 | 2.809e-07 |
| 67 | REGULATION OF CELL ADHESION | 61 | 629 | 4.813e-09 | 3.343e-07 |
| 68 | POSITIVE REGULATION OF CELL COMMUNICATION | 115 | 1532 | 5.944e-09 | 3.951e-07 |
| 69 | SKELETAL SYSTEM DEVELOPMENT | 49 | 455 | 5.92e-09 | 3.951e-07 |
| 70 | REGULATION OF CELL PROLIFERATION | 113 | 1496 | 5.809e-09 | 3.951e-07 |
| 71 | REGULATION OF CELL PROJECTION ORGANIZATION | 56 | 558 | 6.269e-09 | 4.108e-07 |
| 72 | ANGIOGENESIS | 37 | 293 | 6.912e-09 | 4.467e-07 |
| 73 | ACTIN FILAMENT BASED PROCESS | 48 | 450 | 1.152e-08 | 7.342e-07 |
| 74 | UROGENITAL SYSTEM DEVELOPMENT | 37 | 299 | 1.197e-08 | 7.461e-07 |
| 75 | REGULATION OF TRANSPORT | 129 | 1804 | 1.203e-08 | 7.461e-07 |
| 76 | CELL MOTILITY | 73 | 835 | 1.256e-08 | 7.591e-07 |
| 77 | LOCALIZATION OF CELL | 73 | 835 | 1.256e-08 | 7.591e-07 |
| 78 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 104 | 1360 | 1.345e-08 | 8.024e-07 |
| 79 | REGULATION OF STEM CELL DIFFERENTIATION | 21 | 113 | 1.723e-08 | 1.015e-06 |
| 80 | PHOSPHORYLATION | 96 | 1228 | 1.776e-08 | 1.033e-06 |
| 81 | EXTRACELLULAR STRUCTURE ORGANIZATION | 37 | 304 | 1.867e-08 | 1.073e-06 |
| 82 | MORPHOGENESIS OF AN EPITHELIUM | 44 | 400 | 1.9e-08 | 1.078e-06 |
| 83 | REGULATION OF PROTEIN MODIFICATION PROCESS | 123 | 1710 | 1.993e-08 | 1.117e-06 |
| 84 | EPITHELIUM DEVELOPMENT | 79 | 945 | 2.198e-08 | 1.208e-06 |
| 85 | POSITIVE REGULATION OF CELL PROLIFERATION | 71 | 814 | 2.206e-08 | 1.208e-06 |
| 86 | NEURON PROJECTION GUIDANCE | 29 | 205 | 2.401e-08 | 1.299e-06 |
| 87 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 81 | 983 | 2.714e-08 | 1.451e-06 |
| 88 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 84 | 1036 | 3.038e-08 | 1.588e-06 |
| 89 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 84 | 1036 | 3.038e-08 | 1.588e-06 |
| 90 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 82 | 1004 | 3.269e-08 | 1.69e-06 |
| 91 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 44 | 408 | 3.417e-08 | 1.747e-06 |
| 92 | GROWTH | 44 | 410 | 3.946e-08 | 1.996e-06 |
| 93 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 50 | 498 | 4.074e-08 | 2.038e-06 |
| 94 | EMBRYO DEVELOPMENT | 75 | 894 | 4.419e-08 | 2.187e-06 |
| 95 | SINGLE ORGANISM BEHAVIOR | 42 | 384 | 4.704e-08 | 2.304e-06 |
| 96 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 133 | 1929 | 6.124e-08 | 2.968e-06 |
| 97 | NEURON PROJECTION MORPHOGENESIS | 43 | 402 | 6.188e-08 | 2.968e-06 |
| 98 | REGULATION OF OSSIFICATION | 26 | 178 | 6.56e-08 | 3.115e-06 |
| 99 | REGULATION OF POSTSYNAPTIC MEMBRANE POTENTIAL | 14 | 55 | 6.831e-08 | 3.211e-06 |
| 100 | RESPONSE TO LIPID | 74 | 888 | 7.156e-08 | 3.302e-06 |
| 101 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 25 | 167 | 7.169e-08 | 3.302e-06 |
| 102 | SENSORY ORGAN DEVELOPMENT | 49 | 493 | 7.633e-08 | 3.482e-06 |
| 103 | TAXIS | 47 | 464 | 8.001e-08 | 3.58e-06 |
| 104 | EMBRYONIC MORPHOGENESIS | 52 | 539 | 7.986e-08 | 3.58e-06 |
| 105 | OUTFLOW TRACT MORPHOGENESIS | 14 | 56 | 8.745e-08 | 3.875e-06 |
| 106 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 34 | 285 | 1.145e-07 | 5.024e-06 |
| 107 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 40 | 368 | 1.162e-07 | 5.052e-06 |
| 108 | CARDIAC SEPTUM DEVELOPMENT | 17 | 85 | 1.289e-07 | 5.555e-06 |
| 109 | REGULATION OF CYTOSKELETON ORGANIZATION | 49 | 502 | 1.331e-07 | 5.682e-06 |
| 110 | POSITIVE REGULATION OF GENE EXPRESSION | 121 | 1733 | 1.365e-07 | 5.773e-06 |
| 111 | REGULATION OF CELL SUBSTRATE ADHESION | 25 | 173 | 1.447e-07 | 6.065e-06 |
| 112 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 63 | 724 | 1.54e-07 | 6.399e-06 |
| 113 | MUSCLE ORGAN DEVELOPMENT | 33 | 277 | 1.812e-07 | 7.462e-06 |
| 114 | DEVELOPMENTAL GROWTH | 37 | 333 | 1.992e-07 | 8.129e-06 |
| 115 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 88 | 1152 | 2.049e-07 | 8.289e-06 |
| 116 | NEGATIVE REGULATION OF CELL COMMUNICATION | 90 | 1192 | 2.468e-07 | 9.901e-06 |
| 117 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 123 | 1791 | 2.588e-07 | 1.029e-05 |
| 118 | CARDIAC CHAMBER DEVELOPMENT | 22 | 144 | 3.009e-07 | 1.187e-05 |
| 119 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 23 | 156 | 3.141e-07 | 1.228e-05 |
| 120 | SYNAPSE ORGANIZATION | 22 | 145 | 3.403e-07 | 1.32e-05 |
| 121 | APPENDAGE DEVELOPMENT | 24 | 169 | 3.556e-07 | 1.356e-05 |
| 122 | LIMB DEVELOPMENT | 24 | 169 | 3.556e-07 | 1.356e-05 |
| 123 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 29 | 232 | 3.633e-07 | 1.372e-05 |
| 124 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 100 | 1381 | 3.657e-07 | 1.372e-05 |
| 125 | REGULATION OF KINASE ACTIVITY | 65 | 776 | 3.85e-07 | 1.433e-05 |
| 126 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 18 | 103 | 4.691e-07 | 1.732e-05 |
| 127 | CELL CELL SIGNALING | 64 | 767 | 5.422e-07 | 1.987e-05 |
| 128 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 11 | 39 | 5.676e-07 | 2.047e-05 |
| 129 | NEGATIVE CHEMOTAXIS | 11 | 39 | 5.676e-07 | 2.047e-05 |
| 130 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 34 | 306 | 6.188e-07 | 2.212e-05 |
| 131 | POSITIVE REGULATION OF GROWTH | 29 | 238 | 6.227e-07 | 2.212e-05 |
| 132 | CYTOSKELETON ORGANIZATION | 68 | 838 | 6.545e-07 | 2.307e-05 |
| 133 | NEURAL CREST CELL DIFFERENTIATION | 15 | 75 | 7.039e-07 | 2.463e-05 |
| 134 | REGULATION OF BLOOD CIRCULATION | 33 | 295 | 7.749e-07 | 2.691e-05 |
| 135 | REGULATION OF TRANSFERASE ACTIVITY | 74 | 946 | 8.388e-07 | 2.891e-05 |
| 136 | IMMUNE SYSTEM DEVELOPMENT | 52 | 582 | 8.699e-07 | 2.976e-05 |
| 137 | REGULATION OF ORGAN MORPHOGENESIS | 29 | 242 | 8.815e-07 | 2.994e-05 |
| 138 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 14 | 67 | 9.425e-07 | 3.178e-05 |
| 139 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 34 | 312 | 9.685e-07 | 3.242e-05 |
| 140 | CELL PART MORPHOGENESIS | 55 | 633 | 1.008e-06 | 3.352e-05 |
| 141 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 72 | 917 | 1.036e-06 | 3.419e-05 |
| 142 | FOREBRAIN DEVELOPMENT | 37 | 357 | 1.11e-06 | 3.637e-05 |
| 143 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 58 | 684 | 1.121e-06 | 3.647e-05 |
| 144 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 63 | 767 | 1.138e-06 | 3.676e-05 |
| 145 | NEURON MIGRATION | 18 | 110 | 1.278e-06 | 4.101e-05 |
| 146 | DENDRITE DEVELOPMENT | 15 | 79 | 1.414e-06 | 4.446e-05 |
| 147 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 33 | 303 | 1.413e-06 | 4.446e-05 |
| 148 | MUSCLE TISSUE DEVELOPMENT | 31 | 275 | 1.413e-06 | 4.446e-05 |
| 149 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 84 | 1135 | 1.437e-06 | 4.488e-05 |
| 150 | NEURAL CREST CELL MIGRATION | 12 | 51 | 1.479e-06 | 4.587e-05 |
| 151 | RESPONSE TO HORMONE | 70 | 893 | 1.561e-06 | 4.811e-05 |
| 152 | REGULATION OF CELL MATRIX ADHESION | 16 | 90 | 1.593e-06 | 4.876e-05 |
| 153 | REGULATION OF EXTENT OF CELL GROWTH | 17 | 101 | 1.675e-06 | 5.094e-05 |
| 154 | MEMBRANE DEPOLARIZATION | 13 | 61 | 1.816e-06 | 5.487e-05 |
| 155 | REGULATION OF ORGANELLE ORGANIZATION | 86 | 1178 | 1.858e-06 | 5.578e-05 |
| 156 | REGULATION OF SYNAPSE ORGANIZATION | 18 | 113 | 1.911e-06 | 5.701e-05 |
| 157 | KIDNEY EPITHELIUM DEVELOPMENT | 19 | 125 | 2.065e-06 | 6.119e-05 |
| 158 | RETINA VASCULATURE DEVELOPMENT IN CAMERA TYPE EYE | 7 | 16 | 2.404e-06 | 7.08e-05 |
| 159 | CARDIAC CHAMBER MORPHOGENESIS | 17 | 104 | 2.545e-06 | 7.448e-05 |
| 160 | ACTION POTENTIAL | 16 | 94 | 2.892e-06 | 8.411e-05 |
| 161 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 60 | 740 | 3.108e-06 | 8.981e-05 |
| 162 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 15 | 84 | 3.169e-06 | 9.046e-05 |
| 163 | POSITIVE REGULATION OF OSSIFICATION | 15 | 84 | 3.169e-06 | 9.046e-05 |
| 164 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 18 | 117 | 3.194e-06 | 9.063e-05 |
| 165 | REGULATION OF EPITHELIAL CELL MIGRATION | 22 | 166 | 3.498e-06 | 9.864e-05 |
| 166 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 102 | 1492 | 3.782e-06 | 0.000106 |
| 167 | REGULATION OF HYDROLASE ACTIVITY | 93 | 1327 | 3.861e-06 | 0.0001076 |
| 168 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 19 | 131 | 4.215e-06 | 0.0001167 |
| 169 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 20 | 144 | 4.679e-06 | 0.0001288 |
| 170 | REGULATION OF AXON GUIDANCE | 10 | 39 | 4.831e-06 | 0.0001322 |
| 171 | CELLULAR RESPONSE TO LIPID | 42 | 457 | 4.89e-06 | 0.0001331 |
| 172 | REGULATION OF HOMEOSTATIC PROCESS | 41 | 447 | 6.64e-06 | 0.0001776 |
| 173 | MULTICELLULAR ORGANISMAL SIGNALING | 18 | 123 | 6.595e-06 | 0.0001776 |
| 174 | CARDIAC SEPTUM MORPHOGENESIS | 11 | 49 | 6.613e-06 | 0.0001776 |
| 175 | REGULATION OF CELLULAR RESPONSE TO STRESS | 56 | 691 | 6.742e-06 | 0.0001793 |
| 176 | STRIATED MUSCLE CELL DIFFERENTIATION | 22 | 173 | 6.914e-06 | 0.0001828 |
| 177 | REGULATION OF SYNAPSE ASSEMBLY | 14 | 79 | 7.364e-06 | 0.0001925 |
| 178 | CELL FATE COMMITMENT | 26 | 227 | 7.356e-06 | 0.0001925 |
| 179 | POSITIVE REGULATION OF KINASE ACTIVITY | 43 | 482 | 7.99e-06 | 0.0002054 |
| 180 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 102 | 1518 | 7.949e-06 | 0.0002054 |
| 181 | TELENCEPHALON DEVELOPMENT | 26 | 228 | 7.966e-06 | 0.0002054 |
| 182 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 11 | 50 | 8.146e-06 | 0.0002083 |
| 183 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 26 | 229 | 8.621e-06 | 0.0002192 |
| 184 | CELL CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 24 | 204 | 1.039e-05 | 0.0002627 |
| 185 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 29 | 274 | 1.056e-05 | 0.0002641 |
| 186 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 31 | 303 | 1.053e-05 | 0.0002641 |
| 187 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 20 | 153 | 1.183e-05 | 0.0002943 |
| 188 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 20 | 154 | 1.305e-05 | 0.0003212 |
| 189 | AMEBOIDAL TYPE CELL MIGRATION | 20 | 154 | 1.305e-05 | 0.0003212 |
| 190 | REGULATION OF CHEMOTAXIS | 22 | 180 | 1.312e-05 | 0.0003212 |
| 191 | REGULATION OF HEART CONTRACTION | 25 | 221 | 1.361e-05 | 0.0003314 |
| 192 | CONNECTIVE TISSUE DEVELOPMENT | 23 | 194 | 1.391e-05 | 0.0003371 |
| 193 | LOCOMOTORY BEHAVIOR | 22 | 181 | 1.433e-05 | 0.0003455 |
| 194 | REGULATION OF AXONOGENESIS | 21 | 168 | 1.451e-05 | 0.0003479 |
| 195 | REGULATION OF MAPK CASCADE | 53 | 660 | 1.534e-05 | 0.0003647 |
| 196 | EPITHELIAL CELL DIFFERENTIATION | 43 | 495 | 1.536e-05 | 0.0003647 |
| 197 | COGNITION | 27 | 251 | 1.574e-05 | 0.0003717 |
| 198 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 61 | 799 | 1.667e-05 | 0.0003918 |
| 199 | NEUROMUSCULAR JUNCTION DEVELOPMENT | 9 | 36 | 1.81e-05 | 0.000421 |
| 200 | SEMAPHORIN PLEXIN SIGNALING PATHWAY | 9 | 36 | 1.81e-05 | 0.000421 |
| 201 | MUSCLE SYSTEM PROCESS | 29 | 282 | 1.826e-05 | 0.0004226 |
| 202 | REGULATION OF TRANSPORTER ACTIVITY | 23 | 198 | 1.94e-05 | 0.0004469 |
| 203 | GLAND MORPHOGENESIS | 15 | 97 | 1.952e-05 | 0.0004474 |
| 204 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 18 | 133 | 1.982e-05 | 0.0004521 |
| 205 | REGULATION OF CELL GROWTH | 36 | 391 | 2.207e-05 | 0.0005009 |
| 206 | CELLULAR RESPONSE TO HORMONE STIMULUS | 46 | 552 | 2.246e-05 | 0.0005073 |
| 207 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 41 | 472 | 2.415e-05 | 0.0005428 |
| 208 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 46 | 554 | 2.457e-05 | 0.0005497 |
| 209 | SKELETAL SYSTEM MORPHOGENESIS | 23 | 201 | 2.473e-05 | 0.0005506 |
| 210 | COLUMNAR CUBOIDAL EPITHELIAL CELL DIFFERENTIATION | 16 | 111 | 2.57e-05 | 0.0005695 |
| 211 | ACTOMYOSIN STRUCTURE ORGANIZATION | 13 | 77 | 2.664e-05 | 0.0005875 |
| 212 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 45 | 541 | 2.878e-05 | 0.000626 |
| 213 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 16 | 112 | 2.879e-05 | 0.000626 |
| 214 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 45 | 541 | 2.878e-05 | 0.000626 |
| 215 | REGULATION OF ION TRANSPORT | 48 | 592 | 3.02e-05 | 0.0006536 |
| 216 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 22 | 190 | 3.071e-05 | 0.0006615 |
| 217 | NEGATIVE REGULATION OF GENE EXPRESSION | 98 | 1493 | 3.094e-05 | 0.0006634 |
| 218 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 27 | 262 | 3.383e-05 | 0.000722 |
| 219 | REGULATION OF CELL JUNCTION ASSEMBLY | 12 | 68 | 3.426e-05 | 0.0007278 |
| 220 | RESPONSE TO DRUG | 38 | 431 | 3.441e-05 | 0.0007278 |
| 221 | NEGATIVE REGULATION OF LOCOMOTION | 27 | 263 | 3.617e-05 | 0.0007615 |
| 222 | RESPONSE TO STEROID HORMONE | 42 | 497 | 3.669e-05 | 0.0007691 |
| 223 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 23 | 207 | 3.951e-05 | 0.0008243 |
| 224 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 15 | 103 | 4.037e-05 | 0.0008274 |
| 225 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 19 | 153 | 4.01e-05 | 0.0008274 |
| 226 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 18 | 140 | 3.991e-05 | 0.0008274 |
| 227 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 15 | 103 | 4.037e-05 | 0.0008274 |
| 228 | EYE DEVELOPMENT | 31 | 326 | 4.392e-05 | 0.0008964 |
| 229 | NEGATIVE REGULATION OF CELL ADHESION | 24 | 223 | 4.549e-05 | 0.0009202 |
| 230 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 15 | 104 | 4.53e-05 | 0.0009202 |
| 231 | EAR DEVELOPMENT | 22 | 195 | 4.578e-05 | 0.0009221 |
| 232 | METANEPHROS DEVELOPMENT | 13 | 81 | 4.633e-05 | 0.0009292 |
| 233 | NEGATIVE REGULATION OF PHOSPHORYLATION | 37 | 422 | 4.89e-05 | 0.0009723 |
| 234 | EMBRYONIC ORGAN DEVELOPMENT | 36 | 406 | 4.881e-05 | 0.0009723 |
| 235 | ACTIN FILAMENT BASED MOVEMENT | 14 | 93 | 4.982e-05 | 0.0009865 |
| 236 | RESPIRATORY SYSTEM DEVELOPMENT | 22 | 197 | 5.346e-05 | 0.001054 |
| 237 | RESPONSE TO BMP | 14 | 94 | 5.622e-05 | 0.001099 |
| 238 | CELLULAR RESPONSE TO BMP STIMULUS | 14 | 94 | 5.622e-05 | 0.001099 |
| 239 | REGULATION OF TRANSMEMBRANE TRANSPORT | 37 | 426 | 5.962e-05 | 0.001161 |
| 240 | NEGATIVE REGULATION OF CHEMOTAXIS | 10 | 51 | 6.036e-05 | 0.00117 |
| 241 | PEPTIDYL TYROSINE MODIFICATION | 21 | 186 | 6.688e-05 | 0.001291 |
| 242 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 11 | 62 | 6.87e-05 | 0.001321 |
| 243 | REGULATION OF ORGAN GROWTH | 12 | 73 | 7.081e-05 | 0.001356 |
| 244 | CARDIOCYTE DIFFERENTIATION | 14 | 96 | 7.121e-05 | 0.001358 |
| 245 | CARTILAGE DEVELOPMENT | 18 | 147 | 7.636e-05 | 0.00145 |
| 246 | PALATE DEVELOPMENT | 13 | 85 | 7.764e-05 | 0.001468 |
| 247 | REGULATION OF CYTOPLASMIC TRANSPORT | 40 | 481 | 7.936e-05 | 0.001495 |
| 248 | REGENERATION | 19 | 161 | 8.084e-05 | 0.001505 |
| 249 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 48 | 616 | 8.074e-05 | 0.001505 |
| 250 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 48 | 616 | 8.074e-05 | 0.001505 |
| 251 | ACTIN MEDIATED CELL CONTRACTION | 12 | 74 | 8.12e-05 | 0.001505 |
| 252 | DIGESTIVE SYSTEM DEVELOPMENT | 18 | 148 | 8.346e-05 | 0.001535 |
| 253 | POSITIVE REGULATION OF CELL GROWTH | 18 | 148 | 8.346e-05 | 0.001535 |
| 254 | REGULATION OF MORPHOGENESIS OF A BRANCHING STRUCTURE | 10 | 53 | 8.511e-05 | 0.00154 |
| 255 | HEART VALVE DEVELOPMENT | 8 | 34 | 8.49e-05 | 0.00154 |
| 256 | CAMP METABOLIC PROCESS | 8 | 34 | 8.49e-05 | 0.00154 |
| 257 | CELL GROWTH | 17 | 135 | 8.434e-05 | 0.00154 |
| 258 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 24 | 232 | 8.539e-05 | 0.00154 |
| 259 | MUSCLE CONTRACTION | 24 | 233 | 9.135e-05 | 0.001641 |
| 260 | CELL PROLIFERATION | 51 | 672 | 9.496e-05 | 0.001699 |
| 261 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 21 | 191 | 9.815e-05 | 0.00175 |
| 262 | AGING | 26 | 264 | 0.0001001 | 0.00177 |
| 263 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 27 | 279 | 9.971e-05 | 0.00177 |
| 264 | POSITIVE REGULATION OF CELL DEATH | 47 | 605 | 0.0001027 | 0.001809 |
| 265 | NEGATIVE REGULATION OF AXONOGENESIS | 11 | 65 | 0.0001074 | 0.001885 |
| 266 | RESPONSE TO OXYGEN LEVELS | 29 | 311 | 0.0001082 | 0.001893 |
| 267 | REGULATION OF CELLULAR LOCALIZATION | 84 | 1277 | 0.0001096 | 0.00191 |
| 268 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 17 | 138 | 0.0001111 | 0.001929 |
| 269 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 65 | 926 | 0.0001123 | 0.001943 |
| 270 | CELL CELL ADHESION | 47 | 608 | 0.0001154 | 0.001988 |
| 271 | OVULATION CYCLE | 15 | 113 | 0.0001193 | 0.002047 |
| 272 | SPROUTING ANGIOGENESIS | 9 | 45 | 0.0001197 | 0.002047 |
| 273 | CENTRAL NERVOUS SYSTEM NEURON DIFFERENTIATION | 19 | 166 | 0.000122 | 0.002079 |
| 274 | NEGATIVE REGULATION OF AXON GUIDANCE | 7 | 27 | 0.0001224 | 0.002079 |
| 275 | POST EMBRYONIC DEVELOPMENT | 13 | 89 | 0.0001258 | 0.002129 |
| 276 | REGULATION OF BINDING | 27 | 283 | 0.0001265 | 0.002133 |
| 277 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 62 | 876 | 0.0001286 | 0.002152 |
| 278 | HOMOPHILIC CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 18 | 153 | 0.0001284 | 0.002152 |
| 279 | REGULATION OF SENSORY PERCEPTION | 8 | 36 | 0.0001309 | 0.002176 |
| 280 | REGULATION OF SENSORY PERCEPTION OF PAIN | 8 | 36 | 0.0001309 | 0.002176 |
| 281 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 15 | 114 | 0.000132 | 0.002185 |
| 282 | EPITHELIAL TO MESENCHYMAL TRANSITION | 10 | 56 | 0.0001381 | 0.00227 |
| 283 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 10 | 56 | 0.0001381 | 0.00227 |
| 284 | GLAND DEVELOPMENT | 34 | 395 | 0.0001392 | 0.002281 |
| 285 | MESONEPHROS DEVELOPMENT | 13 | 90 | 0.0001413 | 0.002306 |
| 286 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 96 | 1517 | 0.0001452 | 0.002357 |
| 287 | POSITIVE REGULATION OF CELL CYCLE | 30 | 332 | 0.0001454 | 0.002357 |
| 288 | SEMAPHORIN PLEXIN SIGNALING PATHWAY INVOLVED IN NEURON PROJECTION GUIDANCE | 5 | 13 | 0.0001512 | 0.002443 |
| 289 | CARBOHYDRATE DERIVATIVE METABOLIC PROCESS | 71 | 1047 | 0.0001555 | 0.002503 |
| 290 | MELANOCYTE DIFFERENTIATION | 6 | 20 | 0.0001566 | 0.002512 |
| 291 | CYCLIC NUCLEOTIDE METABOLIC PROCESS | 10 | 57 | 0.000161 | 0.002565 |
| 292 | REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 10 | 57 | 0.000161 | 0.002565 |
| 293 | TUBE FORMATION | 16 | 129 | 0.0001616 | 0.002566 |
| 294 | RESPONSE TO ACID CHEMICAL | 29 | 319 | 0.000168 | 0.002659 |
| 295 | REGULATION OF GTPASE ACTIVITY | 50 | 673 | 0.0001854 | 0.002925 |
| 296 | NEGATIVE REGULATION OF CELL PROJECTION ORGANIZATION | 17 | 144 | 0.0001876 | 0.002949 |
| 297 | REGULATION OF CELLULAR COMPONENT SIZE | 30 | 337 | 0.0001889 | 0.002959 |
| 298 | HOMEOSTATIC PROCESS | 86 | 1337 | 0.0001922 | 0.003001 |
| 299 | COLLAGEN FIBRIL ORGANIZATION | 8 | 38 | 0.0001959 | 0.003008 |
| 300 | NEGATIVE REGULATION OF AXON EXTENSION | 8 | 38 | 0.0001959 | 0.003008 |
| 301 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 15 | 118 | 0.0001949 | 0.003008 |
| 302 | REGULATION OF CELL SIZE | 19 | 172 | 0.0001948 | 0.003008 |
| 303 | POSITIVE REGULATION OF ORGAN GROWTH | 8 | 38 | 0.0001959 | 0.003008 |
| 304 | CARDIAC MUSCLE CELL CONTRACTION | 7 | 29 | 0.0001993 | 0.003041 |
| 305 | NEUROBLAST PROLIFERATION | 7 | 29 | 0.0001993 | 0.003041 |
| 306 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 22 | 216 | 0.0002073 | 0.003152 |
| 307 | CARDIAC VENTRICLE DEVELOPMENT | 14 | 106 | 0.0002102 | 0.003186 |
| 308 | SYMPATHETIC NERVOUS SYSTEM DEVELOPMENT | 6 | 21 | 0.0002112 | 0.00319 |
| 309 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 36 | 437 | 0.0002135 | 0.003194 |
| 310 | NEURAL PRECURSOR CELL PROLIFERATION | 11 | 70 | 0.0002128 | 0.003194 |
| 311 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 34 | 404 | 0.0002131 | 0.003194 |
| 312 | VASCULOGENESIS | 10 | 59 | 0.0002164 | 0.003228 |
| 313 | REGULATION OF CELL CYCLE | 65 | 949 | 0.0002239 | 0.003328 |
| 314 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 5 | 14 | 0.0002267 | 0.003339 |
| 315 | MEMBRANE DEPOLARIZATION DURING CARDIAC MUSCLE CELL ACTION POTENTIAL | 5 | 14 | 0.0002267 | 0.003339 |
| 316 | POSITIVE REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 5 | 14 | 0.0002267 | 0.003339 |
| 317 | RESPONSE TO RETINOIC ACID | 14 | 107 | 0.0002323 | 0.00341 |
| 318 | REGULATION OF VASCULATURE DEVELOPMENT | 23 | 233 | 0.0002383 | 0.003487 |
| 319 | CELLULAR RESPONSE TO ACID CHEMICAL | 19 | 175 | 0.0002438 | 0.003544 |
| 320 | EMBRYONIC ORGAN MORPHOGENESIS | 26 | 279 | 0.0002432 | 0.003544 |
| 321 | POSITIVE REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 7 | 30 | 0.0002502 | 0.003583 |
| 322 | STEM CELL PROLIFERATION | 10 | 60 | 0.0002497 | 0.003583 |
| 323 | NEGATIVE REGULATION OF MUSCLE CELL APOPTOTIC PROCESS | 7 | 30 | 0.0002502 | 0.003583 |
| 324 | POSITIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 10 | 60 | 0.0002497 | 0.003583 |
| 325 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 7 | 30 | 0.0002502 | 0.003583 |
| 326 | REGIONALIZATION | 28 | 311 | 0.0002536 | 0.003619 |
| 327 | REPRODUCTIVE SYSTEM DEVELOPMENT | 34 | 408 | 0.000256 | 0.003642 |
| 328 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 15 | 121 | 0.0002577 | 0.003655 |
| 329 | POSITIVE REGULATION OF CELL ADHESION | 32 | 376 | 0.000265 | 0.003736 |
| 330 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 18 | 162 | 0.0002643 | 0.003736 |
| 331 | ADULT BEHAVIOR | 16 | 135 | 0.0002742 | 0.003855 |
| 332 | REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 9 | 50 | 0.0002783 | 0.003885 |
| 333 | POSITIVE REGULATION OF STEM CELL DIFFERENTIATION | 9 | 50 | 0.0002783 | 0.003885 |
| 334 | REGULATION OF RESPONSE TO STRESS | 92 | 1468 | 0.0002797 | 0.003885 |
| 335 | SOMATIC STEM CELL DIVISION | 6 | 22 | 0.0002797 | 0.003885 |
| 336 | DEVELOPMENTAL PIGMENTATION | 8 | 40 | 0.0002851 | 0.003938 |
| 337 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 95 | 1527 | 0.0002852 | 0.003938 |
| 338 | NEGATIVE REGULATION OF GROWTH | 23 | 236 | 0.000287 | 0.00395 |
| 339 | REPRODUCTION | 83 | 1297 | 0.0002932 | 0.004024 |
| 340 | FOREBRAIN CELL MIGRATION | 10 | 62 | 0.0003291 | 0.004491 |
| 341 | CARDIAC VENTRICLE MORPHOGENESIS | 10 | 62 | 0.0003291 | 0.004491 |
| 342 | LEUKOCYTE ACTIVATION | 34 | 414 | 0.0003347 | 0.004547 |
| 343 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 13 | 98 | 0.0003352 | 0.004547 |
| 344 | SENSORY ORGAN MORPHOGENESIS | 23 | 239 | 0.0003441 | 0.004654 |
| 345 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 71 | 1079 | 0.0003721 | 0.005019 |
| 346 | REGULATION OF CARTILAGE DEVELOPMENT | 10 | 63 | 0.0003762 | 0.005044 |
| 347 | NEUROEPITHELIAL CELL DIFFERENTIATION | 10 | 63 | 0.0003762 | 0.005044 |
| 348 | SALIVARY GLAND DEVELOPMENT | 7 | 32 | 0.0003834 | 0.005097 |
| 349 | ENDOCARDIAL CUSHION DEVELOPMENT | 7 | 32 | 0.0003834 | 0.005097 |
| 350 | REGULATION OF CENTROSOME DUPLICATION | 7 | 32 | 0.0003834 | 0.005097 |
| 351 | RESPONSE TO ABIOTIC STIMULUS | 68 | 1024 | 0.0003846 | 0.005099 |
| 352 | PALLIUM DEVELOPMENT | 17 | 153 | 0.0003873 | 0.005119 |
| 353 | REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 11 | 75 | 0.0003952 | 0.005209 |
| 354 | NEGATIVE REGULATION OF CELL PROLIFERATION | 47 | 643 | 0.0004114 | 0.005408 |
| 355 | POSITIVE REGULATION OF MAPK CASCADE | 37 | 470 | 0.0004235 | 0.005551 |
| 356 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 9 | 53 | 0.0004379 | 0.005708 |
| 357 | NEGATIVE REGULATION OF ION TRANSPORT | 15 | 127 | 0.000437 | 0.005708 |
| 358 | MULTI MULTICELLULAR ORGANISM PROCESS | 21 | 213 | 0.000444 | 0.00577 |
| 359 | NEGATIVE REGULATION OF STEM CELL DIFFERENTIATION | 8 | 43 | 0.0004788 | 0.006155 |
| 360 | NEGATIVE REGULATION OF CELL GROWTH | 18 | 170 | 0.0004766 | 0.006155 |
| 361 | CEREBRAL CORTEX CELL MIGRATION | 8 | 43 | 0.0004788 | 0.006155 |
| 362 | MUSCLE CELL DEVELOPMENT | 15 | 128 | 0.0004754 | 0.006155 |
| 363 | REGULATION OF BEHAVIOR | 10 | 65 | 0.000487 | 0.006192 |
| 364 | BONE DEVELOPMENT | 17 | 156 | 0.0004859 | 0.006192 |
| 365 | CELLULAR RESPONSE TO RETINOIC ACID | 10 | 65 | 0.000487 | 0.006192 |
| 366 | REGULATION OF DNA METABOLIC PROCESS | 29 | 340 | 0.0004866 | 0.006192 |
| 367 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 19 | 185 | 0.0004931 | 0.006235 |
| 368 | NEPHRON DEVELOPMENT | 14 | 115 | 0.0004928 | 0.006235 |
| 369 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 13 | 102 | 0.0004971 | 0.006268 |
| 370 | TRANSMISSION OF NERVE IMPULSE | 9 | 54 | 0.0005054 | 0.006356 |
| 371 | REGULATION OF METAL ION TRANSPORT | 28 | 325 | 0.0005157 | 0.006468 |
| 372 | NEGATIVE REGULATION OF TRANSPORT | 36 | 458 | 0.0005181 | 0.006481 |
| 373 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 23 | 247 | 0.000548 | 0.006836 |
| 374 | REGULATION OF MULTICELLULAR ORGANISM GROWTH | 10 | 66 | 0.0005519 | 0.006866 |
| 375 | RENAL TUBULE DEVELOPMENT | 11 | 78 | 0.0005573 | 0.006915 |
| 376 | DNA TEMPLATED TRANSCRIPTION INITIATION | 20 | 202 | 0.0005695 | 0.007048 |
| 377 | IN UTERO EMBRYONIC DEVELOPMENT | 27 | 311 | 0.0005724 | 0.007065 |
| 378 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA PRODUCTION | 6 | 25 | 0.0005934 | 0.007304 |
| 379 | REGULATION OF CYTOSOLIC CALCIUM ION CONCENTRATION | 20 | 203 | 0.0006065 | 0.007446 |
| 380 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 10 | 67 | 0.0006237 | 0.007638 |
| 381 | RESPONSE TO EXTERNAL STIMULUS | 108 | 1821 | 0.0006314 | 0.007691 |
| 382 | RESPONSE TO ALCOHOL | 30 | 362 | 0.0006311 | 0.007691 |
| 383 | MESODERM DEVELOPMENT | 14 | 118 | 0.0006404 | 0.00778 |
| 384 | REGULATION OF PROTEIN LOCALIZATION | 63 | 950 | 0.0006485 | 0.007858 |
| 385 | CEREBRAL CORTEX DEVELOPMENT | 13 | 105 | 0.0006582 | 0.007934 |
| 386 | REGULATION OF RESPONSE TO WOUNDING | 33 | 413 | 0.0006571 | 0.007934 |
| 387 | SEX DIFFERENTIATION | 24 | 266 | 0.0006599 | 0.007934 |
| 388 | REGULATION OF LIPID METABOLIC PROCESS | 25 | 282 | 0.0006659 | 0.007974 |
| 389 | RHYTHMIC PROCESS | 26 | 298 | 0.0006667 | 0.007974 |
| 390 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 36 | 465 | 0.0006836 | 0.008135 |
| 391 | RESPONSE TO MONOAMINE | 7 | 35 | 0.0006834 | 0.008135 |
| 392 | ORGAN GROWTH | 10 | 68 | 0.0007031 | 0.008325 |
| 393 | REGULATION OF NEUROLOGICAL SYSTEM PROCESS | 10 | 68 | 0.0007031 | 0.008325 |
| 394 | NEPHRON EPITHELIUM DEVELOPMENT | 12 | 93 | 0.0007227 | 0.008534 |
| 395 | REGULATION OF MACROPHAGE ACTIVATION | 6 | 26 | 0.0007431 | 0.008754 |
| 396 | MORPHOGENESIS OF EMBRYONIC EPITHELIUM | 15 | 134 | 0.0007722 | 0.009074 |
| 397 | REGULATION OF NEURON APOPTOTIC PROCESS | 19 | 192 | 0.0007784 | 0.009123 |
| 398 | NEURAL TUBE FORMATION | 12 | 94 | 0.0007961 | 0.009308 |
| 399 | CORTICAL CYTOSKELETON ORGANIZATION | 7 | 36 | 0.0008167 | 0.009524 |
| 400 | REGULATION OF CATION TRANSMEMBRANE TRANSPORT | 20 | 208 | 0.0008241 | 0.009586 |
| 401 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 36 | 470 | 0.0008288 | 0.009617 |
| 402 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 5 | 18 | 0.0008387 | 0.00966 |
| 403 | POLYOL CATABOLIC PROCESS | 5 | 18 | 0.0008387 | 0.00966 |
| 404 | PHARYNGEAL SYSTEM DEVELOPMENT | 5 | 18 | 0.0008387 | 0.00966 |
| 405 | REGULATION OF MAP KINASE ACTIVITY | 27 | 319 | 0.0008419 | 0.009673 |
| 406 | CARDIAC CONDUCTION | 11 | 82 | 0.0008565 | 0.009816 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | GROWTH FACTOR BINDING | 23 | 123 | 3.197e-09 | 2.97e-06 |
| 2 | PROTEIN KINASE ACTIVITY | 60 | 640 | 2.243e-08 | 1.042e-05 |
| 3 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 17 | 81 | 6.112e-08 | 1.893e-05 |
| 4 | CYTOSKELETAL PROTEIN BINDING | 69 | 819 | 1.339e-07 | 2.871e-05 |
| 5 | ENZYME BINDING | 121 | 1737 | 1.545e-07 | 2.871e-05 |
| 6 | KINASE ACTIVITY | 69 | 842 | 3.766e-07 | 5.831e-05 |
| 7 | PROTEIN COMPLEX BINDING | 74 | 935 | 5.39e-07 | 7.154e-05 |
| 8 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 35 | 328 | 1.101e-06 | 0.0001278 |
| 9 | CYTOKINE BINDING | 16 | 92 | 2.156e-06 | 0.0001989 |
| 10 | RECEPTOR BINDING | 102 | 1476 | 2.355e-06 | 0.0001989 |
| 11 | MACROMOLECULAR COMPLEX BINDING | 98 | 1399 | 2.115e-06 | 0.0001989 |
| 12 | MOLECULAR FUNCTION REGULATOR | 95 | 1353 | 2.783e-06 | 0.0002155 |
| 13 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 13 | 64 | 3.218e-06 | 0.00023 |
| 14 | ACTIN BINDING | 38 | 393 | 4.254e-06 | 0.0002823 |
| 15 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 74 | 992 | 4.747e-06 | 0.000294 |
| 16 | PROTEIN TYROSINE KINASE ACTIVITY | 22 | 176 | 9.141e-06 | 0.0005307 |
| 17 | INTEGRIN BINDING | 16 | 105 | 1.26e-05 | 0.0006884 |
| 18 | CHEMOREPELLENT ACTIVITY | 8 | 27 | 1.362e-05 | 0.0007031 |
| 19 | REGULATORY REGION NUCLEIC ACID BINDING | 62 | 818 | 1.77e-05 | 0.0008656 |
| 20 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 25 | 226 | 1.999e-05 | 0.0009286 |
| 21 | ZINC ION BINDING | 80 | 1155 | 2.882e-05 | 0.001217 |
| 22 | PROTEIN DOMAIN SPECIFIC BINDING | 50 | 624 | 2.82e-05 | 0.001217 |
| 23 | DOUBLE STRANDED DNA BINDING | 58 | 764 | 3.158e-05 | 0.001276 |
| 24 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 50 | 629 | 3.468e-05 | 0.001324 |
| 25 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 82 | 1199 | 3.564e-05 | 0.001324 |
| 26 | TRANSFORMING GROWTH FACTOR BETA BINDING | 6 | 16 | 3.76e-05 | 0.001343 |
| 27 | CELL ADHESION MOLECULE BINDING | 21 | 186 | 6.688e-05 | 0.002301 |
| 28 | MICROTUBULE BINDING | 22 | 201 | 7.236e-05 | 0.002401 |
| 29 | SULFUR COMPOUND BINDING | 24 | 234 | 9.768e-05 | 0.003129 |
| 30 | EXTRACELLULAR MATRIX STRUCTURAL CONSTITUENT | 12 | 76 | 0.000106 | 0.003282 |
| 31 | VOLTAGE GATED SODIUM CHANNEL ACTIVITY | 6 | 20 | 0.0001566 | 0.004693 |
| 32 | TUBULIN BINDING | 26 | 273 | 0.0001723 | 0.005001 |
| 33 | HEPARIN BINDING | 18 | 157 | 0.0001784 | 0.005022 |
| 34 | SEQUENCE SPECIFIC DNA BINDING | 70 | 1037 | 0.0001977 | 0.005249 |
| 35 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 19 | 172 | 0.0001948 | 0.005249 |
| 36 | KINASE BINDING | 46 | 606 | 0.0002058 | 0.00531 |
| 37 | VOLTAGE GATED ION CHANNEL ACTIVITY | 20 | 190 | 0.0002564 | 0.006437 |
| 38 | ENZYME REGULATOR ACTIVITY | 65 | 959 | 0.0002983 | 0.007021 |
| 39 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 36 | 445 | 0.0003023 | 0.007021 |
| 40 | CATION CHANNEL ACTIVITY | 27 | 298 | 0.0002933 | 0.007021 |
| 41 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 28 | 315 | 0.0003125 | 0.00708 |
| 42 | PROTEIN DIMERIZATION ACTIVITY | 75 | 1149 | 0.0003238 | 0.007162 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEURON PROJECTION | 86 | 942 | 6.555e-11 | 2.052e-08 |
| 2 | CELL PROJECTION | 136 | 1786 | 7.026e-11 | 2.052e-08 |
| 3 | NEURON PART | 99 | 1265 | 9.815e-09 | 1.496e-06 |
| 4 | POSTSYNAPSE | 43 | 378 | 1.025e-08 | 1.496e-06 |
| 5 | SYNAPSE | 67 | 754 | 2.745e-08 | 3.206e-06 |
| 6 | SYNAPSE PART | 55 | 610 | 3.135e-07 | 3.051e-05 |
| 7 | PROTEINACEOUS EXTRACELLULAR MATRIX | 38 | 356 | 3.811e-07 | 3.18e-05 |
| 8 | SOMATODENDRITIC COMPARTMENT | 57 | 650 | 4.749e-07 | 3.467e-05 |
| 9 | EXTRACELLULAR MATRIX | 42 | 426 | 8.069e-07 | 5.236e-05 |
| 10 | CELL JUNCTION | 85 | 1151 | 1.352e-06 | 7.896e-05 |
| 11 | EXCITATORY SYNAPSE | 25 | 197 | 1.735e-06 | 9.211e-05 |
| 12 | CYTOSKELETON | 128 | 1967 | 2.519e-06 | 0.0001226 |
| 13 | AXON | 39 | 418 | 7.455e-06 | 0.000311 |
| 14 | RECEPTOR COMPLEX | 33 | 327 | 7.38e-06 | 0.000311 |
| 15 | CELL PROJECTION PART | 70 | 946 | 1.133e-05 | 0.0004411 |
| 16 | VOLTAGE GATED SODIUM CHANNEL COMPLEX | 6 | 14 | 1.52e-05 | 0.0005549 |
| 17 | NEURON SPINE | 17 | 121 | 2.043e-05 | 0.0007018 |
| 18 | EXTRACELLULAR MATRIX COMPONENT | 17 | 125 | 3.138e-05 | 0.001018 |
| 19 | SODIUM CHANNEL COMPLEX | 6 | 17 | 5.596e-05 | 0.00172 |
| 20 | SITE OF POLARIZED GROWTH | 18 | 149 | 9.113e-05 | 0.002534 |
| 21 | DENDRITE | 38 | 451 | 9.051e-05 | 0.002534 |
| 22 | INTRACELLULAR VESICLE | 83 | 1259 | 0.000112 | 0.002974 |
| 23 | CELL BODY | 40 | 494 | 0.0001403 | 0.003563 |
| 24 | CELL SURFACE | 55 | 757 | 0.0001592 | 0.003832 |
| 25 | MEMBRANE MICRODOMAIN | 27 | 288 | 0.0001689 | 0.003832 |
| 26 | CYTOPLASMIC VESICLE PART | 46 | 601 | 0.0001706 | 0.003832 |
| 27 | ENDOPLASMIC RETICULUM LUMEN | 21 | 201 | 0.0002017 | 0.004362 |
| 28 | FILOPODIUM | 13 | 94 | 0.0002206 | 0.0046 |
| 29 | ACTIN CYTOSKELETON | 36 | 444 | 0.0002896 | 0.005833 |
| 30 | ACTOMYOSIN | 10 | 62 | 0.0003291 | 0.006407 |
| 31 | CELL CELL JUNCTION | 32 | 383 | 0.0003669 | 0.006439 |
| 32 | SARCOLEMMA | 15 | 125 | 0.000368 | 0.006439 |
| 33 | ACTIN BASED CELL PROJECTION | 19 | 181 | 0.0003749 | 0.006439 |
| 34 | PLASMA MEMBRANE RAFT | 12 | 86 | 0.0003505 | 0.006439 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
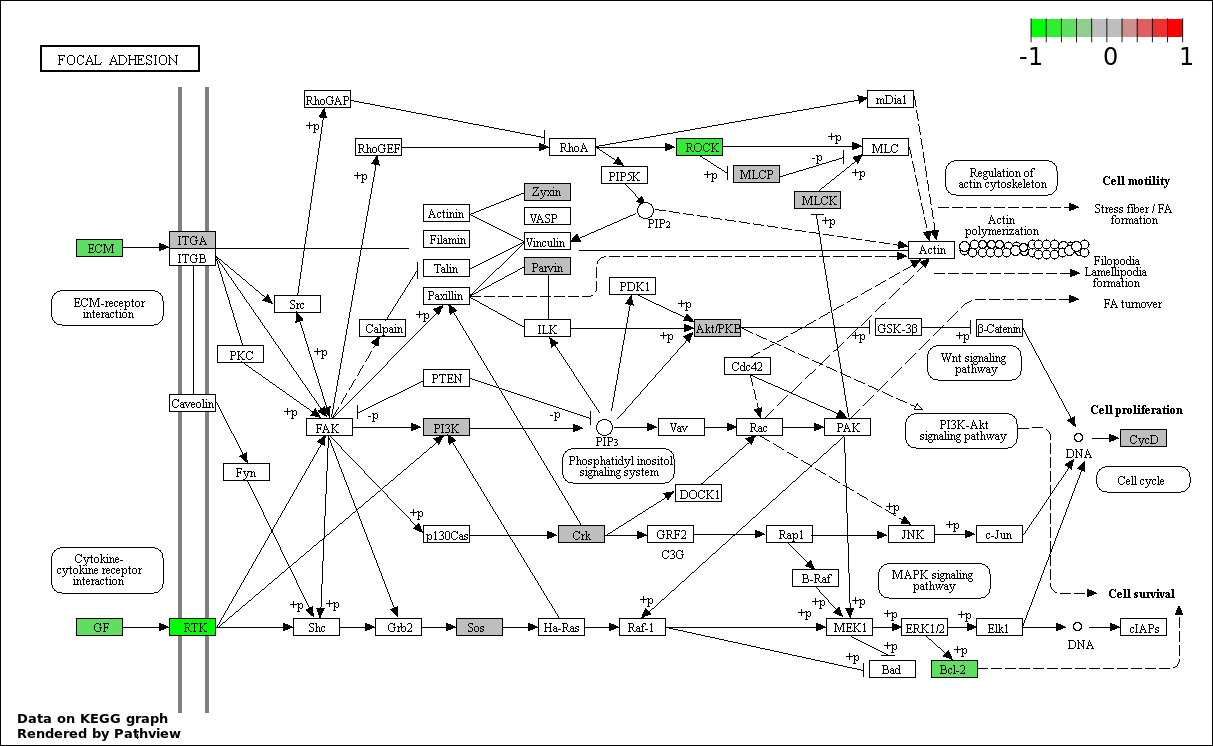
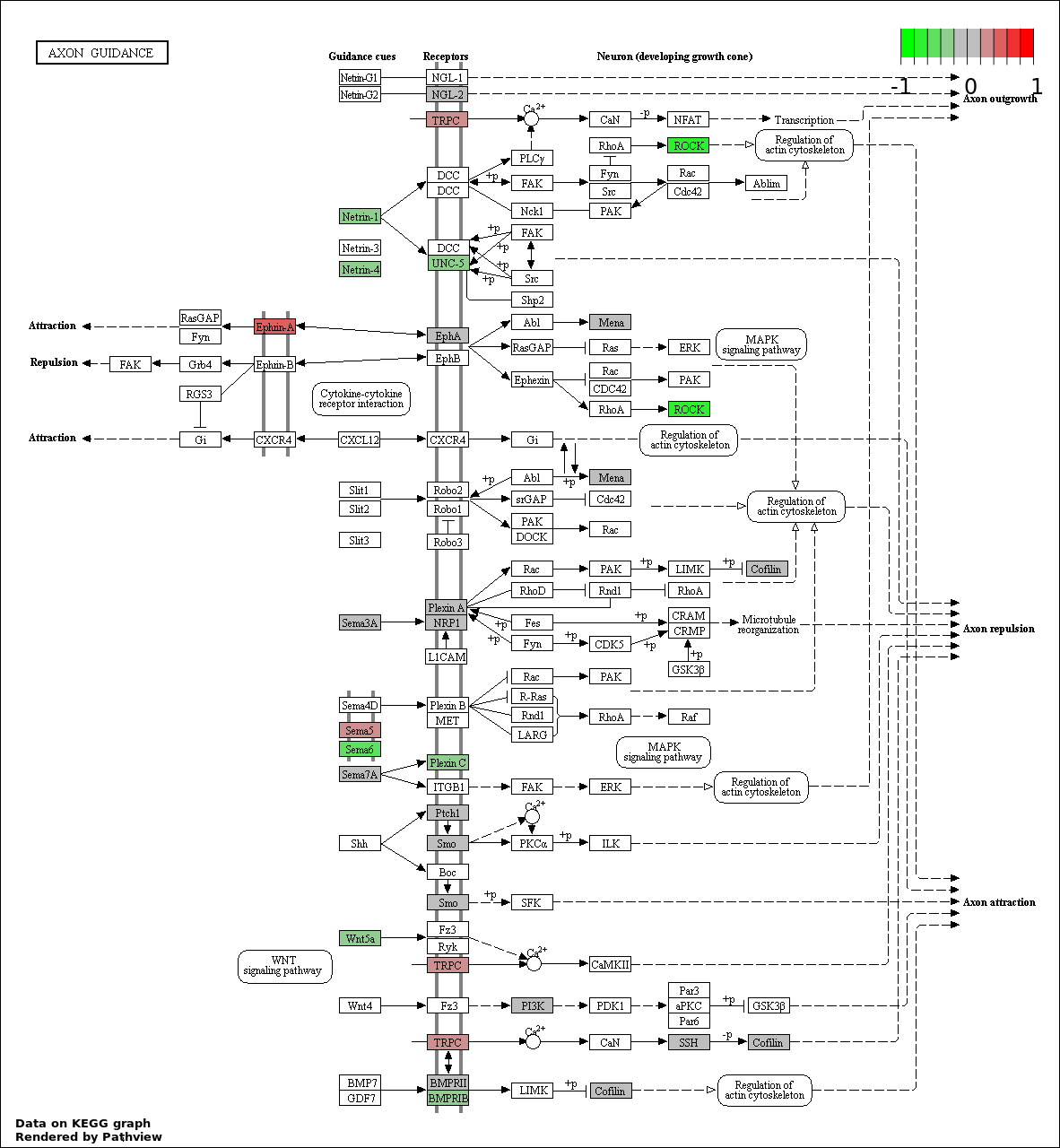
| 1 | hsa04510_Focal_adhesion | 27 | 200 | 1.926e-07 | 3.467e-05 | |
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 37 | 351 | 7.359e-07 | 6.623e-05 | |
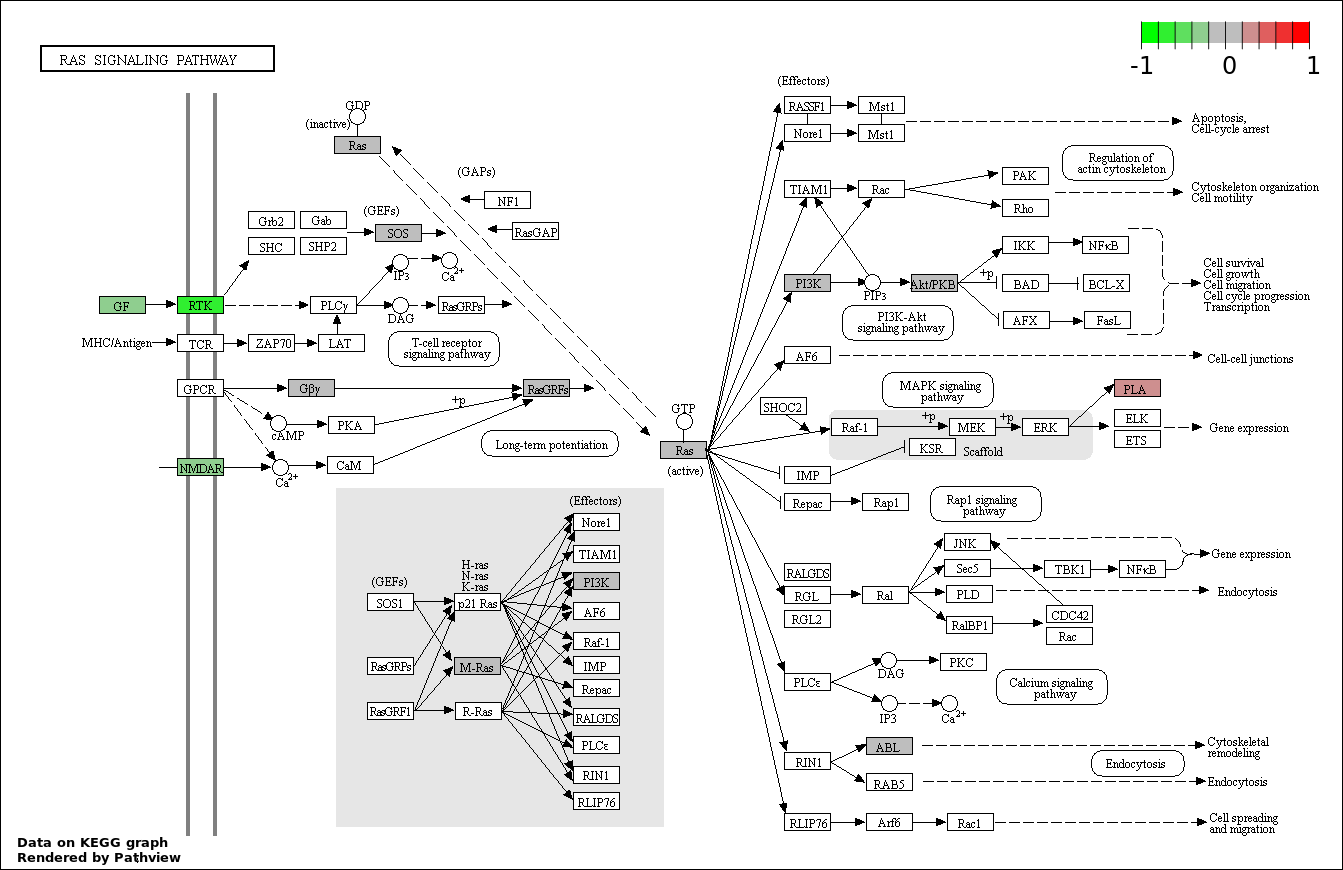
| 3 | hsa04014_Ras_signaling_pathway | 25 | 236 | 4.146e-05 | 0.002488 | |
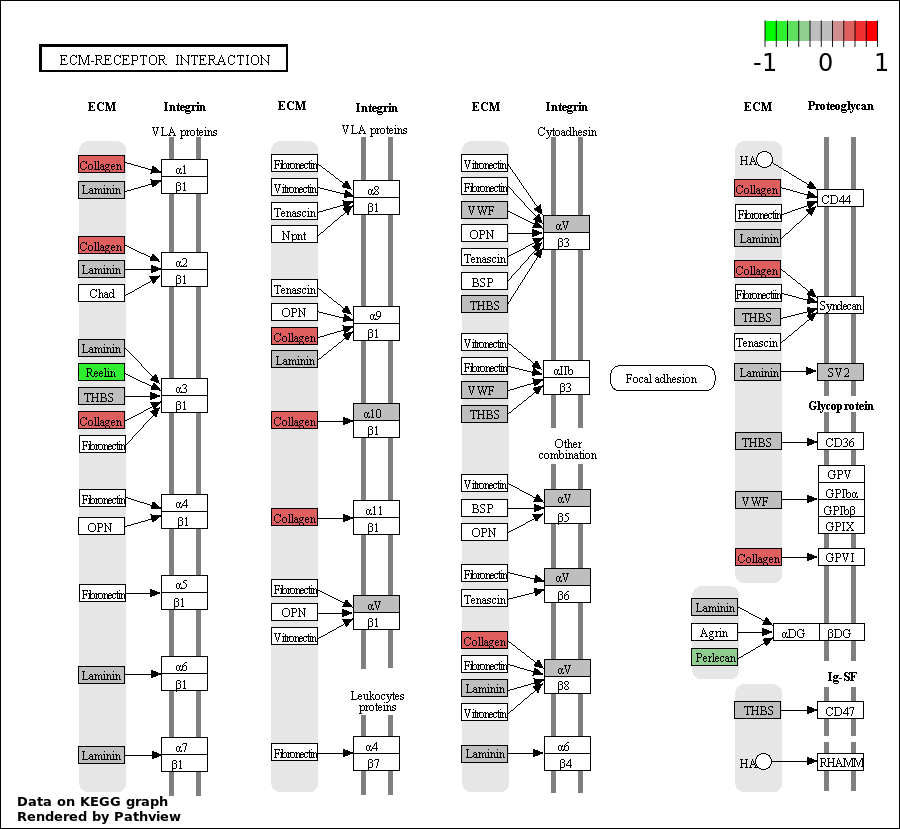
| 4 | hsa04512_ECM.receptor_interaction | 13 | 85 | 7.764e-05 | 0.003494 | |
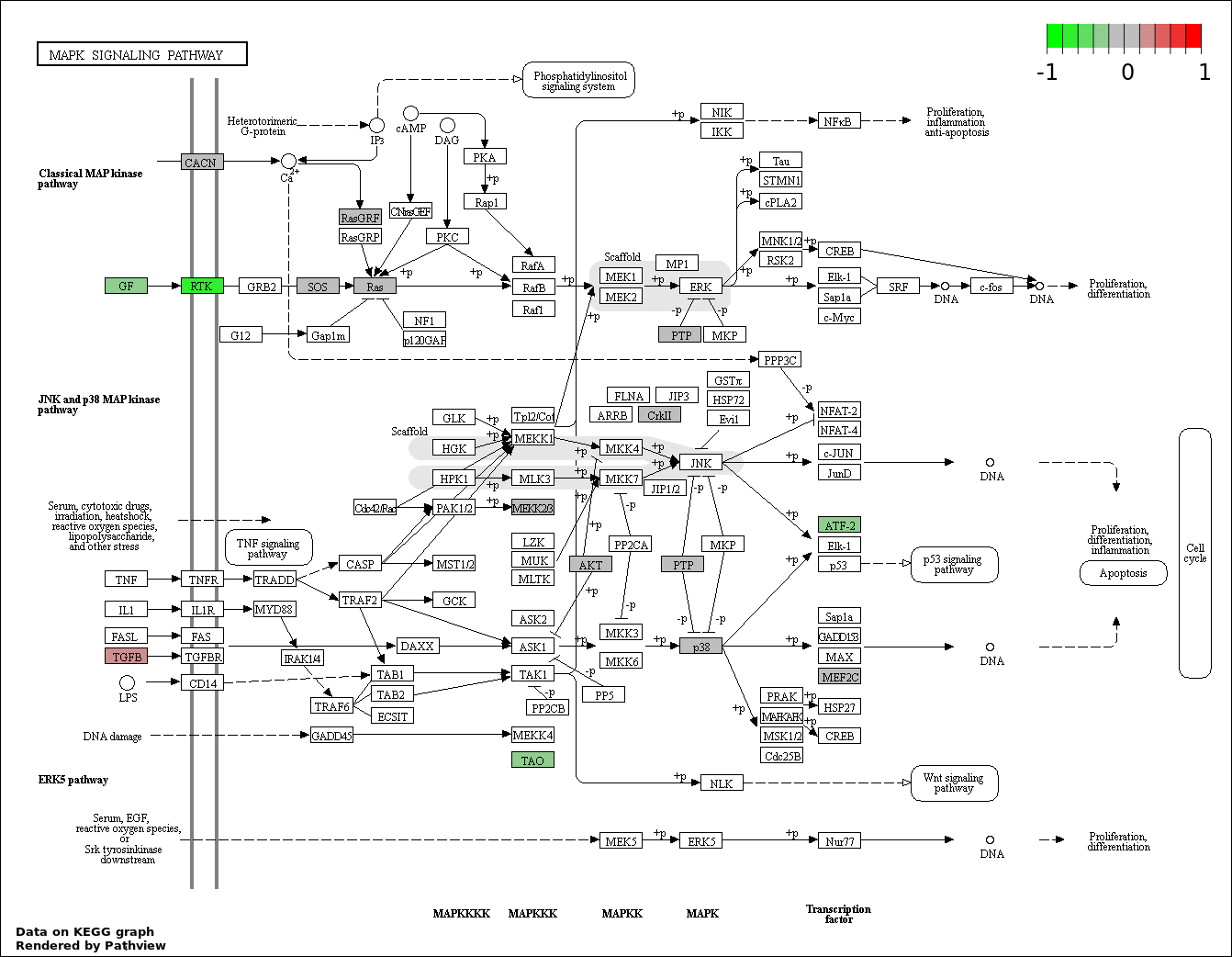
| 5 | hsa04010_MAPK_signaling_pathway | 26 | 268 | 0.0001279 | 0.004604 | |
| 6 | hsa04360_Axon_guidance | 16 | 130 | 0.0001769 | 0.005308 | |
| 7 | hsa04810_Regulation_of_actin_cytoskeleton | 21 | 214 | 0.0004725 | 0.01215 | |
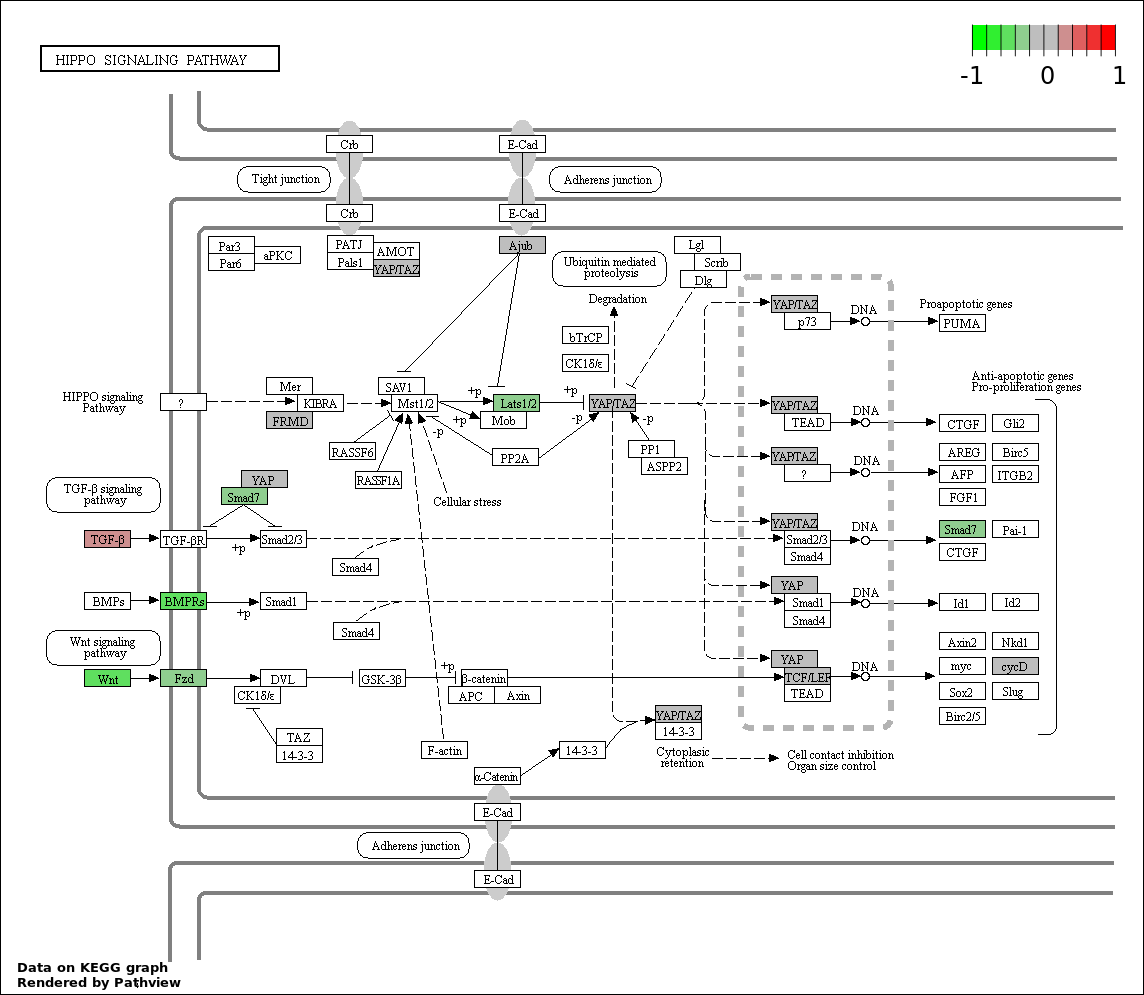
| 8 | hsa04390_Hippo_signaling_pathway | 16 | 154 | 0.001177 | 0.02649 | |
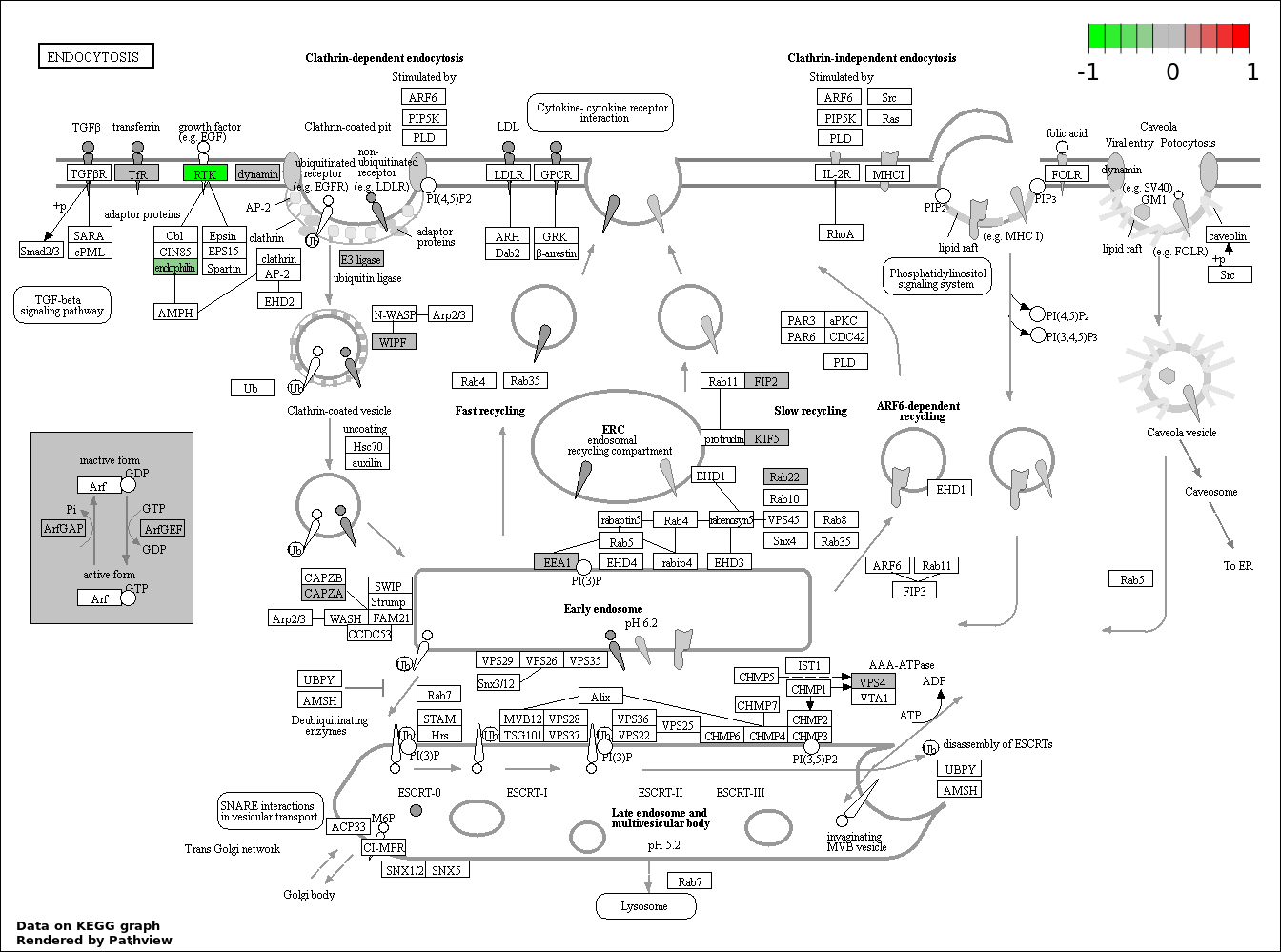
| 9 | hsa04144_Endocytosis | 19 | 203 | 0.00151 | 0.0302 | |
| 10 | hsa04020_Calcium_signaling_pathway | 17 | 177 | 0.001986 | 0.03574 | |
| 11 | hsa04514_Cell_adhesion_molecules_.CAMs. | 14 | 136 | 0.002538 | 0.04153 | |
| 12 | hsa04340_Hedgehog_signaling_pathway | 8 | 56 | 0.002847 | 0.04271 | |
| 13 | hsa04350_TGF.beta_signaling_pathway | 10 | 85 | 0.003894 | 0.05392 | |
| 14 | hsa04310_Wnt_signaling_pathway | 14 | 151 | 0.006486 | 0.08339 | |
| 15 | hsa00230_Purine_metabolism | 14 | 162 | 0.01172 | 0.1338 | |
| 16 | hsa04270_Vascular_smooth_muscle_contraction | 11 | 116 | 0.01268 | 0.1338 | |
| 17 | hsa04916_Melanogenesis | 10 | 101 | 0.01286 | 0.1338 | |
| 18 | hsa04914_Progesterone.mediated_oocyte_maturation | 9 | 87 | 0.01364 | 0.1338 | |
| 19 | hsa04520_Adherens_junction | 8 | 73 | 0.01412 | 0.1338 | |
| 20 | hsa04970_Salivary_secretion | 9 | 89 | 0.01566 | 0.1409 | |
| 21 | hsa04540_Gap_junction | 9 | 90 | 0.01675 | 0.1436 | |
| 22 | hsa04910_Insulin_signaling_pathway | 12 | 138 | 0.01794 | 0.1467 | |
| 23 | hsa03022_Basal_transcription_factors | 5 | 37 | 0.02155 | 0.1686 | |
| 24 | hsa04730_Long.term_depression | 7 | 70 | 0.03284 | 0.2463 | |
| 25 | hsa00562_Inositol_phosphate_metabolism | 6 | 57 | 0.03751 | 0.2701 | |
| 26 | hsa00670_One_carbon_pool_by_folate | 3 | 18 | 0.04158 | 0.2878 | |
| 27 | hsa00531_Glycosaminoglycan_degradation | 3 | 19 | 0.04782 | 0.3188 | |
| 28 | hsa04114_Oocyte_meiosis | 9 | 114 | 0.06203 | 0.3951 | |
| 29 | hsa04974_Protein_digestion_and_absorption | 7 | 81 | 0.06365 | 0.3951 | |
| 30 | hsa00051_Fructose_and_mannose_metabolism | 4 | 36 | 0.07066 | 0.4239 | |
| 31 | hsa04912_GnRH_signaling_pathway | 8 | 101 | 0.07461 | 0.4245 | |
| 32 | hsa04710_Circadian_rhythm_._mammal | 3 | 23 | 0.07695 | 0.4245 | |
| 33 | hsa04630_Jak.STAT_signaling_pathway | 11 | 155 | 0.07782 | 0.4245 | |
| 34 | hsa04115_p53_signaling_pathway | 6 | 69 | 0.08058 | 0.4266 | |
| 35 | hsa04722_Neurotrophin_signaling_pathway | 9 | 127 | 0.1045 | 0.5373 | |
| 36 | hsa04260_Cardiac_muscle_contraction | 6 | 77 | 0.1201 | 0.5798 | |
| 37 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 7 | 95 | 0.1219 | 0.5798 | |
| 38 | hsa04062_Chemokine_signaling_pathway | 12 | 189 | 0.1256 | 0.5798 | |
| 39 | hsa04070_Phosphatidylinositol_signaling_system | 6 | 78 | 0.1256 | 0.5798 | |
| 40 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 3 | 30 | 0.1416 | 0.6374 | |
| 41 | hsa04972_Pancreatic_secretion | 7 | 101 | 0.1531 | 0.6721 | |
| 42 | hsa04012_ErbB_signaling_pathway | 6 | 87 | 0.1803 | 0.7727 | |
| 43 | hsa04720_Long.term_potentiation | 5 | 70 | 0.1908 | 0.7744 | |
| 44 | hsa04150_mTOR_signaling_pathway | 4 | 52 | 0.1915 | 0.7744 | |
| 45 | hsa04210_Apoptosis | 6 | 89 | 0.1936 | 0.7744 | |
| 46 | hsa04976_Bile_secretion | 5 | 71 | 0.1984 | 0.7765 | |
| 47 | hsa04971_Gastric_acid_secretion | 5 | 74 | 0.222 | 0.8503 | |
| 48 | hsa04370_VEGF_signaling_pathway | 5 | 76 | 0.2382 | 0.8932 | |
| 49 | hsa04145_Phagosome | 9 | 156 | 0.2434 | 0.8941 | |
| 50 | hsa04664_Fc_epsilon_RI_signaling_pathway | 5 | 79 | 0.263 | 0.9468 | |
| 51 | hsa00760_Nicotinate_and_nicotinamide_metabolism | 2 | 24 | 0.2825 | 0.988 | |
| 52 | hsa04320_Dorso.ventral_axis_formation | 2 | 25 | 0.2989 | 0.988 | |
| 53 | hsa00280_Valine._leucine_and_isoleucine_degradation | 3 | 44 | 0.3019 | 0.988 | |
| 54 | hsa02010_ABC_transporters | 3 | 44 | 0.3019 | 0.988 | |
| 55 | hsa04973_Carbohydrate_digestion_and_absorption | 3 | 44 | 0.3019 | 0.988 | |
| 56 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 2 | 26 | 0.3153 | 0.9984 | |
| 57 | hsa00010_Glycolysis_._Gluconeogenesis | 4 | 65 | 0.3162 | 0.9984 | |
| 58 | hsa04975_Fat_digestion_and_absorption | 3 | 46 | 0.3261 | 1 | |
| 59 | hsa04660_T_cell_receptor_signaling_pathway | 6 | 108 | 0.3337 | 1 | |
| 60 | hsa04920_Adipocytokine_signaling_pathway | 4 | 68 | 0.3462 | 1 | |
| 61 | hsa00510_N.Glycan_biosynthesis | 3 | 49 | 0.3624 | 1 | |
| 62 | hsa04530_Tight_junction | 7 | 133 | 0.3627 | 1 | |
| 63 | hsa00020_Citrate_cycle_.TCA_cycle. | 2 | 30 | 0.3796 | 1 | |
| 64 | hsa00640_Propanoate_metabolism | 2 | 32 | 0.4108 | 1 | |
| 65 | hsa04662_B_cell_receptor_signaling_pathway | 4 | 75 | 0.4161 | 1 | |
| 66 | hsa04142_Lysosome | 6 | 121 | 0.4356 | 1 | |
| 67 | hsa00564_Glycerophospholipid_metabolism | 4 | 80 | 0.4651 | 1 | |
| 68 | hsa00565_Ether_lipid_metabolism | 2 | 36 | 0.4707 | 1 | |
| 69 | hsa03030_DNA_replication | 2 | 36 | 0.4707 | 1 | |
| 70 | hsa04110_Cell_cycle | 6 | 128 | 0.4896 | 1 | |
| 71 | hsa04380_Osteoclast_differentiation | 6 | 128 | 0.4896 | 1 | |
| 72 | hsa03015_mRNA_surveillance_pathway | 4 | 83 | 0.4938 | 1 | |
| 73 | hsa00600_Sphingolipid_metabolism | 2 | 40 | 0.5268 | 1 | |
| 74 | hsa04640_Hematopoietic_cell_lineage | 4 | 88 | 0.54 | 1 | |
| 75 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 2 | 42 | 0.5533 | 1 | |
| 76 | hsa00310_Lysine_degradation | 2 | 44 | 0.5788 | 1 | |
| 77 | hsa04962_Vasopressin.regulated_water_reabsorption | 2 | 44 | 0.5788 | 1 | |
| 78 | hsa04670_Leukocyte_transendothelial_migration | 5 | 117 | 0.5839 | 1 | |
| 79 | hsa03420_Nucleotide_excision_repair | 2 | 45 | 0.5911 | 1 | |
| 80 | hsa00514_Other_types_of_O.glycan_biosynthesis | 2 | 46 | 0.6031 | 1 | |
| 81 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 7 | 168 | 0.6049 | 1 | |
| 82 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 3 | 71 | 0.6052 | 1 | |
| 83 | hsa03018_RNA_degradation | 3 | 71 | 0.6052 | 1 | |
| 84 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.6149 | 1 | |
| 85 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 2 | 48 | 0.6264 | 1 | |
| 86 | hsa00240_Pyrimidine_metabolism | 4 | 99 | 0.6334 | 1 | |
| 87 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.6486 | 1 | |
| 88 | hsa00500_Starch_and_sucrose_metabolism | 2 | 54 | 0.6899 | 1 | |
| 89 | hsa04120_Ubiquitin_mediated_proteolysis | 5 | 139 | 0.7317 | 1 | |
| 90 | hsa04621_NOD.like_receptor_signaling_pathway | 2 | 59 | 0.7357 | 1 | |
| 91 | hsa04610_Complement_and_coagulation_cascades | 2 | 69 | 0.8102 | 1 | |
| 92 | hsa03320_PPAR_signaling_pathway | 2 | 70 | 0.8166 | 1 | |
| 93 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.8286 | 1 | |
| 94 | hsa00982_Drug_metabolism_._cytochrome_P450 | 2 | 73 | 0.8344 | 1 | |
| 95 | hsa04146_Peroxisome | 2 | 79 | 0.8654 | 1 | |
| 96 | hsa03013_RNA_transport | 4 | 152 | 0.9036 | 1 | |
| 97 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 2 | 136 | 0.9837 | 1 | |
| 98 | hsa04740_Olfactory_transduction | 3 | 388 | 1 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|