Browse ATF2 in pancancer
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00170 bZIP transcription factor |
||||||||||
| Function |
Transcriptional activator which regulates the transcription of various genes, including those involved in anti-apoptosis, cell growth, and DNA damage response. Dependent on its binding partner, binds to CRE (cAMP response element) consensus sequences (5'-TGACGTCA-3') or to AP-1 (activator protein 1) consensus sequences (5'-TGACTCA-3'). In the nucleus, contributes to global transcription and the DNA damage response, in addition to specific transcriptional activities that are related to cell development, proliferation and death. In the cytoplasm, interacts with and perturbs HK1- and VDAC1-containing complexes at the mitochondrial outer membrane, thereby impairing mitochondrial membrane potential, inducing mitochondrial leakage and promoting cell death. The phosphorylated form (mediated by ATM) plays a role in the DNA damage response and is involved in the ionizing radiation (IR)-induced S phase checkpoint control and in the recruitment of the MRN complex into the IR-induced foci (IRIF). Exhibits histone acetyltransferase (HAT) activity which specifically acetylates histones H2B and H4 in vitro. In concert with CUL3 and RBX1, promotes the degradation of KAT5 thereby attenuating its ability to acetylate and activate ATM. Can elicit oncogenic or tumor suppressor activities depending on the tissue or cell type. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000077 DNA damage checkpoint GO:0001101 response to acid chemical GO:0001819 positive regulation of cytokine production GO:0003007 heart morphogenesis GO:0003151 outflow tract morphogenesis GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006839 mitochondrial transport GO:0006970 response to osmotic stress GO:0007006 mitochondrial membrane organization GO:0007093 mitotic cell cycle checkpoint GO:0007346 regulation of mitotic cell cycle GO:0007507 heart development GO:0008637 apoptotic mitochondrial changes GO:0009414 response to water deprivation GO:0009415 response to water GO:0010035 response to inorganic substance GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0031570 DNA integrity checkpoint GO:0031573 intra-S DNA damage checkpoint GO:0032906 transforming growth factor beta2 production GO:0032909 regulation of transforming growth factor beta2 production GO:0032915 positive regulation of transforming growth factor beta2 production GO:0035794 positive regulation of mitochondrial membrane permeability GO:0042475 odontogenesis of dentin-containing tooth GO:0042476 odontogenesis GO:0043523 regulation of neuron apoptotic process GO:0043525 positive regulation of neuron apoptotic process GO:0043543 protein acylation GO:0044773 mitotic DNA damage checkpoint GO:0044774 mitotic DNA integrity checkpoint GO:0045444 fat cell differentiation GO:0045786 negative regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0046902 regulation of mitochondrial membrane permeability GO:0050673 epithelial cell proliferation GO:0050678 regulation of epithelial cell proliferation GO:0050680 negative regulation of epithelial cell proliferation GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051402 neuron apoptotic process GO:0060612 adipose tissue development GO:0061448 connective tissue development GO:0070997 neuron death GO:0071604 transforming growth factor beta production GO:0071634 regulation of transforming growth factor beta production GO:0071636 positive regulation of transforming growth factor beta production GO:0090559 regulation of membrane permeability GO:0097186 amelogenesis GO:1901214 regulation of neuron death GO:1901216 positive regulation of neuron death GO:1902108 regulation of mitochondrial membrane permeability involved in apoptotic process GO:1902110 positive regulation of mitochondrial membrane permeability involved in apoptotic process GO:1902686 mitochondrial outer membrane permeabilization involved in programmed cell death |
| Molecular Function |
GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000982 transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001077 transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0001085 RNA polymerase II transcription factor binding GO:0001102 RNA polymerase II activating transcription factor binding GO:0001158 enhancer sequence-specific DNA binding GO:0001228 transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding GO:0003682 chromatin binding GO:0003705 transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0008140 cAMP response element binding protein binding GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0033613 activating transcription factor binding GO:0034212 peptide N-acetyltransferase activity GO:0035326 enhancer binding GO:0035497 cAMP response element binding GO:0046982 protein heterodimerization activity GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0005741 mitochondrial outer membrane GO:0019867 outer membrane GO:0031968 organelle outer membrane GO:0035861 site of double-strand break |
| KEGG |
hsa04010 MAPK signaling pathway hsa04022 cGMP-PKG signaling pathway hsa04151 PI3K-Akt signaling pathway hsa04261 Adrenergic signaling in cardiomyocytes hsa04668 TNF signaling pathway hsa04728 Dopaminergic synapse hsa04911 Insulin secretion hsa04915 Estrogen signaling pathway hsa04918 Thyroid hormone synthesis hsa04922 Glucagon signaling pathway |
| Reactome |
R-HSA-166054: Activated TLR4 signalling R-HSA-450341: Activation of the AP-1 family of transcription factors R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-168256: Immune System R-HSA-168249: Innate Immune System R-HSA-450294: MAP kinase activation in TLR cascade R-HSA-450282: MAPK targets/ Nuclear events mediated by MAP kinases R-HSA-1592230: Mitochondrial biogenesis R-HSA-975871: MyD88 cascade initiated on plasma membrane R-HSA-975155: MyD88 dependent cascade initiated on endosome R-HSA-166166: MyD88-independent TLR3/TLR4 cascade R-HSA-166058: MyD88 R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-6796648: TP53 Regulates Transcription of DNA Repair Genes R-HSA-168180: TRAF6 Mediated Induction of proinflammatory cytokines R-HSA-975138: TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation R-HSA-937061: TRIF-mediated TLR3/TLR4 signaling R-HSA-168142: Toll Like Receptor 10 (TLR10) Cascade R-HSA-181438: Toll Like Receptor 2 (TLR2) Cascade R-HSA-168164: Toll Like Receptor 3 (TLR3) Cascade R-HSA-166016: Toll Like Receptor 4 (TLR4) Cascade R-HSA-168176: Toll Like Receptor 5 (TLR5) Cascade R-HSA-168181: Toll Like Receptor 7/8 (TLR7/8) Cascade R-HSA-168138: Toll Like Receptor 9 (TLR9) Cascade R-HSA-168179: Toll Like Receptor TLR1 R-HSA-168188: Toll Like Receptor TLR6 R-HSA-168898: Toll-Like Receptors Cascades R-HSA-3700989: Transcriptional Regulation by TP53 R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis |
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for ATF2. |
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
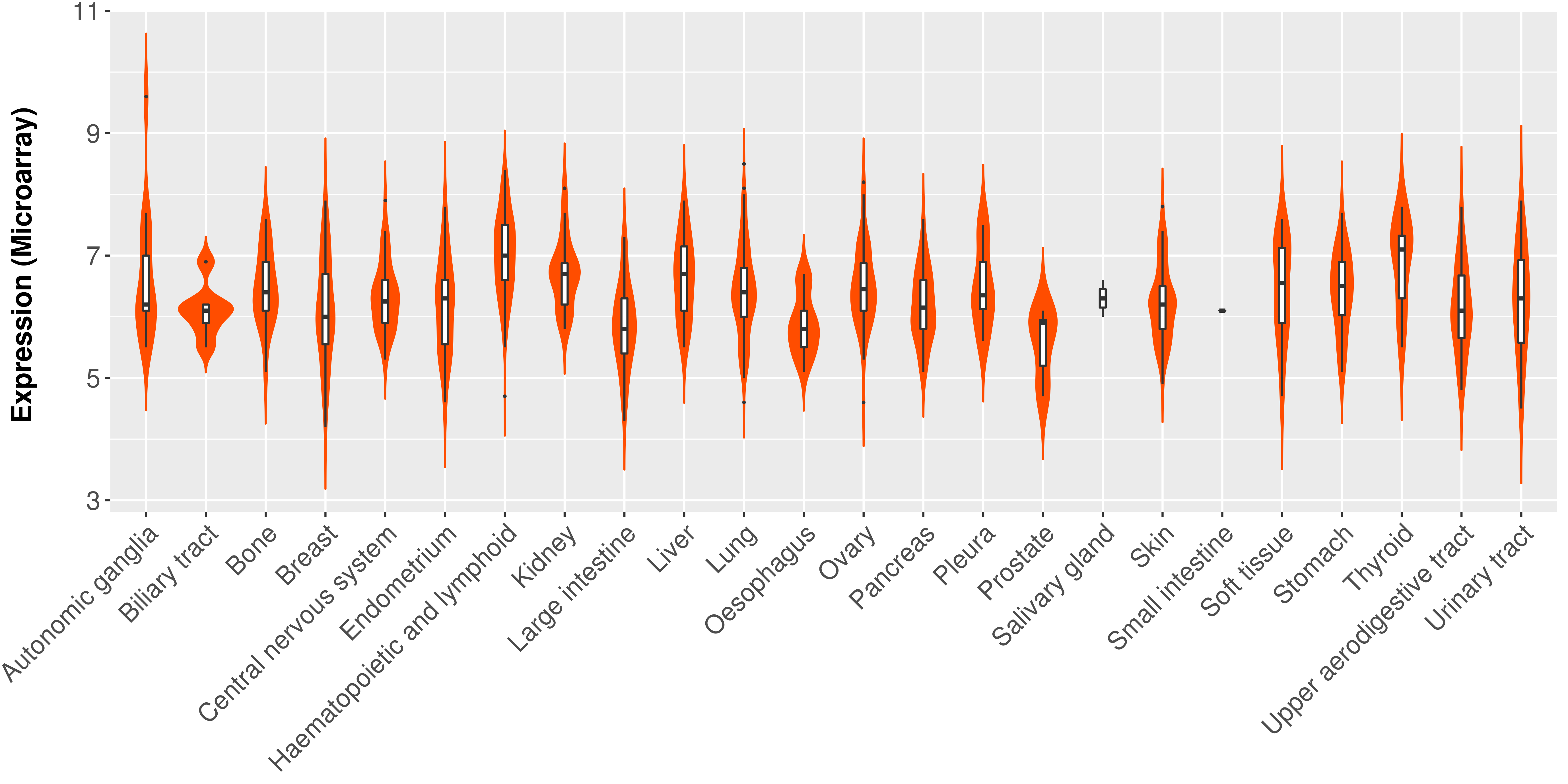
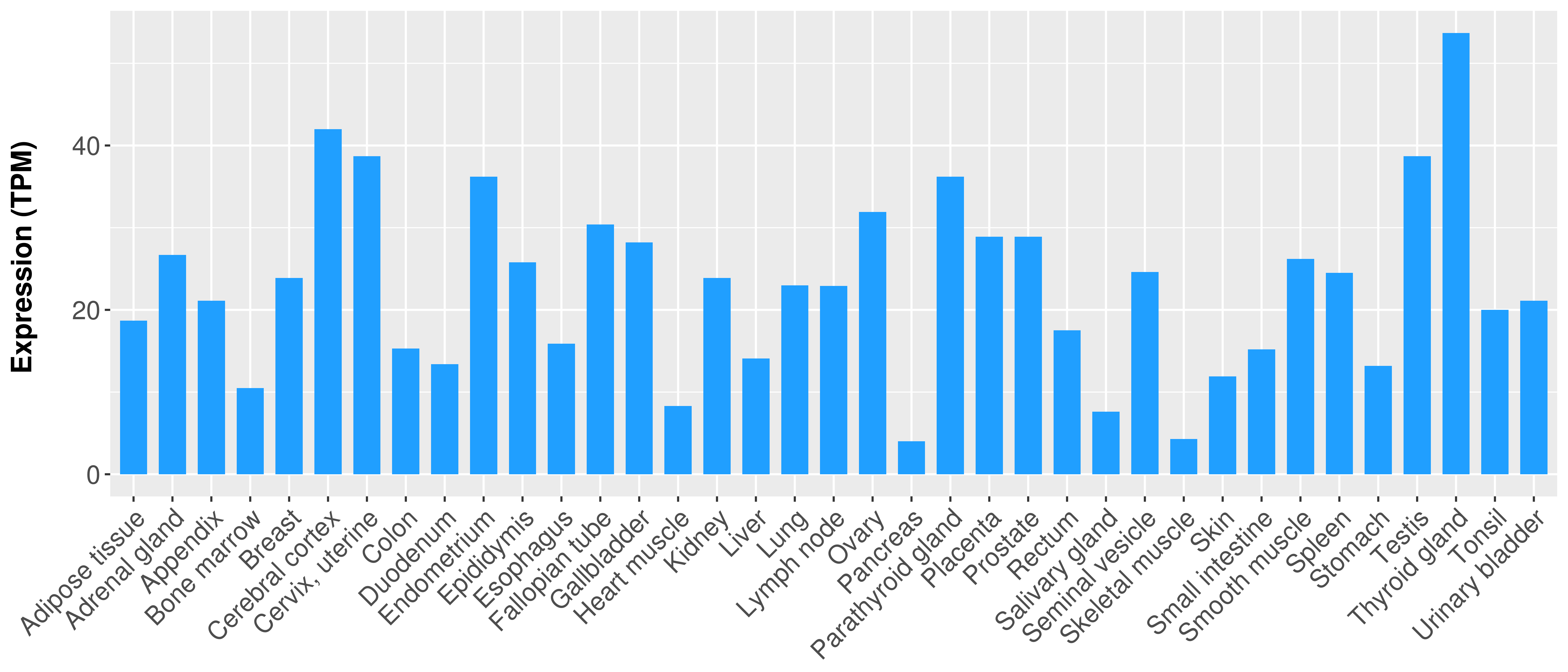
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
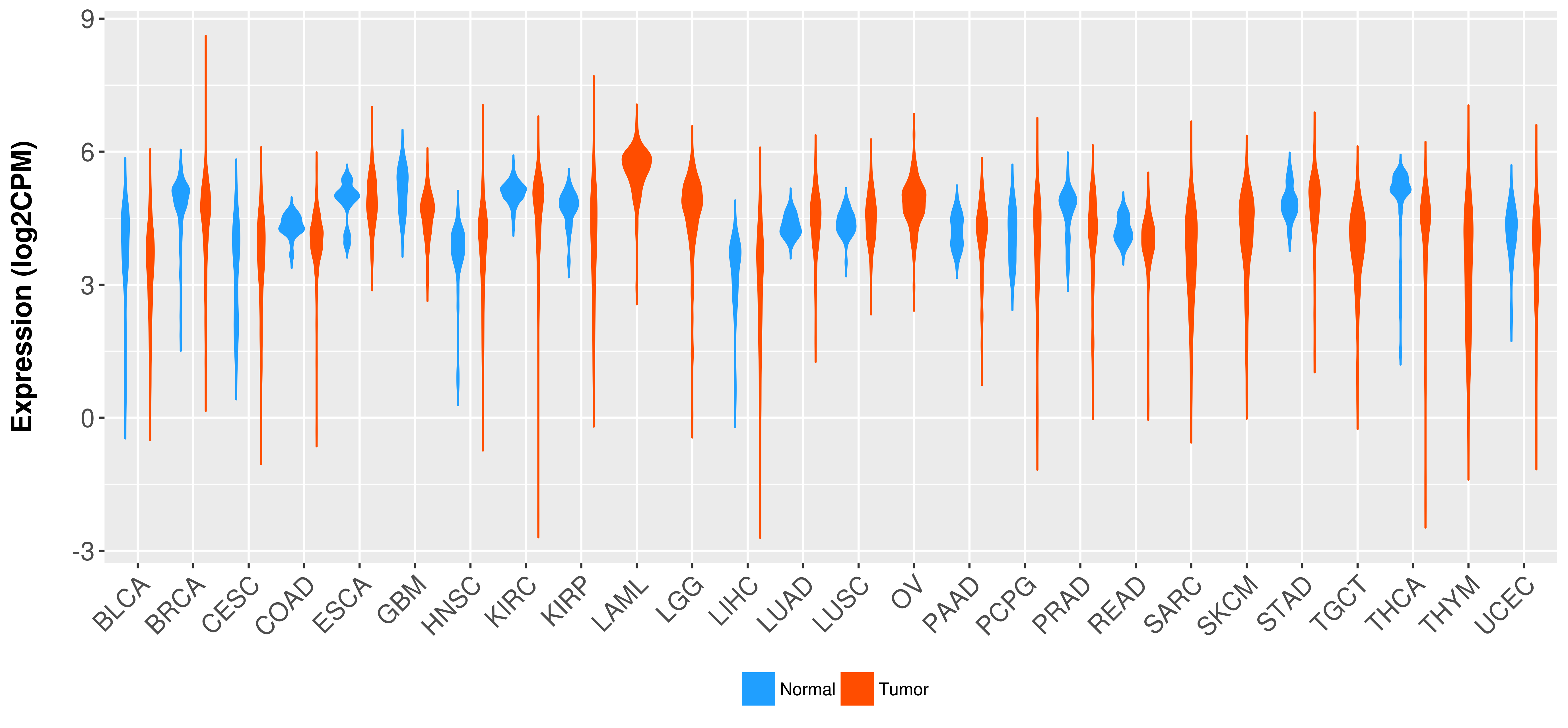
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
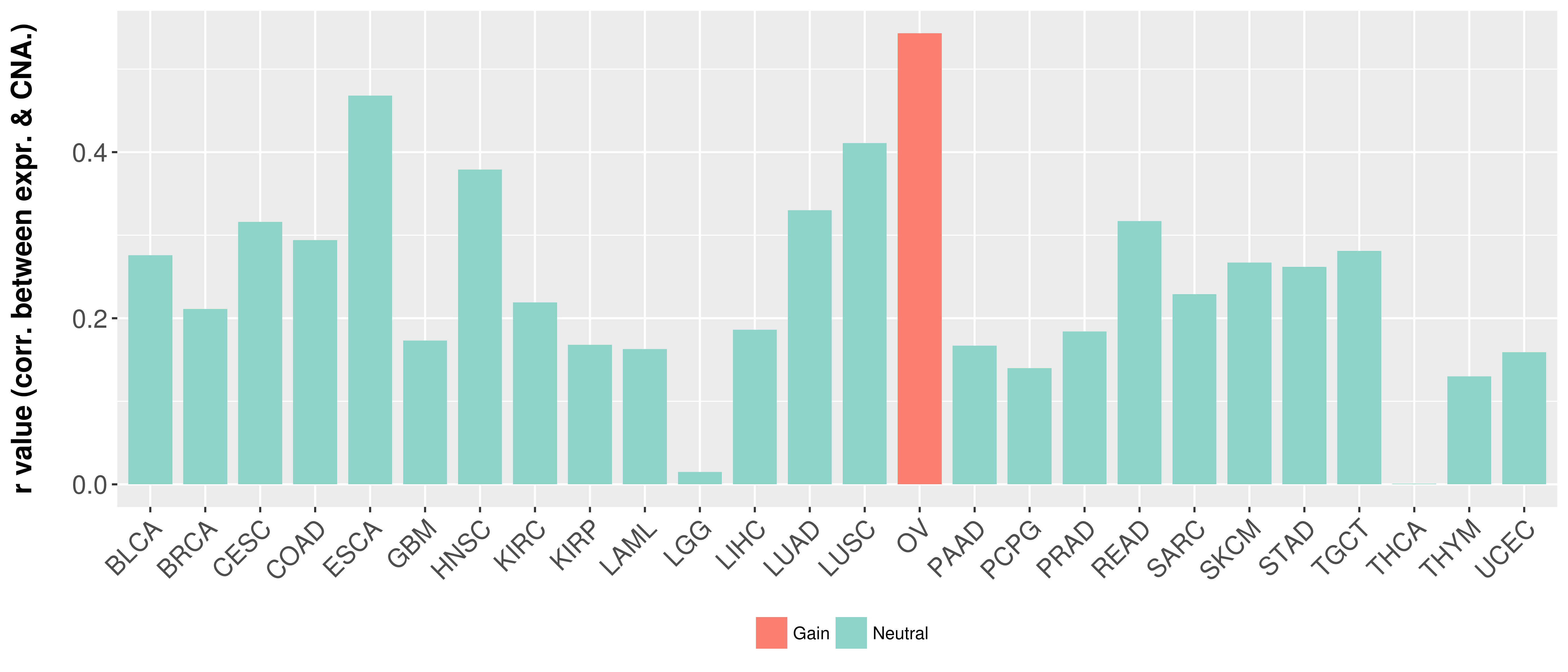
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
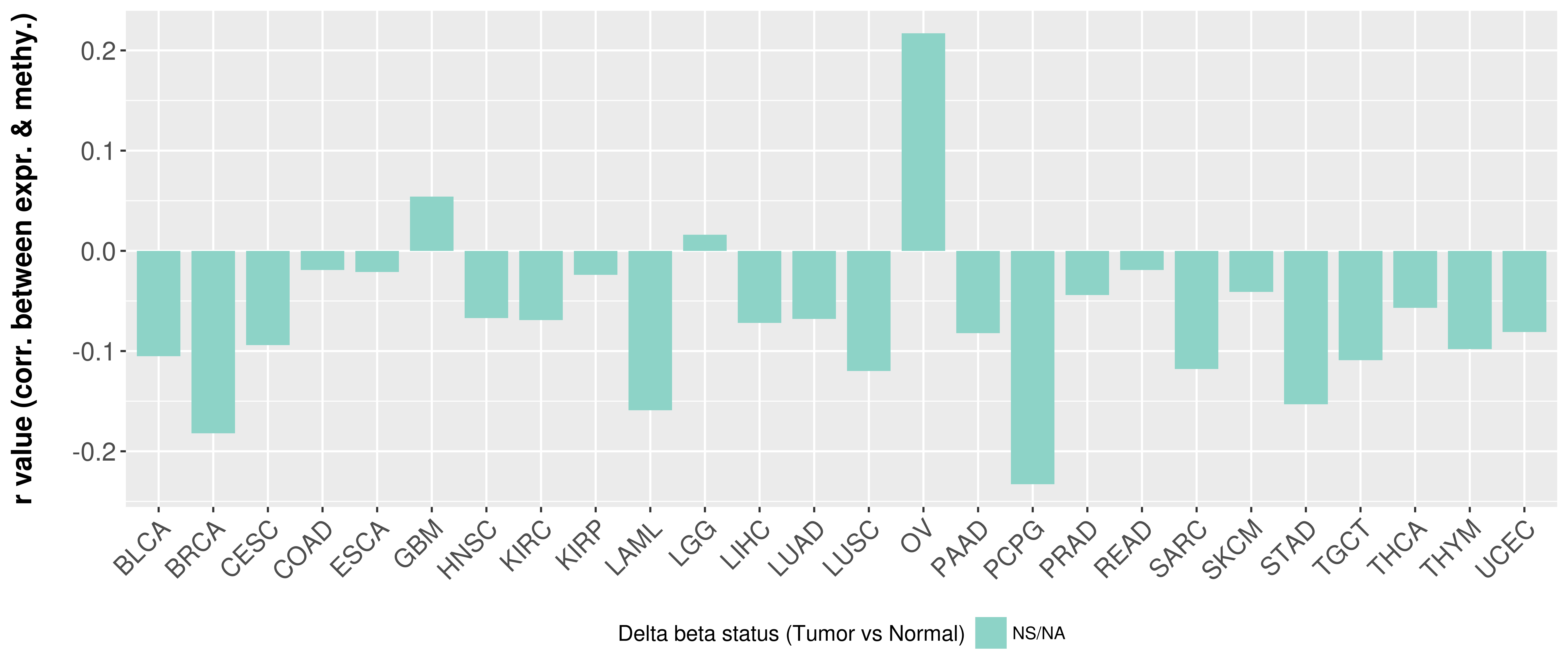
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
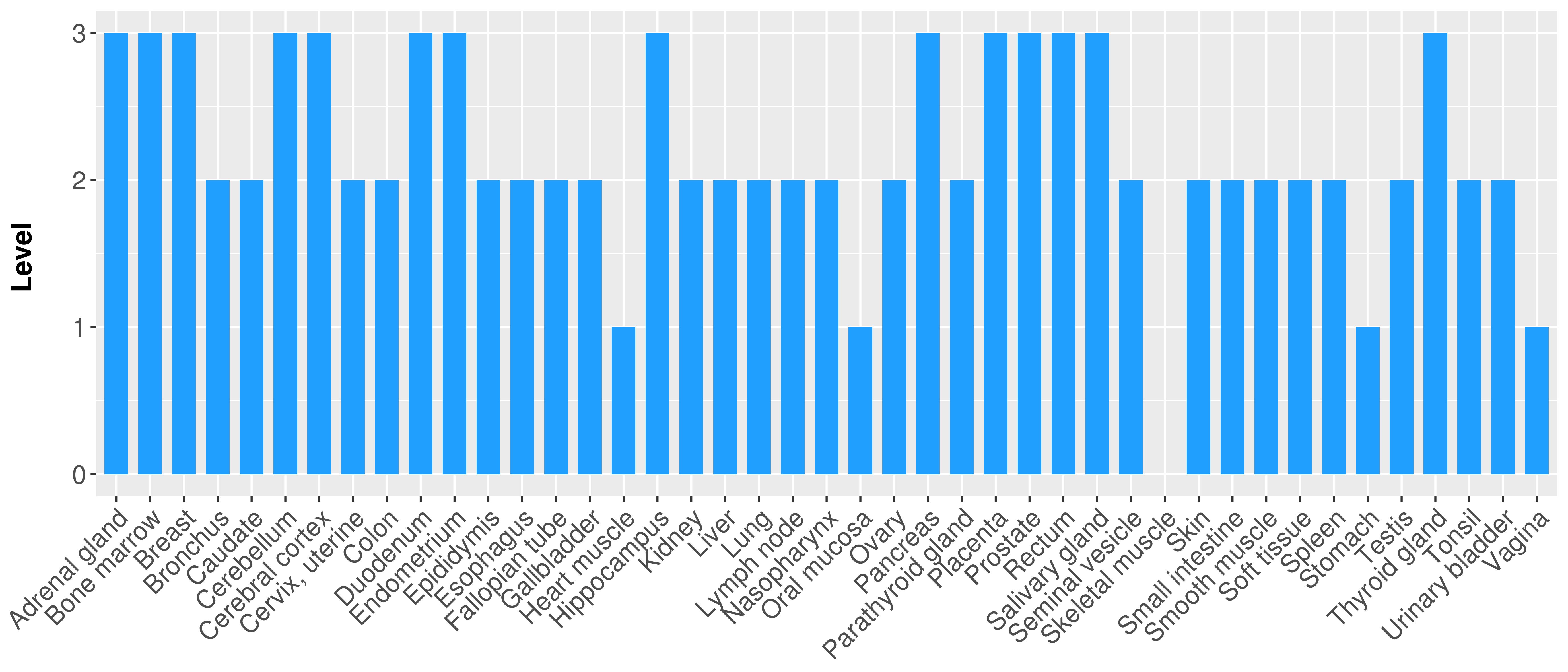
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
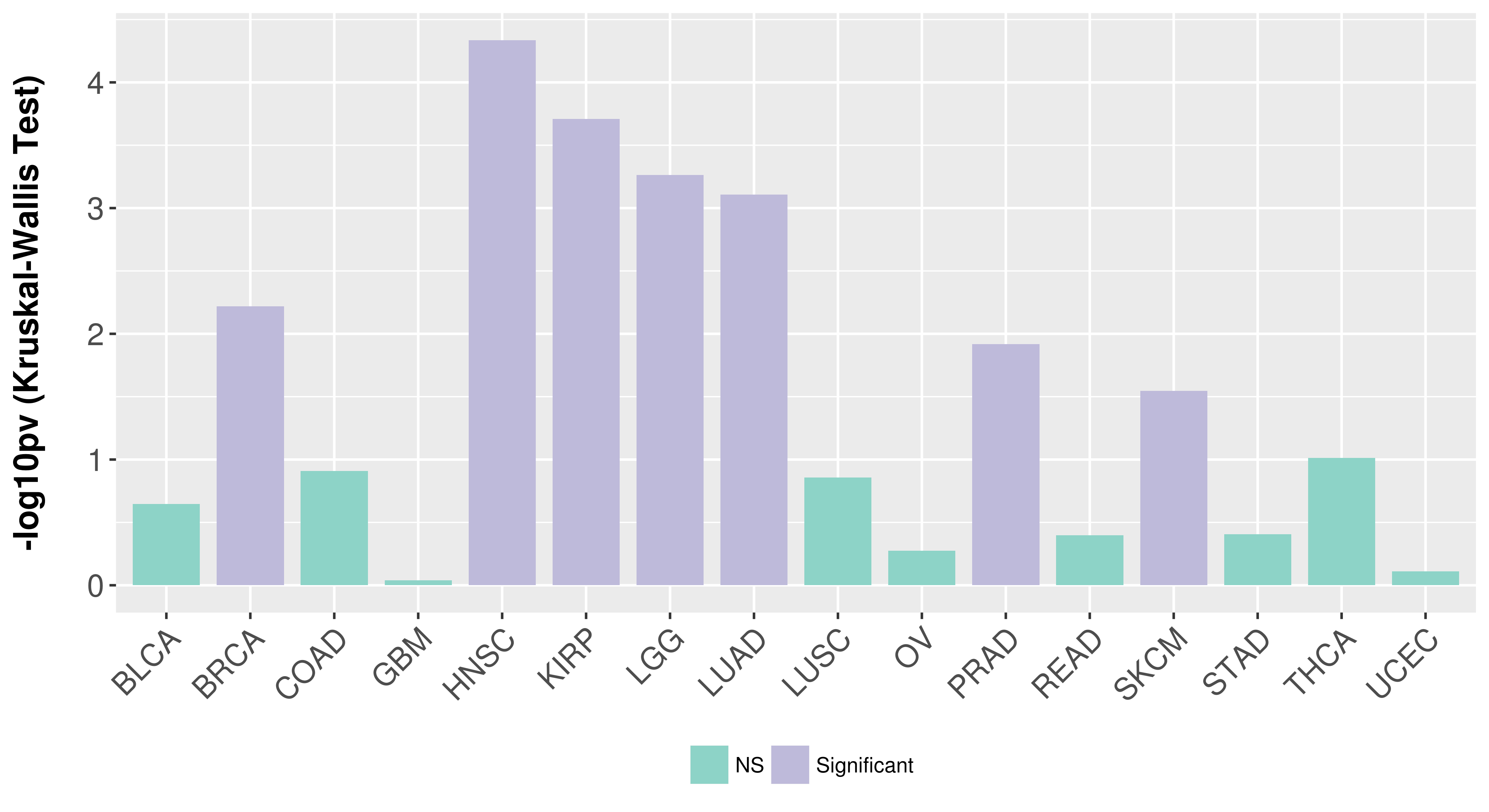
Association between expresson and subtype.
|
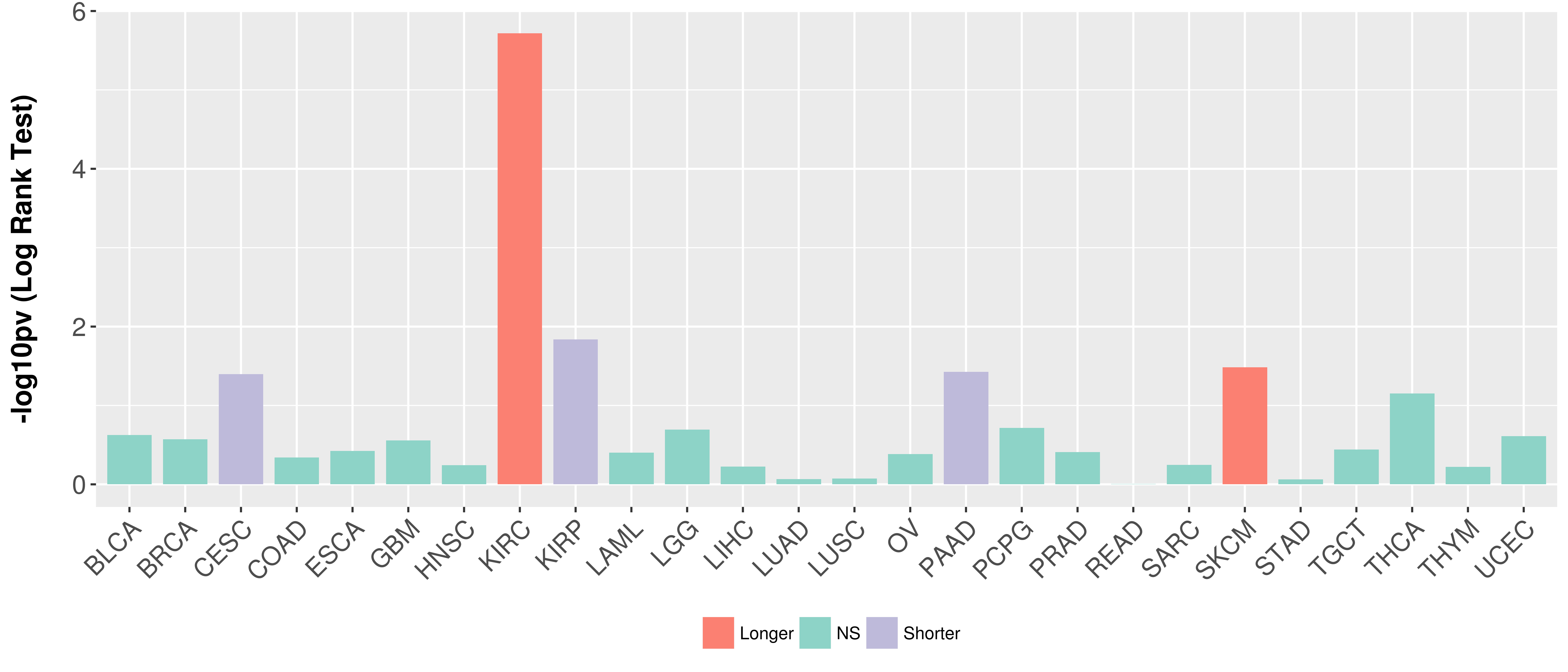
Overall survival analysis based on expression.
|
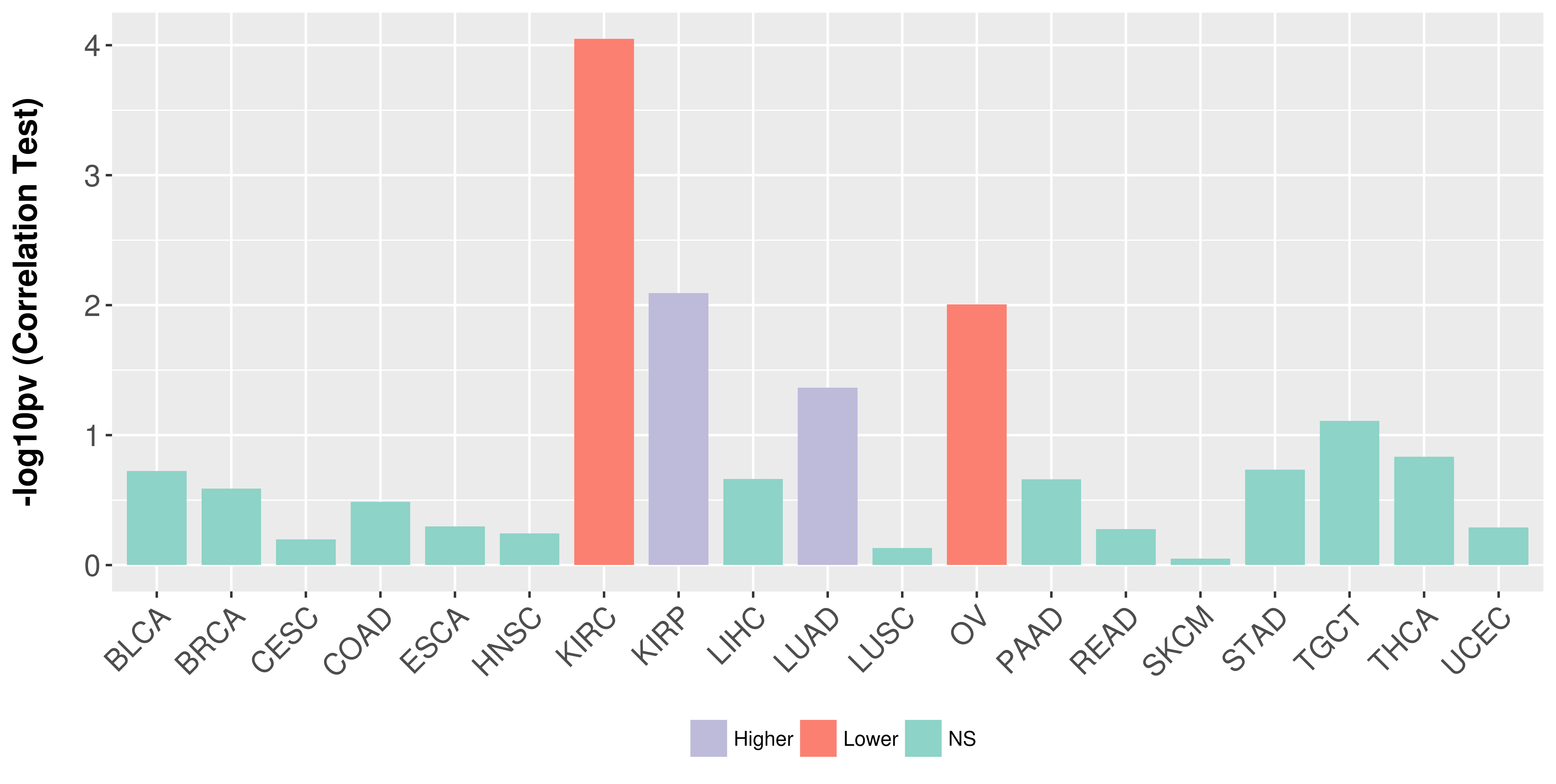
Association between expresson and stage.
|
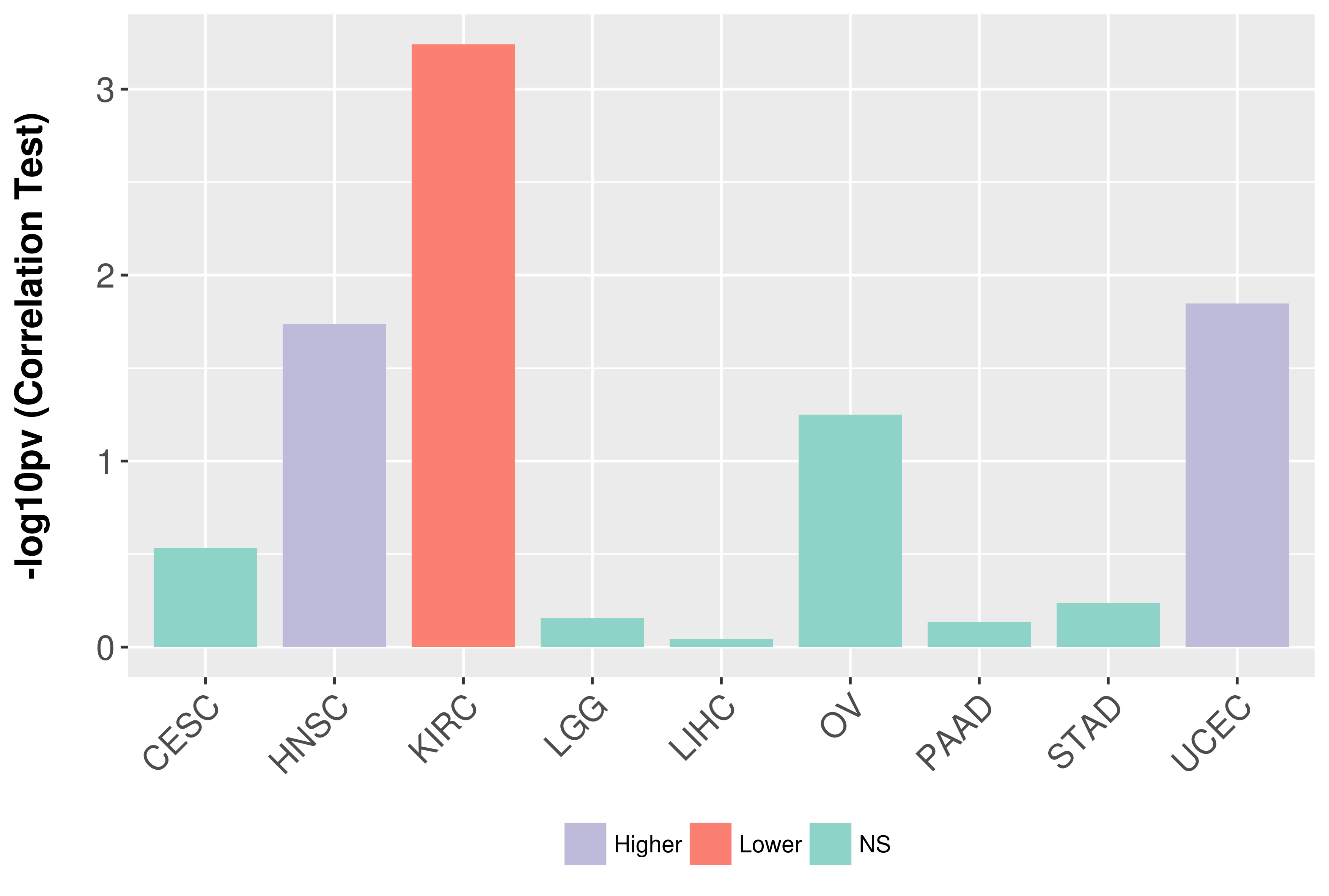
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
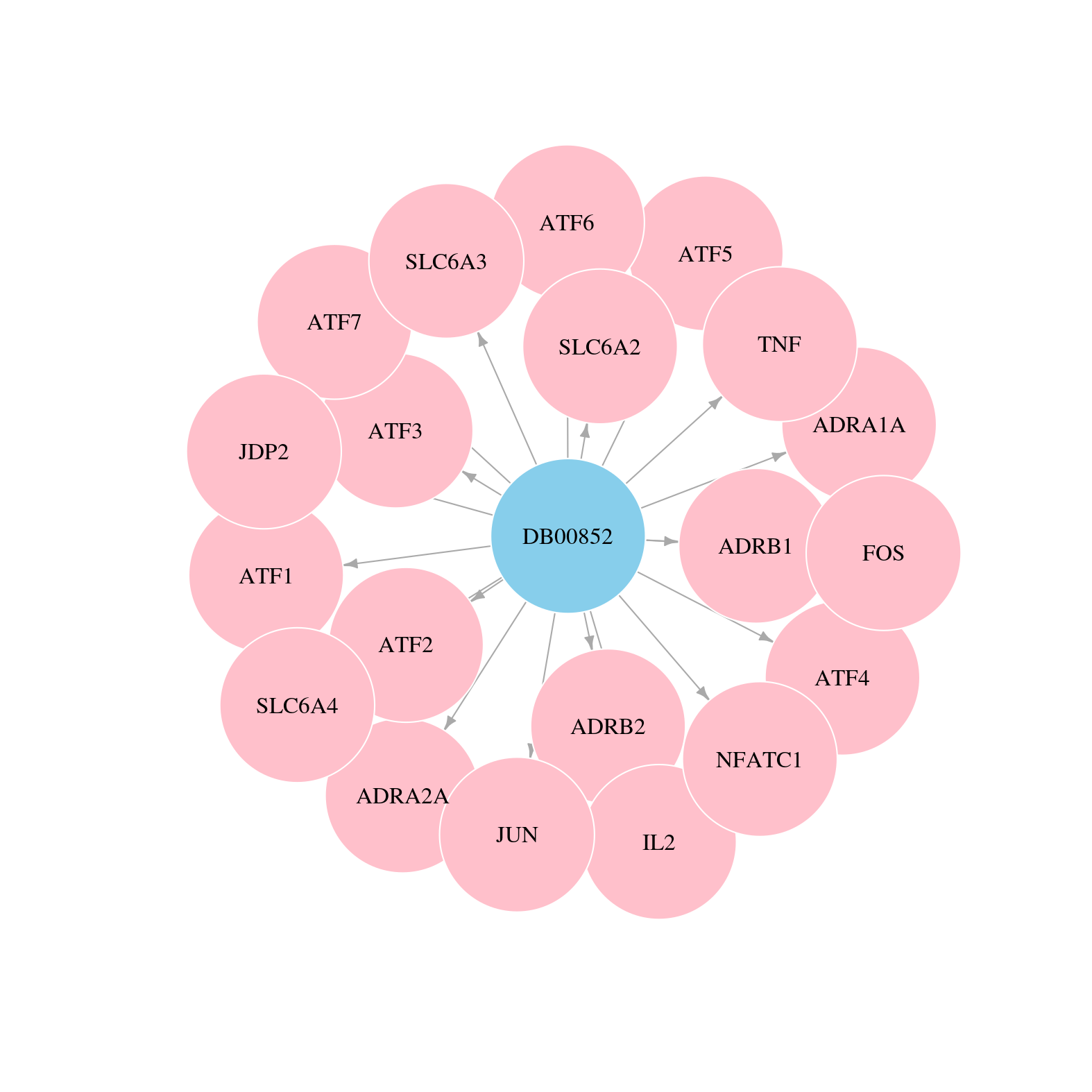
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
|
| Summary | |
|---|---|
| Symbol | ATF2 |
| Name | activating transcription factor 2 |
| Aliases | TREB7; CRE-BP1; HB16; cAMP responsive element binding protein 2; activating transcription factor 2 splice va ...... |
| Location | 2q31.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|