Browse BAG6 in pancancer
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF12057 Domain of unknown function (DUF3538) PF00240 Ubiquitin family |
||||||||||
| Function |
ATP-independent molecular chaperone preventing the aggregation of misfolded and hydrophobic patches-containing proteins (PubMed:21636303). Functions as part of a cytosolic protein quality control complex, the BAG6/BAT3 complex, which maintains these client proteins in a soluble state and participates to their proper delivery to the endoplasmic reticulum or alternatively can promote their sorting to the proteasome where they undergo degradation (PubMed:20516149, PubMed:21636303, PubMed:21743475, PubMed:28104892). The BAG6/BAT3 complex is involved in the post-translational delivery of tail-anchored/type II transmembrane proteins to the endoplasmic reticulum membrane. Recruited to ribosomes, it interacts with the transmembrane region of newly synthesized tail-anchored proteins and together with SGTA and ASNA1 mediates their delivery to the endoplasmic reticulum (PubMed:20516149, PubMed:20676083, PubMed:28104892). Client proteins that cannot be properly delivered to the endoplasmic reticulum are ubiquitinated by RNF126, an E3 ubiquitin-protein ligase associated with BAG6 and are sorted to the proteasome (PubMed:24981174, PubMed:28104892, PubMed:27193484). SGTA which prevents the recruitment of RNF126 to BAG6 may negatively regulate the ubiquitination and the proteasomal degradation of client proteins (PubMed:23129660, PubMed:25179605, PubMed:27193484). Similarly, the BAG6/BAT3 complex also functions as a sorting platform for proteins of the secretory pathway that are mislocalized to the cytosol either delivering them to the proteasome for degradation or to the endoplasmic reticulum (PubMed:21743475). The BAG6/BAT3 complex also plays a role in the endoplasmic reticulum-associated degradation (ERAD), a quality control mechanism that eliminates unwanted proteins of the endoplasmic reticulum through their retrotranslocation to the cytosol and their targeting to the proteasome. It maintains these retrotranslocated proteins in an unfolded yet soluble state condition in the cytosol to ensure their proper delivery to the proteasome (PubMed:21636303). BAG6 is also required for selective ubiquitin-mediated degradation of defective nascent chain polypeptides by the proteasome. In this context, it may participate to the production of antigenic peptides and play a role in antigen presentation in immune response (By similarity). BAG6 is also involved in endoplasmic reticulum stress-induced pre-emptive quality control, a mechanism that selectively attenuates the translocation of newly synthesized proteins into the endoplasmic reticulum and reroutes them to the cytosol for proteasomal degradation. BAG6 may ensure the proper degradation of these proteins and thereby protects the endoplasmic reticulum from protein overload upon stress (PubMed:26565908). By inhibiting the polyubiquitination and subsequent proteasomal degradation of HSPA2 it may also play a role in the assembly of the synaptonemal complex during spermatogenesis (By similarity). Also positively regulates apoptosis by interacting with and stabilizing the proapoptotic factor AIFM1 (By similarity). ; FUNCTION: Involved in DNA damage-induced apoptosis: following DNA damage, accumulates in the nucleus and forms a complex with p300/EP300, enhancing p300/EP300-mediated p53/TP53 acetylation leading to increase p53/TP53 transcriptional activity (PubMed:17403783). When nuclear, may also act as a component of some chromatin regulator complex that regulates histone 3 'Lys-4' dimethylation (H3K4me2) (PubMed:18765639). ; FUNCTION: Released extracellularly via exosomes, it is a ligand of the natural killer/NK cells receptor NCR3 and stimulates NK cells cytotoxicity. It may thereby trigger NK cells cytotoxicity against neighboring tumor cells and immature myeloid dendritic cells (DC). ; FUNCTION: Mediates ricin-induced apoptosis. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001655 urogenital system development GO:0001822 kidney development GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0007029 endoplasmic reticulum organization GO:0007059 chromosome segregation GO:0007126 meiotic nuclear division GO:0007127 meiosis I GO:0007129 synapsis GO:0007130 synaptonemal complex assembly GO:0007283 spermatogenesis GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0009894 regulation of catabolic process GO:0009895 negative regulation of catabolic process GO:0009896 positive regulation of catabolic process GO:0010498 proteasomal protein catabolic process GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0030323 respiratory tube development GO:0030324 lung development GO:0031329 regulation of cellular catabolic process GO:0031330 negative regulation of cellular catabolic process GO:0031647 regulation of protein stability GO:0032434 regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0032435 negative regulation of proteasomal ubiquitin-dependent protein catabolic process GO:0034976 response to endoplasmic reticulum stress GO:0036503 ERAD pathway GO:0036506 maintenance of unfolded protein GO:0042176 regulation of protein catabolic process GO:0042177 negative regulation of protein catabolic process GO:0042771 intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043543 protein acylation GO:0045048 protein insertion into ER membrane GO:0045132 meiotic chromosome segregation GO:0045143 homologous chromosome segregation GO:0045732 positive regulation of protein catabolic process GO:0045861 negative regulation of proteolysis GO:0048232 male gamete generation GO:0050821 protein stabilization GO:0051205 protein insertion into membrane GO:0051321 meiotic cell cycle GO:0060541 respiratory system development GO:0061136 regulation of proteasomal protein catabolic process GO:0070059 intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress GO:0070192 chromosome organization involved in meiotic cell cycle GO:0070193 synaptonemal complex organization GO:0071816 tail-anchored membrane protein insertion into ER membrane GO:0072001 renal system development GO:0072331 signal transduction by p53 class mediator GO:0072332 intrinsic apoptotic signaling pathway by p53 class mediator GO:0072657 protein localization to membrane GO:0090150 establishment of protein localization to membrane GO:0097193 intrinsic apoptotic signaling pathway GO:0098813 nuclear chromosome segregation GO:1901799 negative regulation of proteasomal protein catabolic process GO:1903046 meiotic cell cycle process GO:1903050 regulation of proteolysis involved in cellular protein catabolic process GO:1903051 negative regulation of proteolysis involved in cellular protein catabolic process GO:1903362 regulation of cellular protein catabolic process GO:1903363 negative regulation of cellular protein catabolic process GO:1904292 regulation of ERAD pathway GO:1904294 positive regulation of ERAD pathway GO:1904327 protein localization to cytosolic proteasome complex GO:1904378 maintenance of unfolded protein involved in ERAD pathway GO:1904379 protein localization to cytosolic proteasome complex involved in ERAD pathway |
| Molecular Function |
GO:0002020 protease binding GO:0030544 Hsp70 protein binding GO:0031072 heat shock protein binding GO:0031593 polyubiquitin binding GO:0031625 ubiquitin protein ligase binding GO:0032182 ubiquitin-like protein binding GO:0043021 ribonucleoprotein complex binding GO:0043022 ribosome binding GO:0043130 ubiquitin binding GO:0044389 ubiquitin-like protein ligase binding GO:0051787 misfolded protein binding GO:0070628 proteasome binding GO:1990381 ubiquitin-specific protease binding |
| Cellular Component |
GO:0044445 cytosolic part GO:0071818 BAT3 complex GO:0072379 ER membrane insertion complex |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
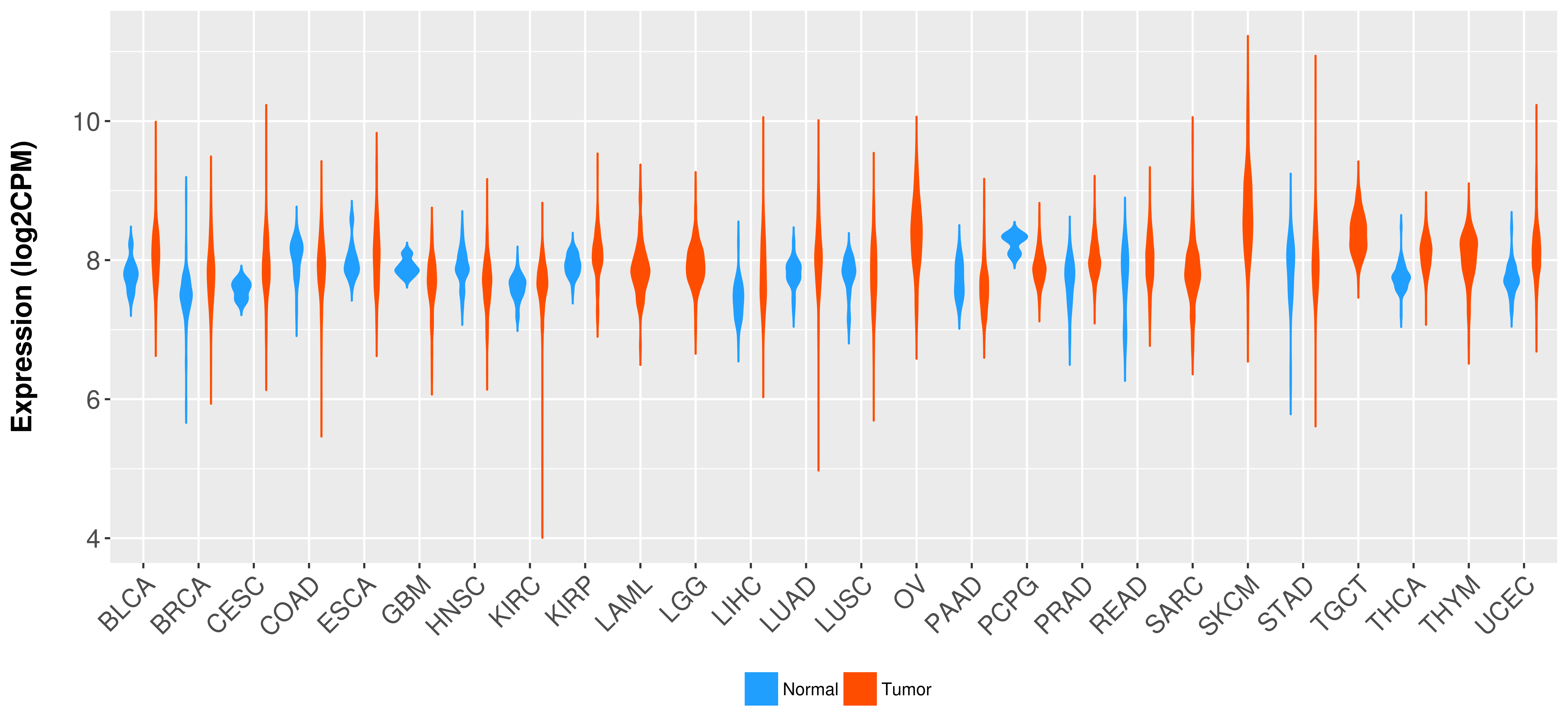
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
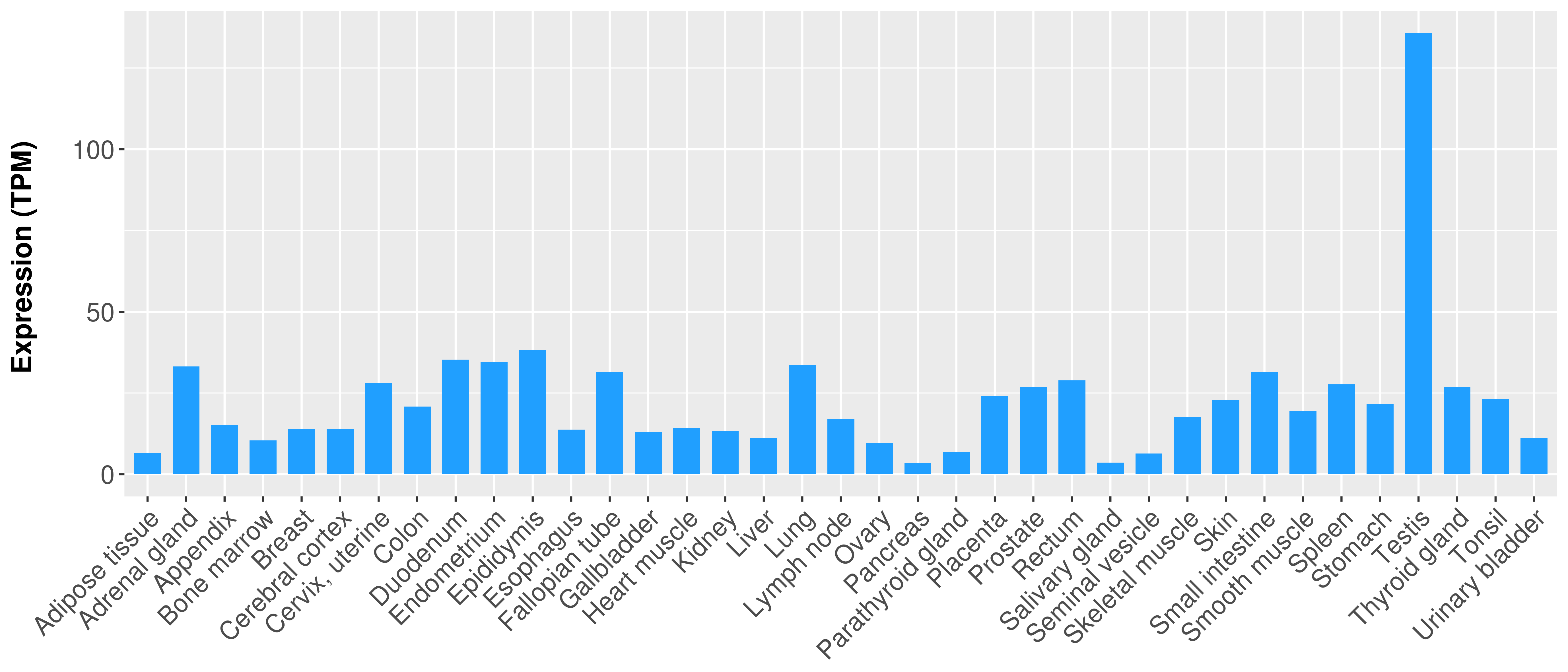
|
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
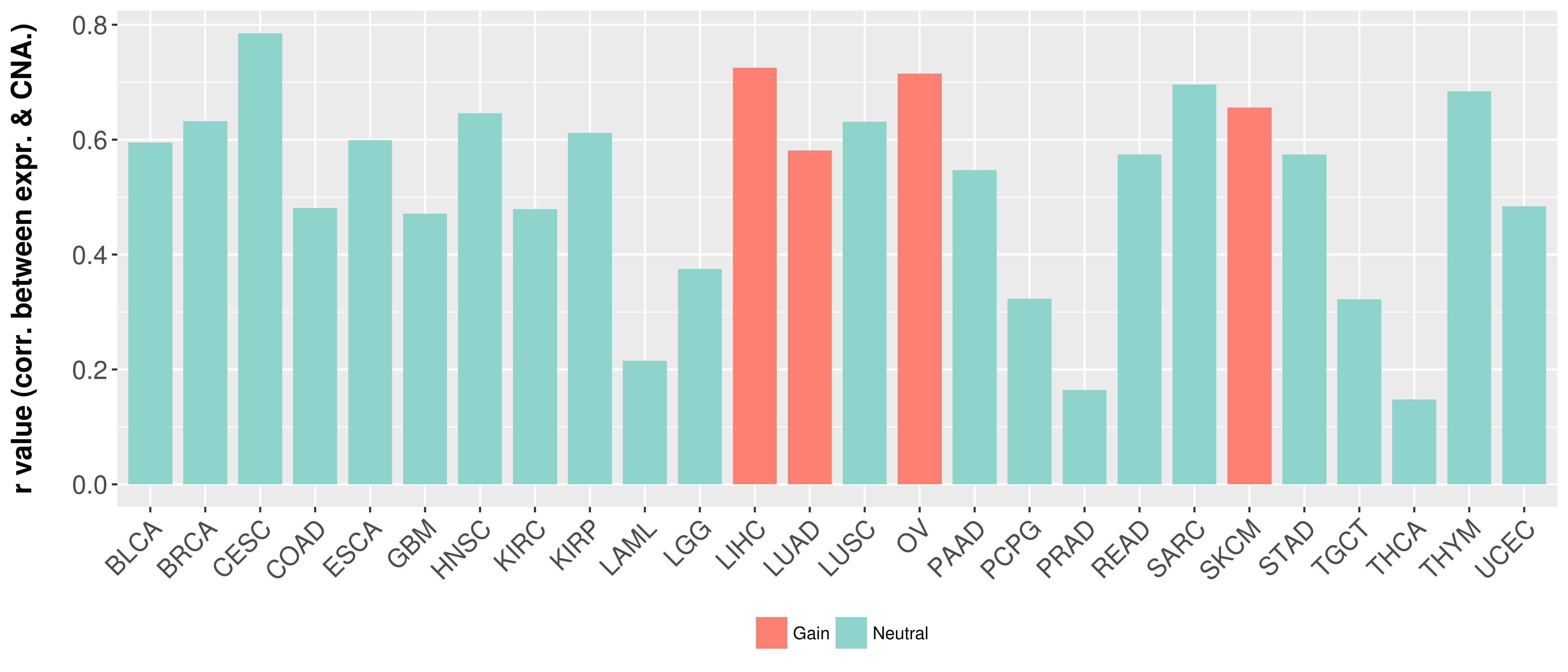
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
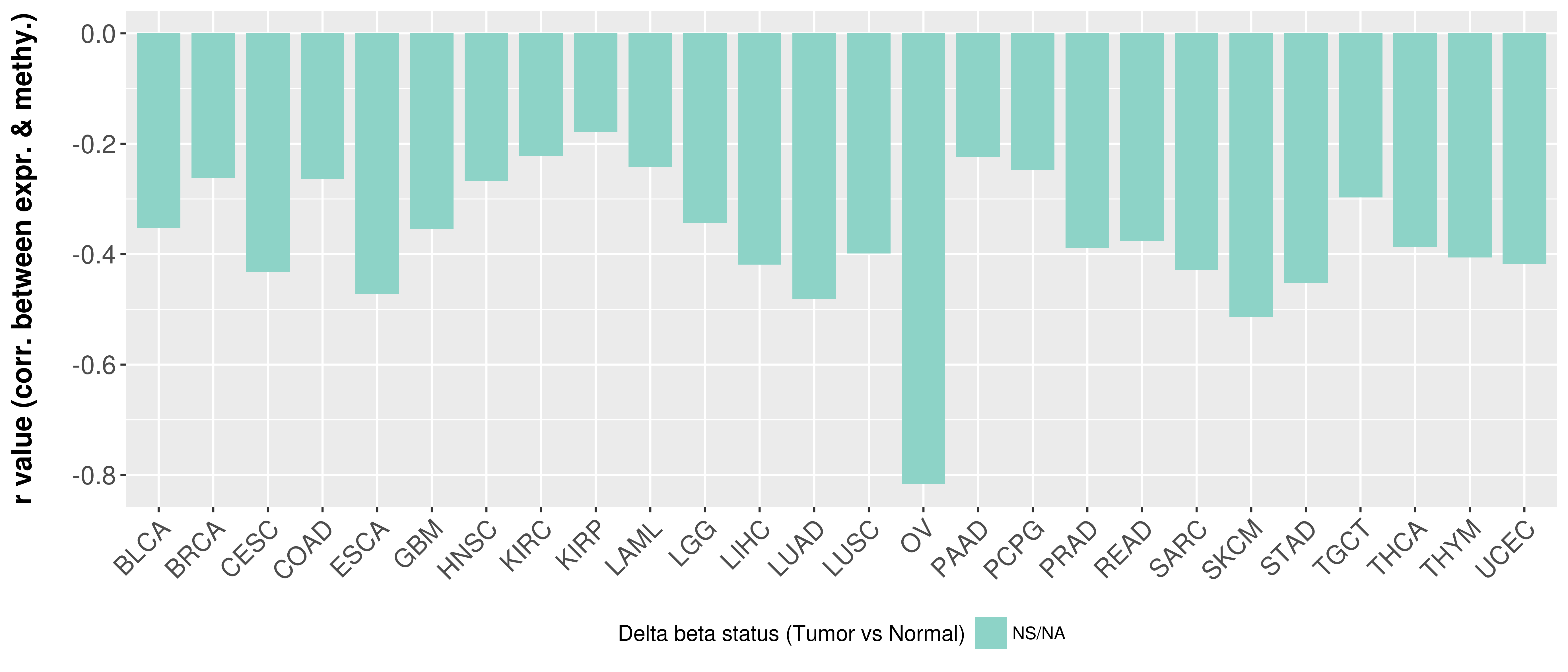
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
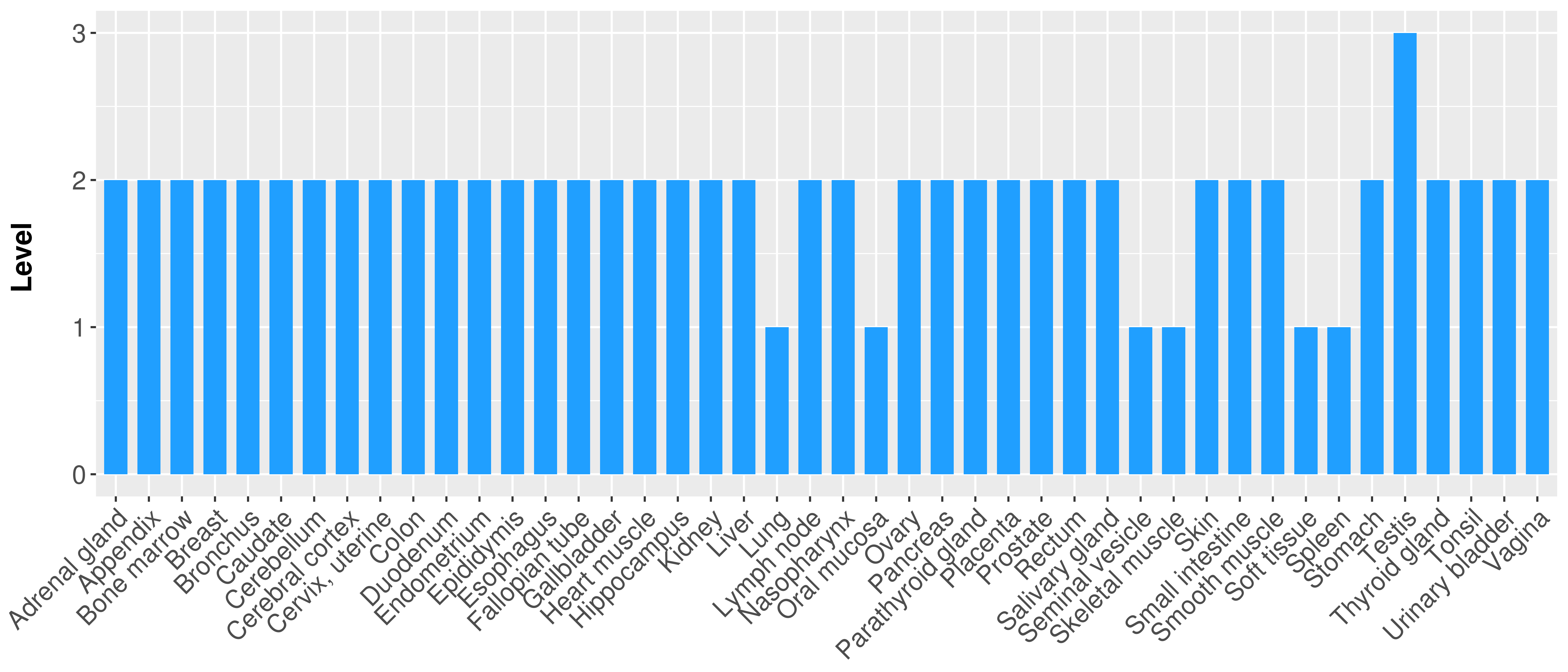
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
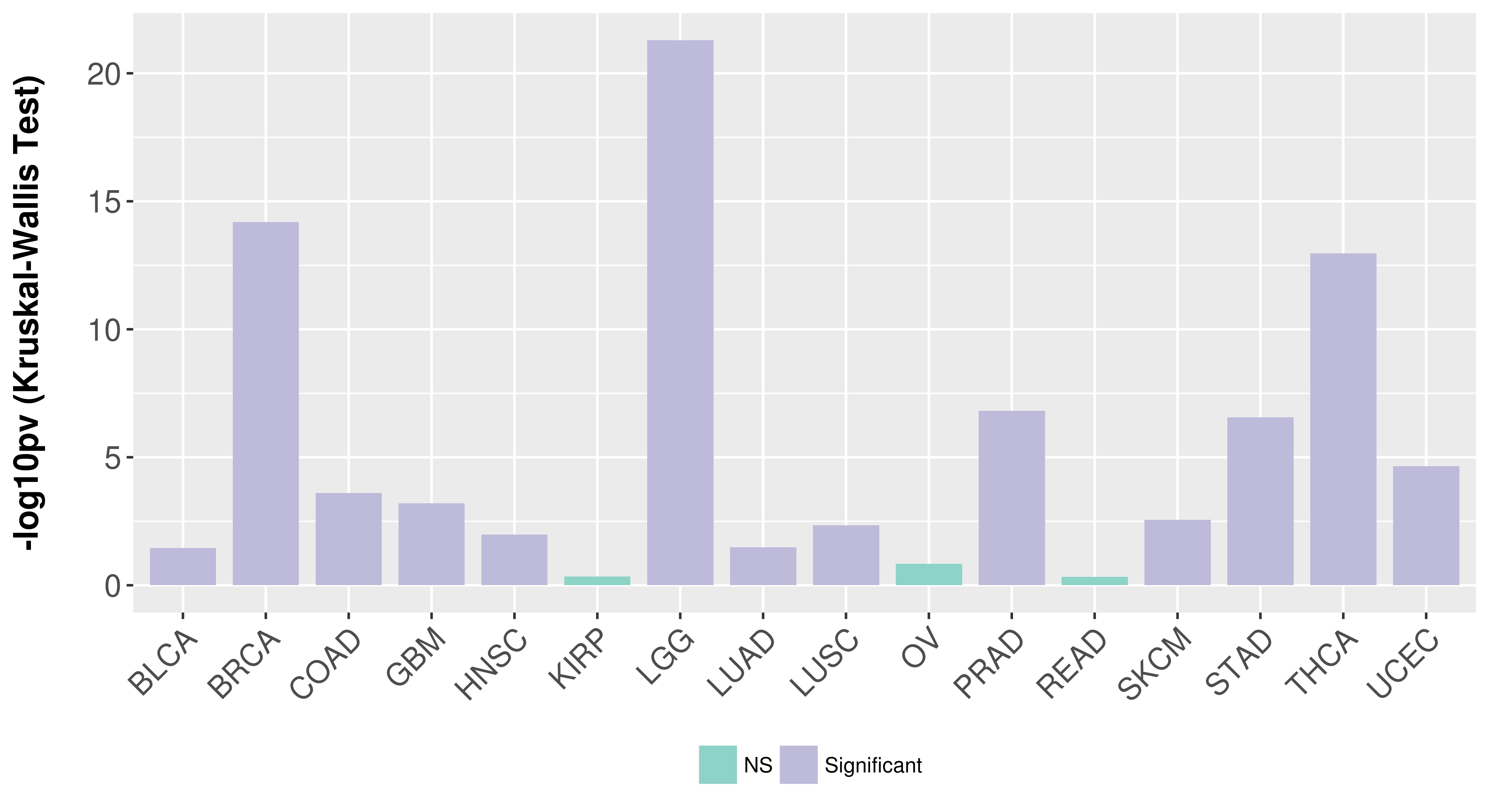
Association between expresson and subtype.
|
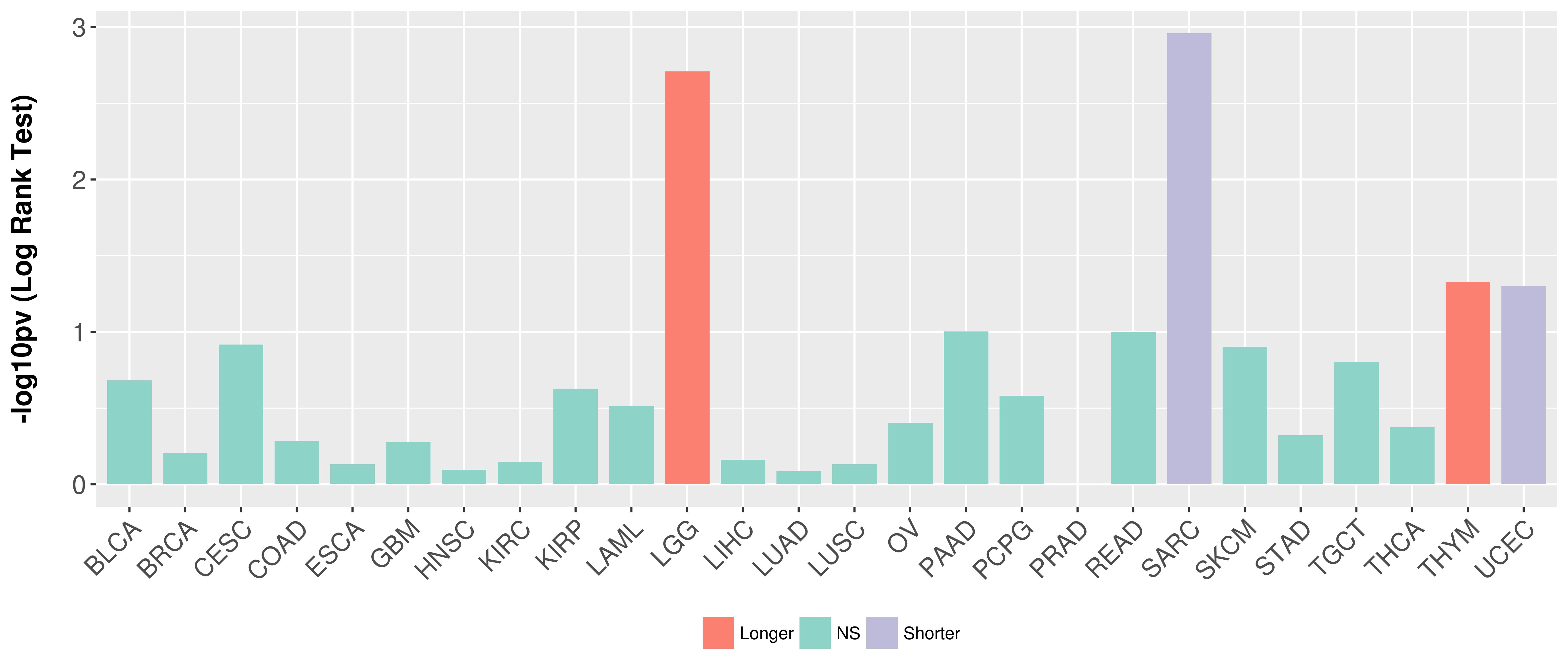
Overall survival analysis based on expression.
|
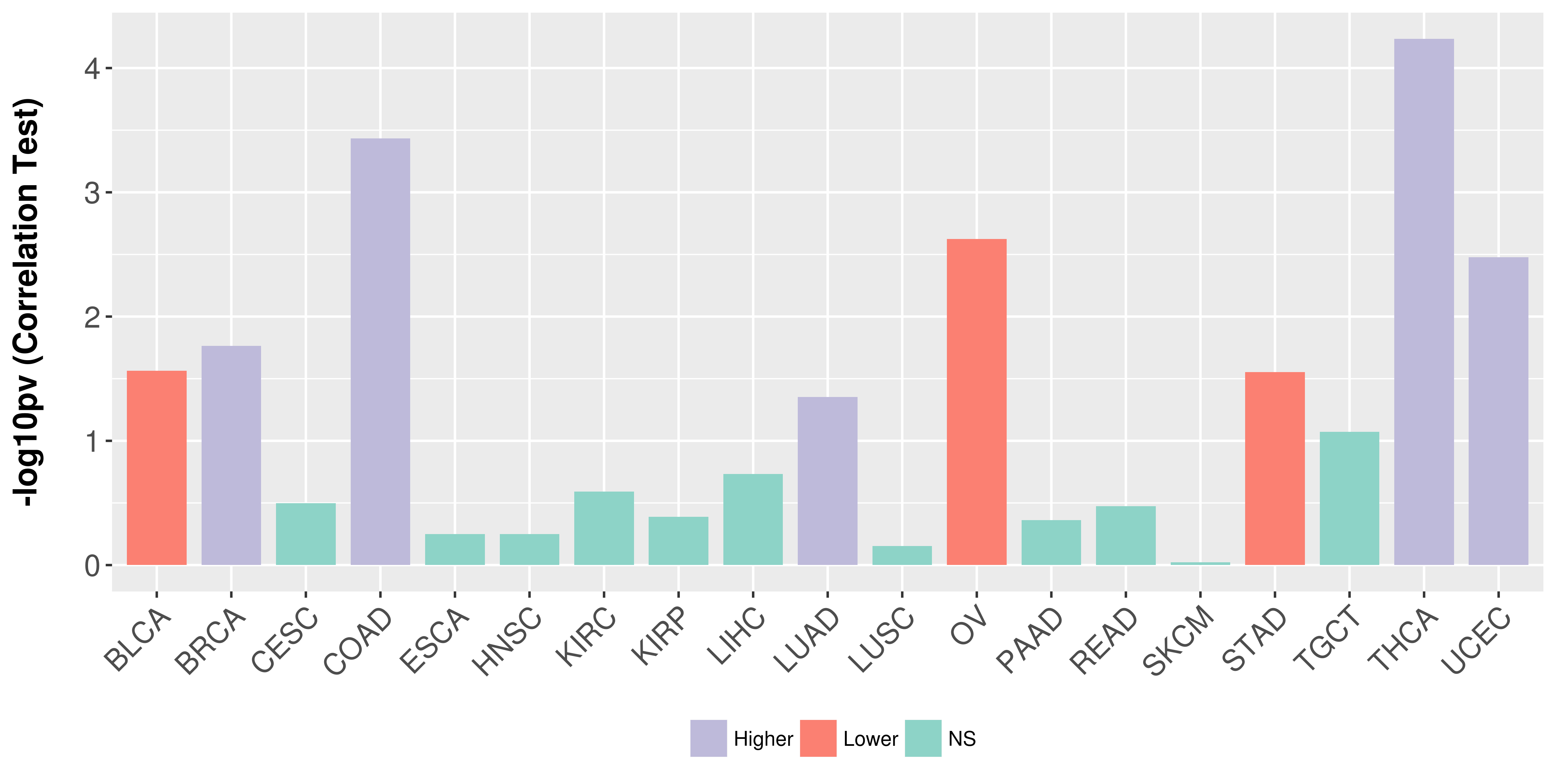
Association between expresson and stage.
|
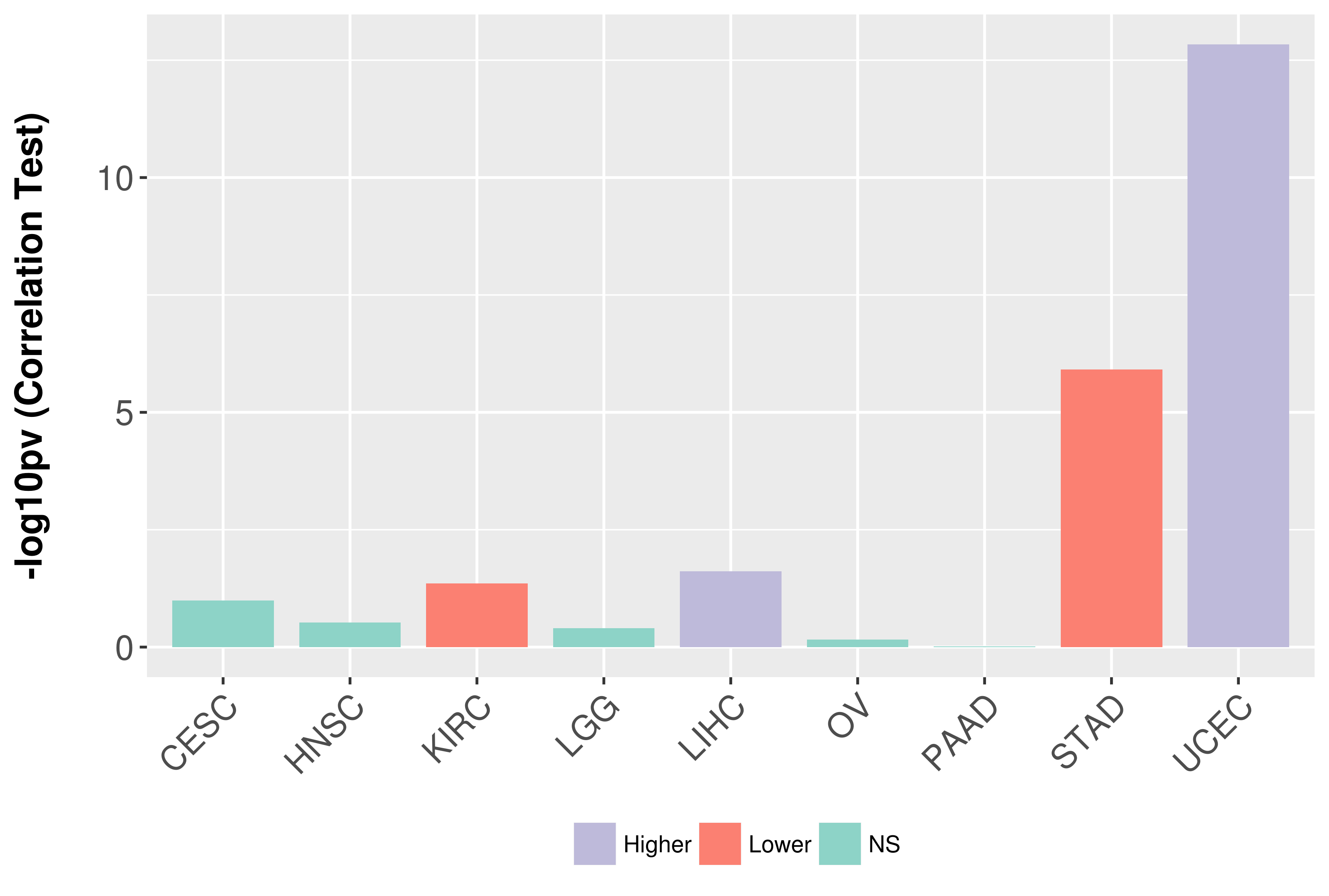
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for BAG6. |
| Summary | |
|---|---|
| Symbol | BAG6 |
| Name | BCL2 associated athanogene 6 |
| Aliases | G3; D6S52E; BAT3; HLA-B associated transcript 3; BAG-6; BAG family molecular chaperone regulator 6; HLA-B as ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for BAG6. |
| There is no record for BAG6. |