Browse DNMT3B in pancancer
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00145 C-5 cytosine-specific DNA methylase PF00855 PWWP domain |
||||||||||
| Function |
Required for genome-wide de novo methylation and is essential for the establishment of DNA methylation patterns during development. DNA methylation is coordinated with methylation of histones. May preferentially methylates nucleosomal DNA within the nucleosome core region. May function as transcriptional co-repressor by associating with CBX4 and independently of DNA methylation. Seems to be involved in gene silencing (By similarity). In association with DNMT1 and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Isoforms 4 and 5 are probably not functional due to the deletion of two conserved methyltransferase motifs. Function as transcriptional corepressor by associating with ZHX1. Required for DUX4 silencing in somatic cells (PubMed:27153398). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001666 response to hypoxia GO:0006304 DNA modification GO:0006305 DNA alkylation GO:0006306 DNA methylation GO:0006479 protein methylation GO:0006790 sulfur compound metabolic process GO:0007584 response to nutrient GO:0008213 protein alkylation GO:0009116 nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009314 response to radiation GO:0009636 response to toxic substance GO:0009991 response to extracellular stimulus GO:0010212 response to ionizing radiation GO:0010424 DNA methylation on cytosine within a CG sequence GO:0010720 positive regulation of cell development GO:0014074 response to purine-containing compound GO:0014823 response to activity GO:0016570 histone modification GO:0016571 histone methylation GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0031000 response to caffeine GO:0031056 regulation of histone modification GO:0031057 negative regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031060 regulation of histone methylation GO:0031061 negative regulation of histone methylation GO:0031062 positive regulation of histone methylation GO:0031667 response to nutrient levels GO:0031960 response to corticosteroid GO:0032259 methylation GO:0032355 response to estradiol GO:0032776 DNA methylation on cytosine GO:0033189 response to vitamin A GO:0033273 response to vitamin GO:0034968 histone lysine methylation GO:0036293 response to decreased oxygen levels GO:0036295 cellular response to increased oxygen levels GO:0036296 response to increased oxygen levels GO:0040029 regulation of gene expression, epigenetic GO:0042220 response to cocaine GO:0042278 purine nucleoside metabolic process GO:0042493 response to drug GO:0043279 response to alkaloid GO:0043414 macromolecule methylation GO:0044728 DNA methylation or demethylation GO:0045666 positive regulation of neuron differentiation GO:0045814 negative regulation of gene expression, epigenetic GO:0046128 purine ribonucleoside metabolic process GO:0046498 S-adenosylhomocysteine metabolic process GO:0046499 S-adenosylmethioninamine metabolic process GO:0048545 response to steroid hormone GO:0050769 positive regulation of neurogenesis GO:0051384 response to glucocorticoid GO:0051567 histone H3-K9 methylation GO:0051568 histone H3-K4 methylation GO:0051569 regulation of histone H3-K4 methylation GO:0051570 regulation of histone H3-K9 methylation GO:0051571 positive regulation of histone H3-K4 methylation GO:0051573 negative regulation of histone H3-K9 methylation GO:0051962 positive regulation of nervous system development GO:0055093 response to hyperoxia GO:0061647 histone H3-K9 modification GO:0070482 response to oxygen levels GO:0071383 cellular response to steroid hormone stimulus GO:0071384 cellular response to corticosteroid stimulus GO:0071385 cellular response to glucocorticoid stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071453 cellular response to oxygen levels GO:0071455 cellular response to hyperoxia GO:0071548 response to dexamethasone GO:0071549 cellular response to dexamethasone stimulus GO:0090116 C-5 methylation of cytosine GO:1901605 alpha-amino acid metabolic process GO:1901654 response to ketone GO:1901655 cellular response to ketone GO:1901657 glycosyl compound metabolic process GO:1902275 regulation of chromatin organization GO:1905268 negative regulation of chromatin organization GO:1905269 positive regulation of chromatin organization |
| Molecular Function |
GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0003886 DNA (cytosine-5-)-methyltransferase activity GO:0008168 methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0009008 DNA-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0042826 histone deacetylase binding GO:0045322 unmethylated CpG binding GO:0051718 DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates |
| Cellular Component | - |
| KEGG |
hsa00270 Cysteine and methionine metabolism hsa01100 Metabolic pathways |
| Reactome |
R-HSA-5334118: DNA methylation R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-5250941: Negative epigenetic regulation of rRNA expression R-HSA-427413: NoRC negatively regulates rRNA expression R-HSA-212300: PRC2 methylates histones and DNA |
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
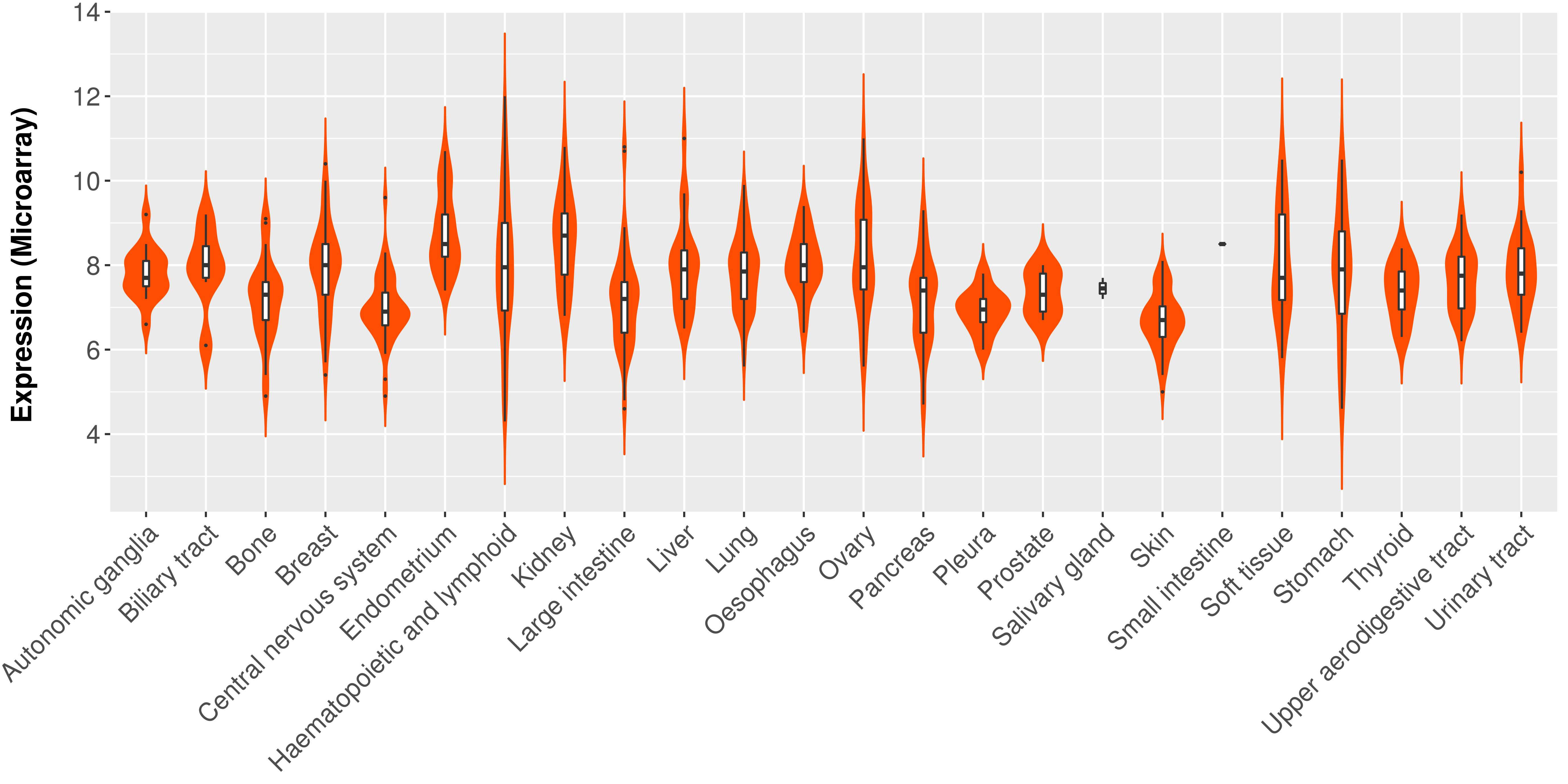
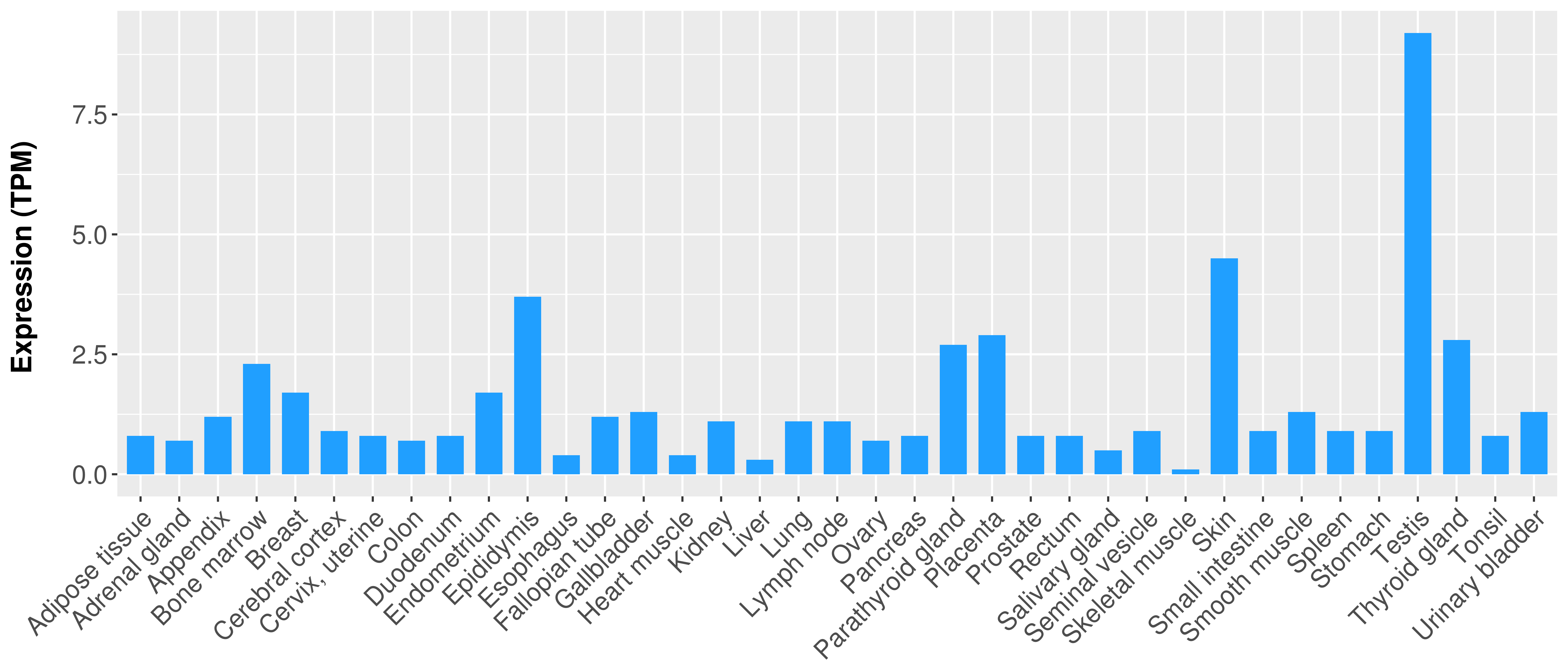
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
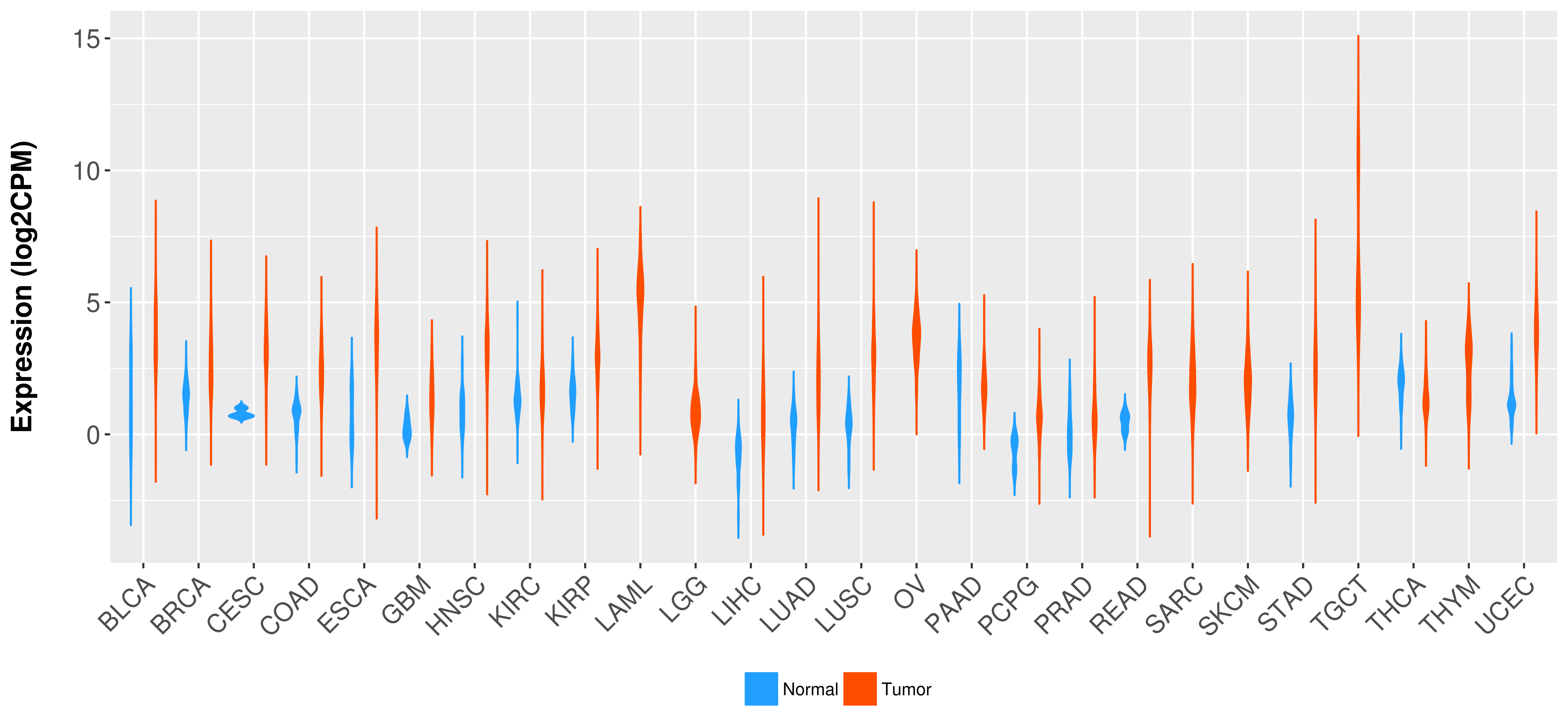
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
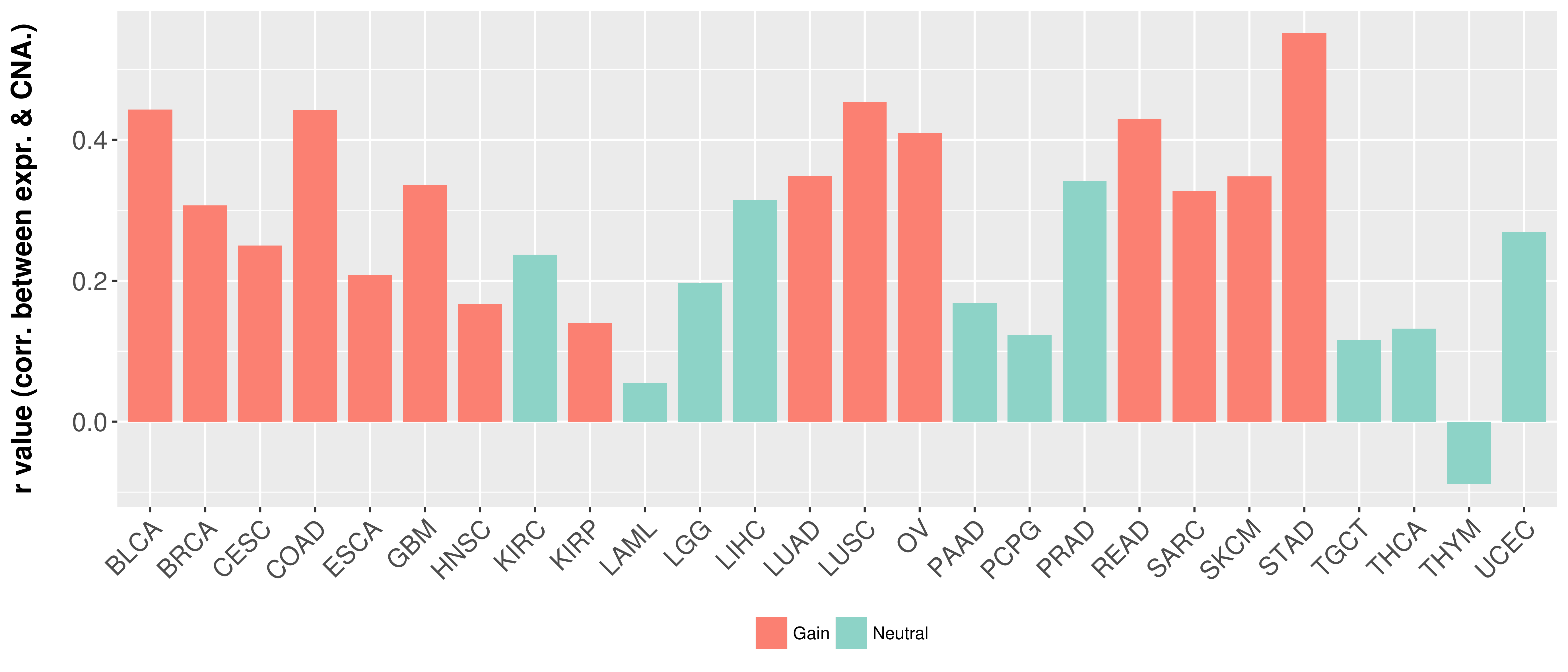
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
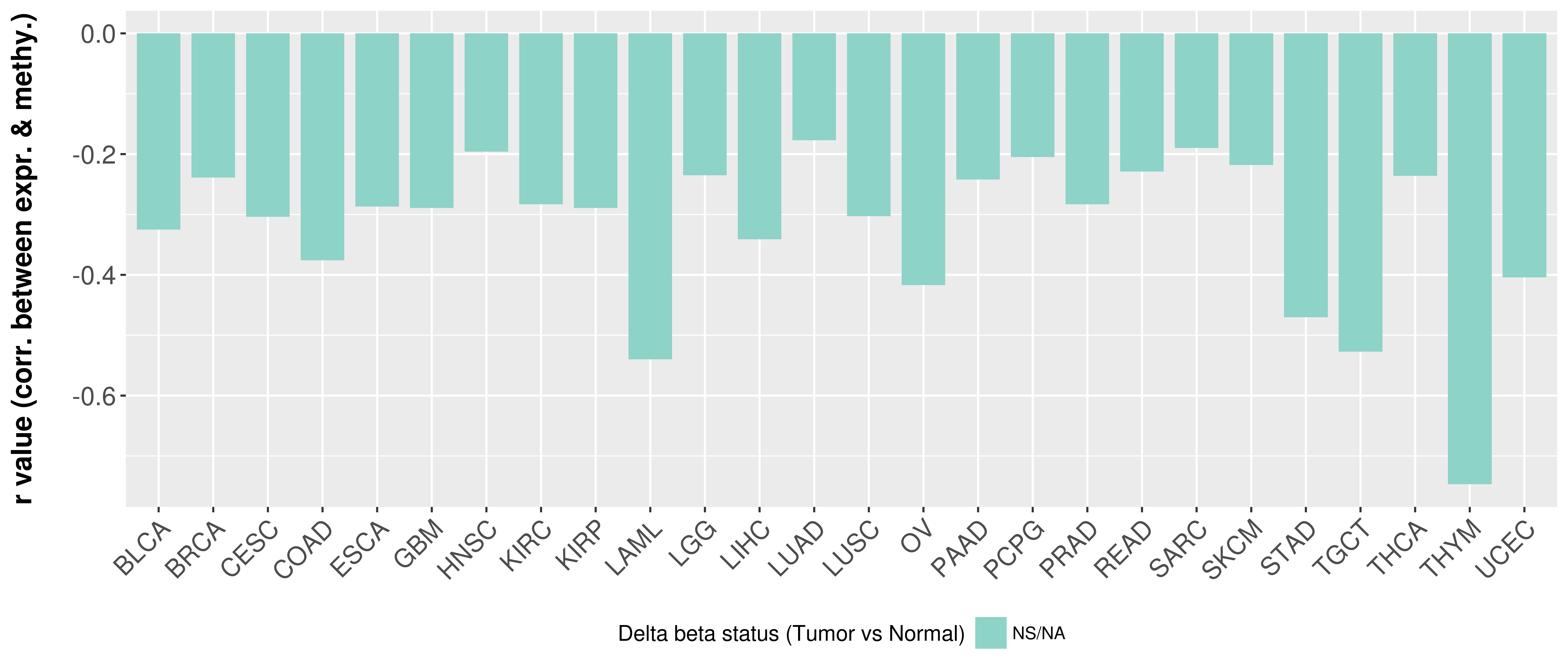
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
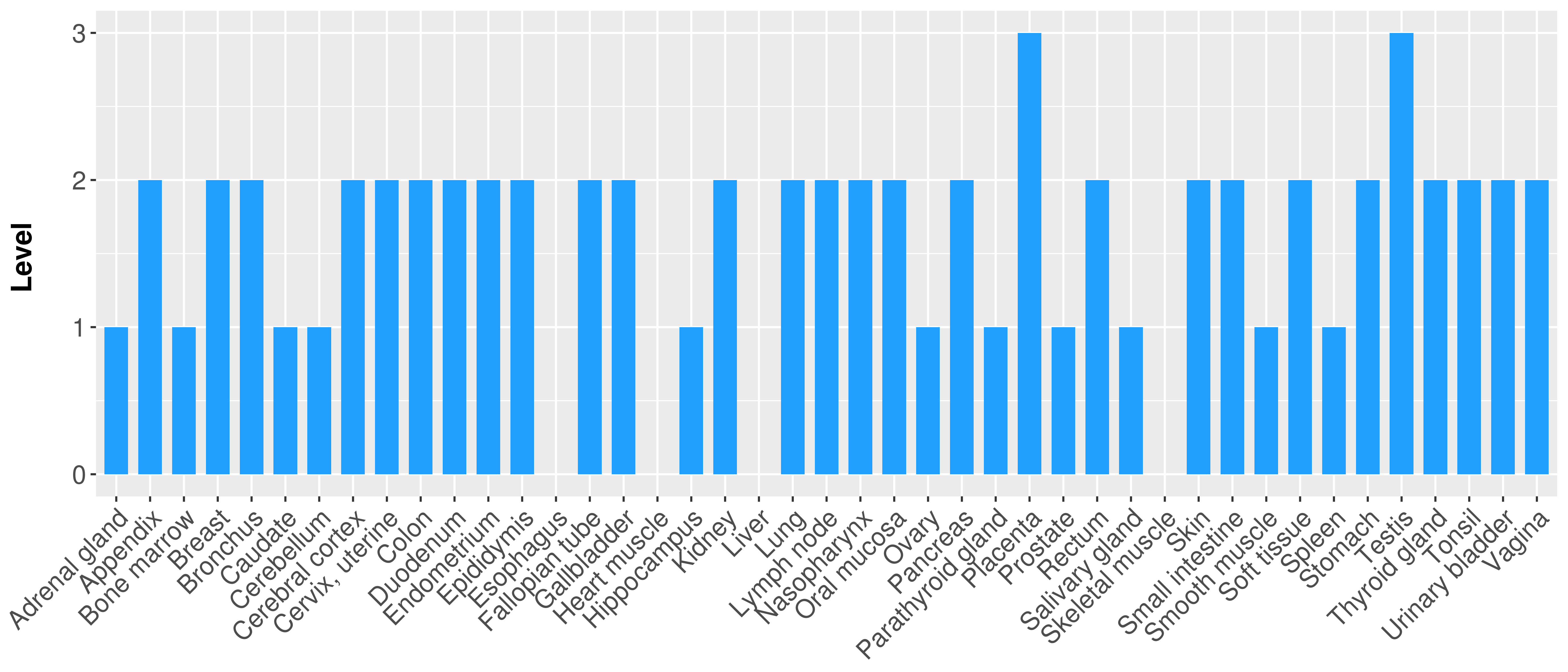
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
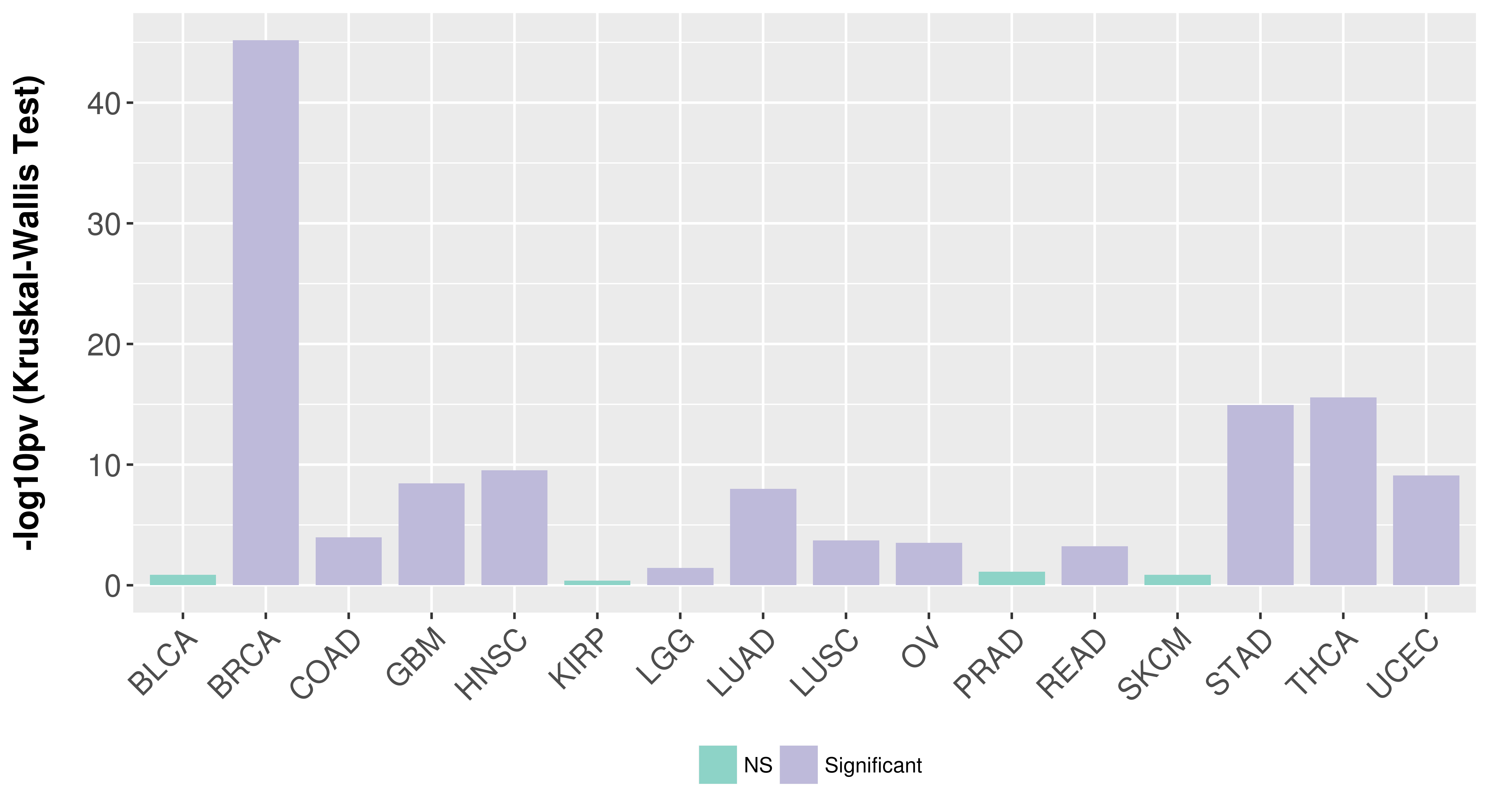
Association between expresson and subtype.
|
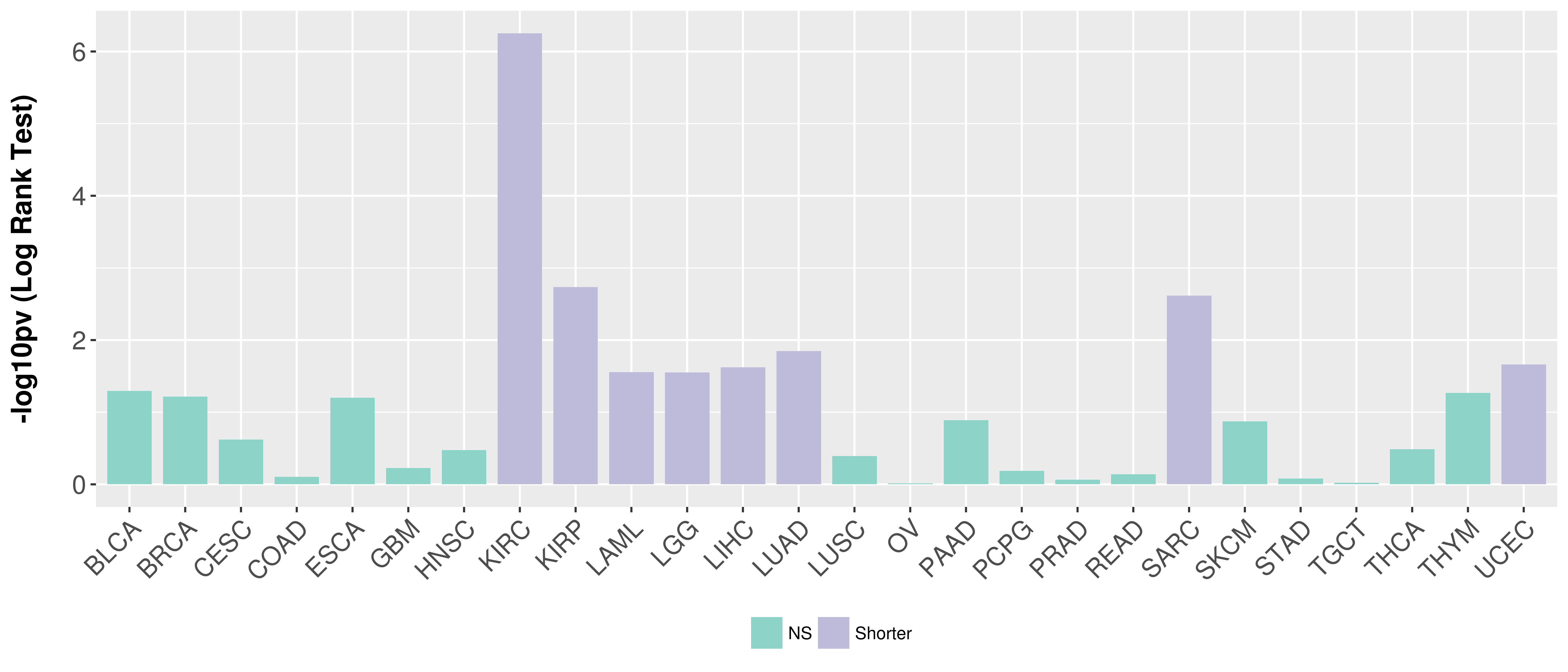
Overall survival analysis based on expression.
|
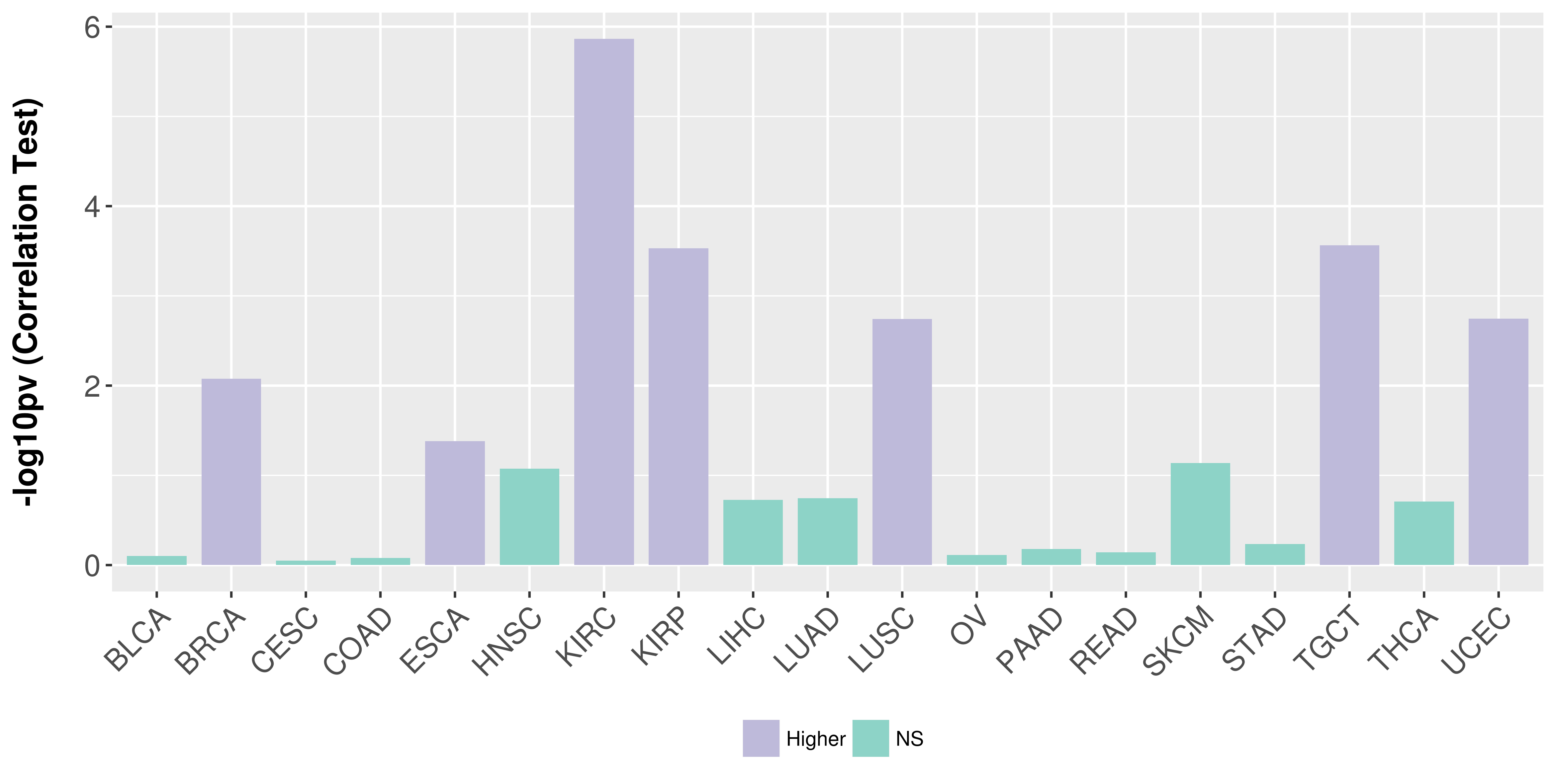
Association between expresson and stage.
|
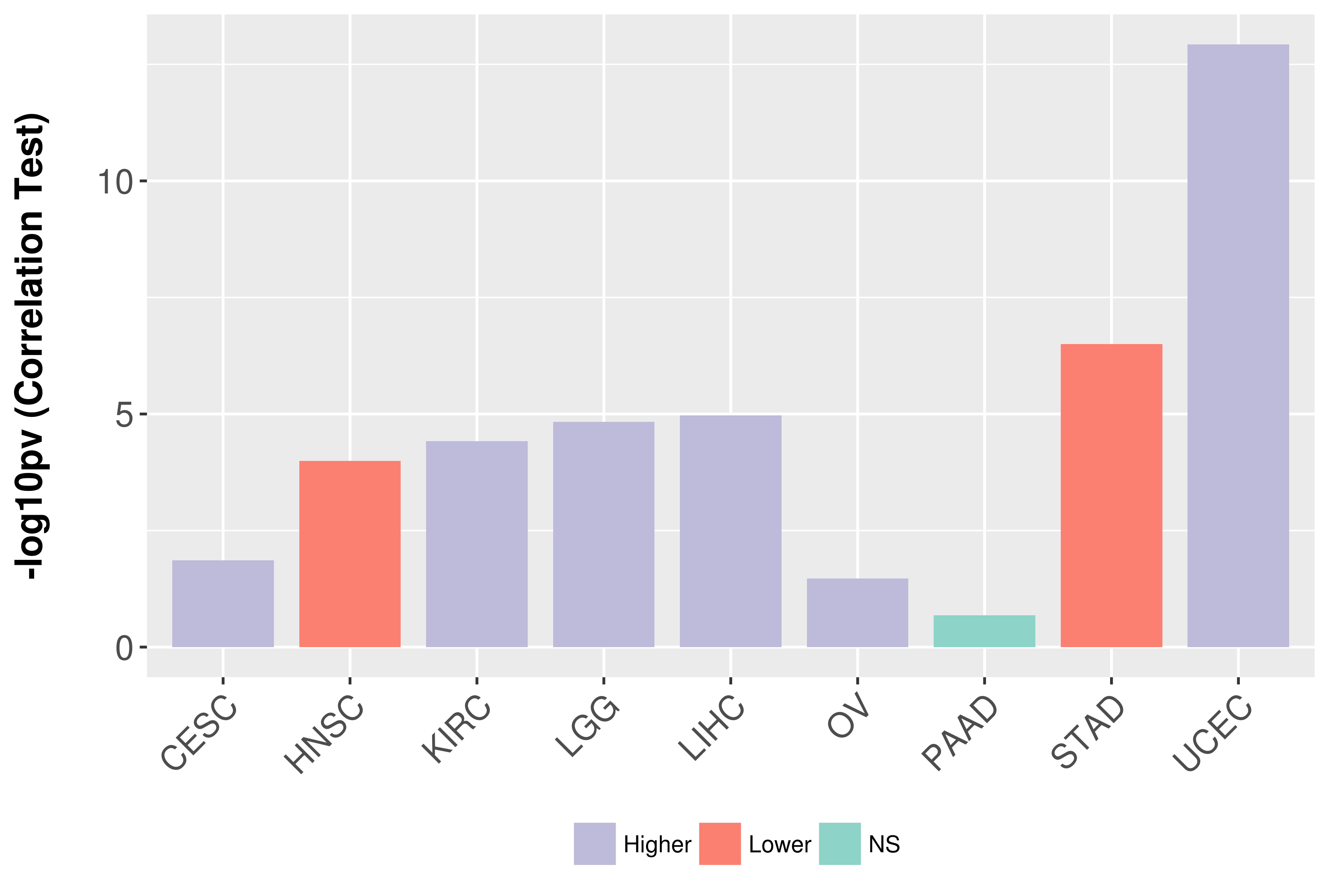
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for DNMT3B. |
| Summary | |
|---|---|
| Symbol | DNMT3B |
| Name | DNA methyltransferase 3 beta |
| Aliases | ICF; ICF1; M.HsaIIIB; DNA MTase HsaIIIB; DNA methyltransferase HsaIIIB; DNA (cytosine-5)-methyltransferase 3 ...... |
| Location | 20q11.21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|