Browse ERCC6 in pancancer
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00271 Helicase conserved C-terminal domain PF00176 SNF2 family N-terminal domain |
||||||||||
| Function |
Essential factor involved in transcription-coupled nucleotide excision repair which allows RNA polymerase II-blocking lesions to be rapidly removed from the transcribed strand of active genes. Upon DNA-binding, it locally modifies DNA conformation by wrapping the DNA around itself, thereby modifying the interface between stalled RNA polymerase II and DNA. It is required for transcription-coupled repair complex formation. It recruits the CSA complex (DCX(ERCC8) complex), nucleotide excision repair proteins and EP300 to the at sites of RNA polymerase II-blocking lesions. ; FUNCTION: Isoform 2: Like isoform 1, it is involved in repair of DNA damage, acting on its own or synergistically with isoform 1. May bind to PiggyBac transposable elements known as MER85 scattered in the genome and may therefore regulate the expression of nearby genes. Able to stimulate the antiviral response and modulate the expression of genes involved in metabolism regulation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000186 activation of MAPKK activity GO:0000187 activation of MAPK activity GO:0000302 response to reactive oxygen species GO:0000303 response to superoxide GO:0000305 response to oxygen radical GO:0001894 tissue homeostasis GO:0001895 retina homeostasis GO:0006283 transcription-coupled nucleotide-excision repair GO:0006284 base-excision repair GO:0006289 nucleotide-excision repair GO:0006290 pyrimidine dimer repair GO:0006354 DNA-templated transcription, elongation GO:0006360 transcription from RNA polymerase I promoter GO:0006362 transcription elongation from RNA polymerase I promoter GO:0006979 response to oxidative stress GO:0007254 JNK cascade GO:0007256 activation of JNKK activity GO:0007257 activation of JUN kinase activity GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0009636 response to toxic substance GO:0010035 response to inorganic substance GO:0010165 response to X-ray GO:0010212 response to ionizing radiation GO:0010224 response to UV-B GO:0010332 response to gamma radiation GO:0018108 peptidyl-tyrosine phosphorylation GO:0018212 peptidyl-tyrosine modification GO:0031098 stress-activated protein kinase signaling cascade GO:0032147 activation of protein kinase activity GO:0032784 regulation of DNA-templated transcription, elongation GO:0032786 positive regulation of DNA-templated transcription, elongation GO:0032872 regulation of stress-activated MAPK cascade GO:0032874 positive regulation of stress-activated MAPK cascade GO:0033674 positive regulation of kinase activity GO:0035264 multicellular organism growth GO:0040029 regulation of gene expression, epigenetic GO:0043405 regulation of MAP kinase activity GO:0043406 positive regulation of MAP kinase activity GO:0043410 positive regulation of MAPK cascade GO:0043506 regulation of JUN kinase activity GO:0043507 positive regulation of JUN kinase activity GO:0045494 photoreceptor cell maintenance GO:0045815 positive regulation of gene expression, epigenetic GO:0045860 positive regulation of protein kinase activity GO:0046328 regulation of JNK cascade GO:0046330 positive regulation of JNK cascade GO:0048871 multicellular organismal homeostasis GO:0050730 regulation of peptidyl-tyrosine phosphorylation GO:0050731 positive regulation of peptidyl-tyrosine phosphorylation GO:0051403 stress-activated MAPK cascade GO:0060249 anatomical structure homeostasis GO:0061097 regulation of protein tyrosine kinase activity GO:0061098 positive regulation of protein tyrosine kinase activity GO:0070302 regulation of stress-activated protein kinase signaling cascade GO:0070304 positive regulation of stress-activated protein kinase signaling cascade GO:0071900 regulation of protein serine/threonine kinase activity GO:0071902 positive regulation of protein serine/threonine kinase activity GO:0097193 intrinsic apoptotic signaling pathway |
| Molecular Function |
GO:0003682 chromatin binding GO:0008022 protein C-terminus binding GO:0008047 enzyme activator activity GO:0008094 DNA-dependent ATPase activity GO:0016887 ATPase activity GO:0019207 kinase regulator activity GO:0019209 kinase activator activity GO:0019887 protein kinase regulator activity GO:0030295 protein kinase activator activity GO:0030296 protein tyrosine kinase activator activity GO:0042623 ATPase activity, coupled GO:0047485 protein N-terminus binding |
| Cellular Component |
GO:0008023 transcription elongation factor complex |
| KEGG |
hsa03420 Nucleotide excision repair |
| Reactome |
R-HSA-5250924: B-WICH complex positively regulates rRNA expression R-HSA-73894: DNA Repair R-HSA-6782135: Dual incision in TC-NER R-HSA-427389: ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression R-HSA-212165: Epigenetic regulation of gene expression R-HSA-6781823: Formation of TC-NER Pre-Incision Complex R-HSA-6782210: Gap-filling DNA repair synthesis and ligation in TC-NER R-HSA-74160: Gene Expression R-HSA-5696398: Nucleotide Excision Repair R-HSA-5250913: Positive epigenetic regulation of rRNA expression R-HSA-73854: RNA Polymerase I Promoter Clearance R-HSA-73864: RNA Polymerase I Transcription R-HSA-73762: RNA Polymerase I Transcription Initiation R-HSA-504046: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription R-HSA-6781827: Transcription-Coupled Nucleotide Excision Repair (TC-NER) |
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
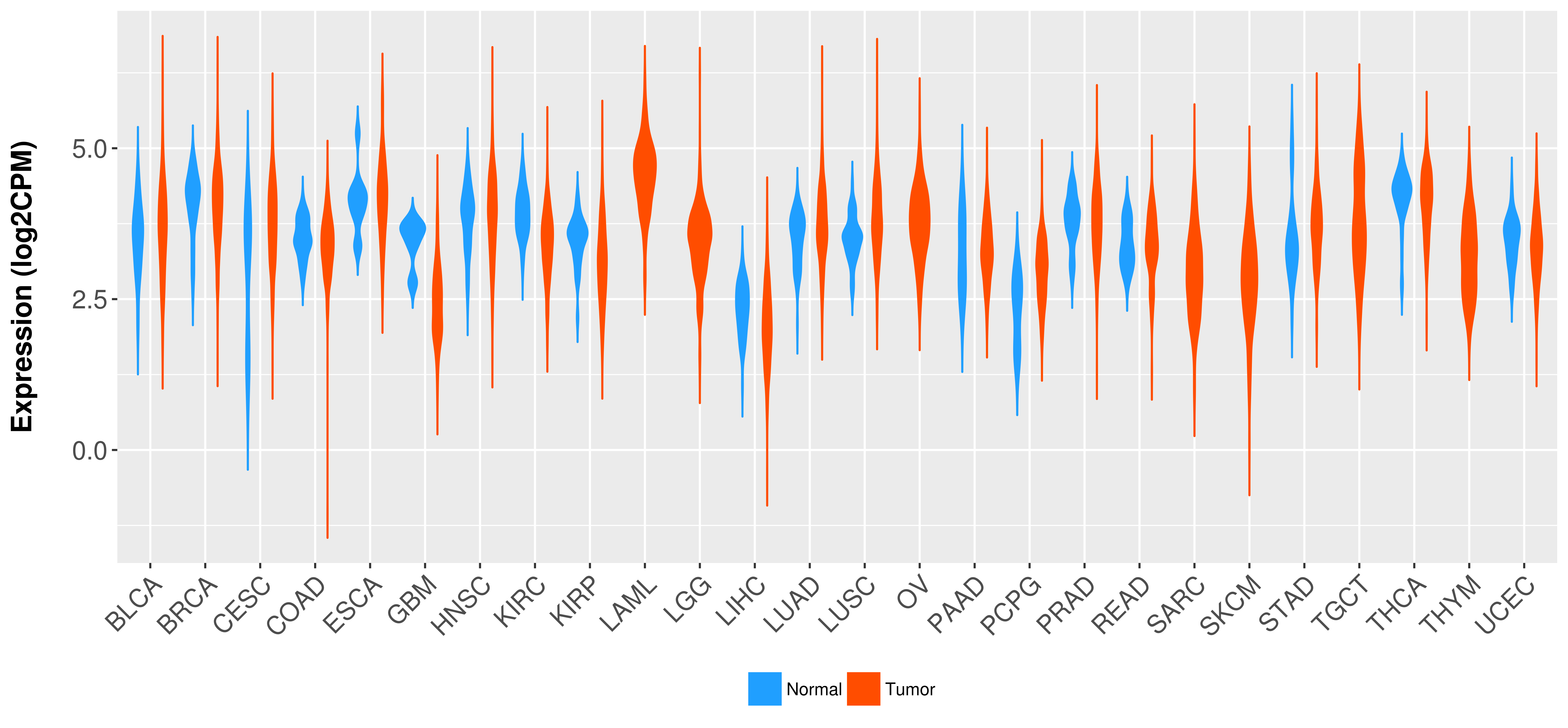
Differential expression analysis for cancers with more than 10 normal samples
|
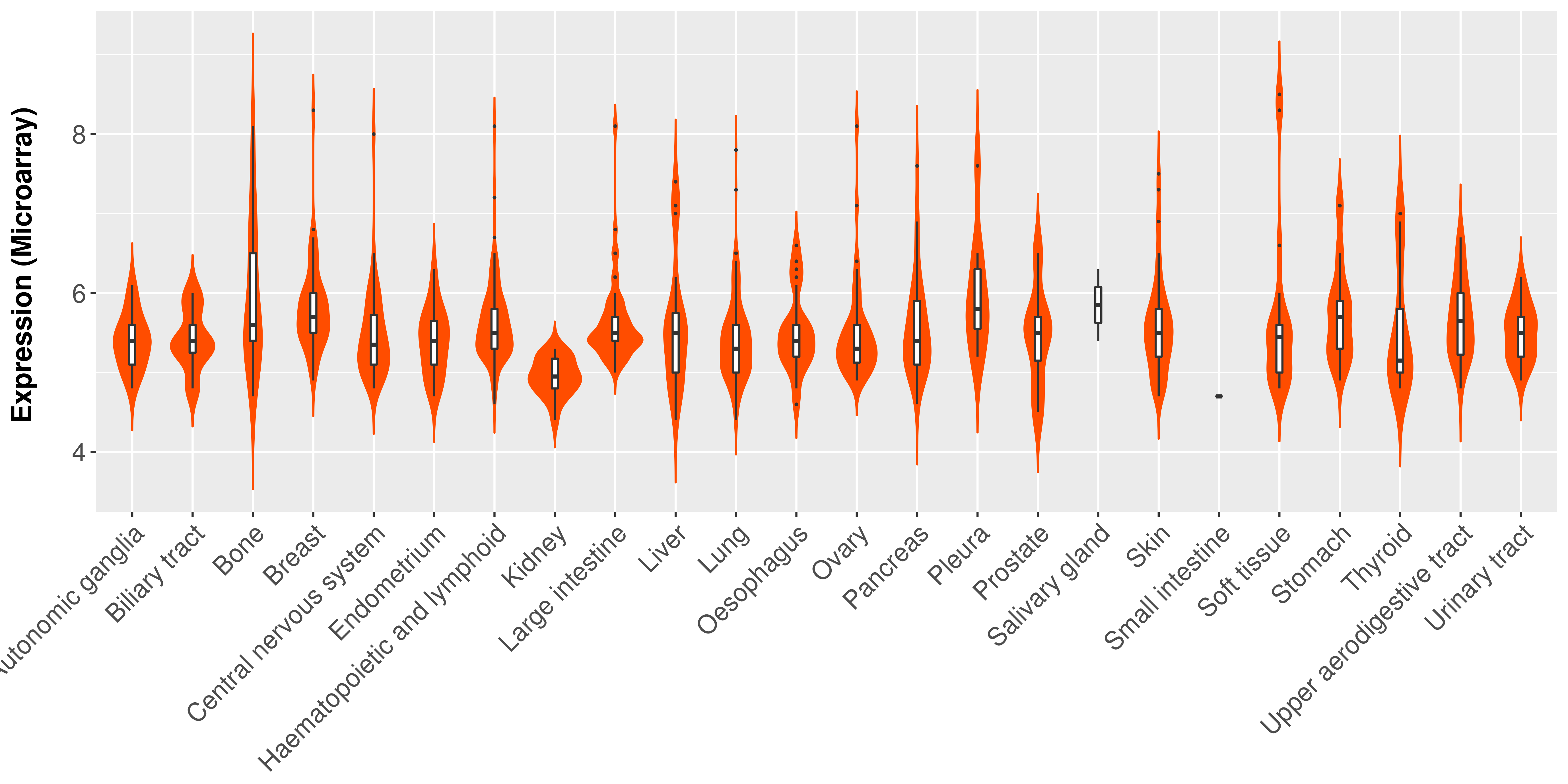
|

|
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
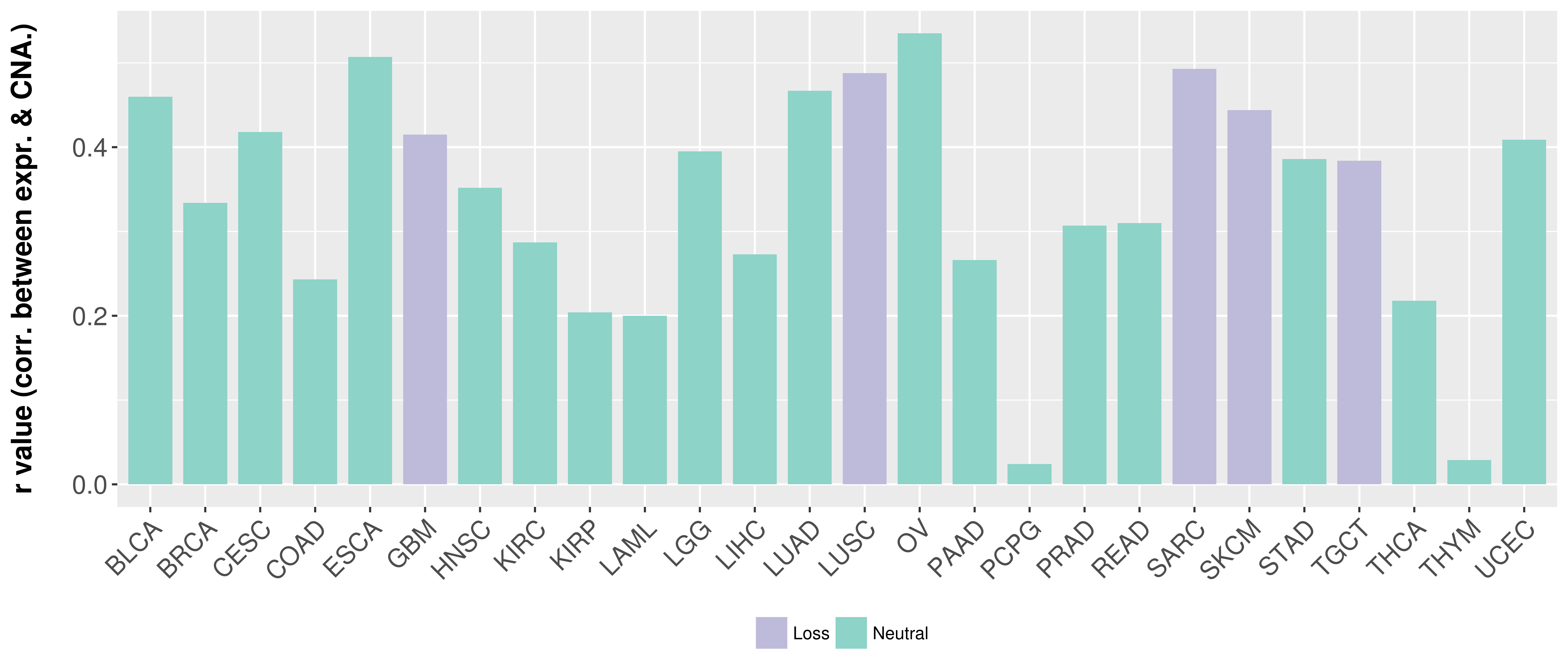
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
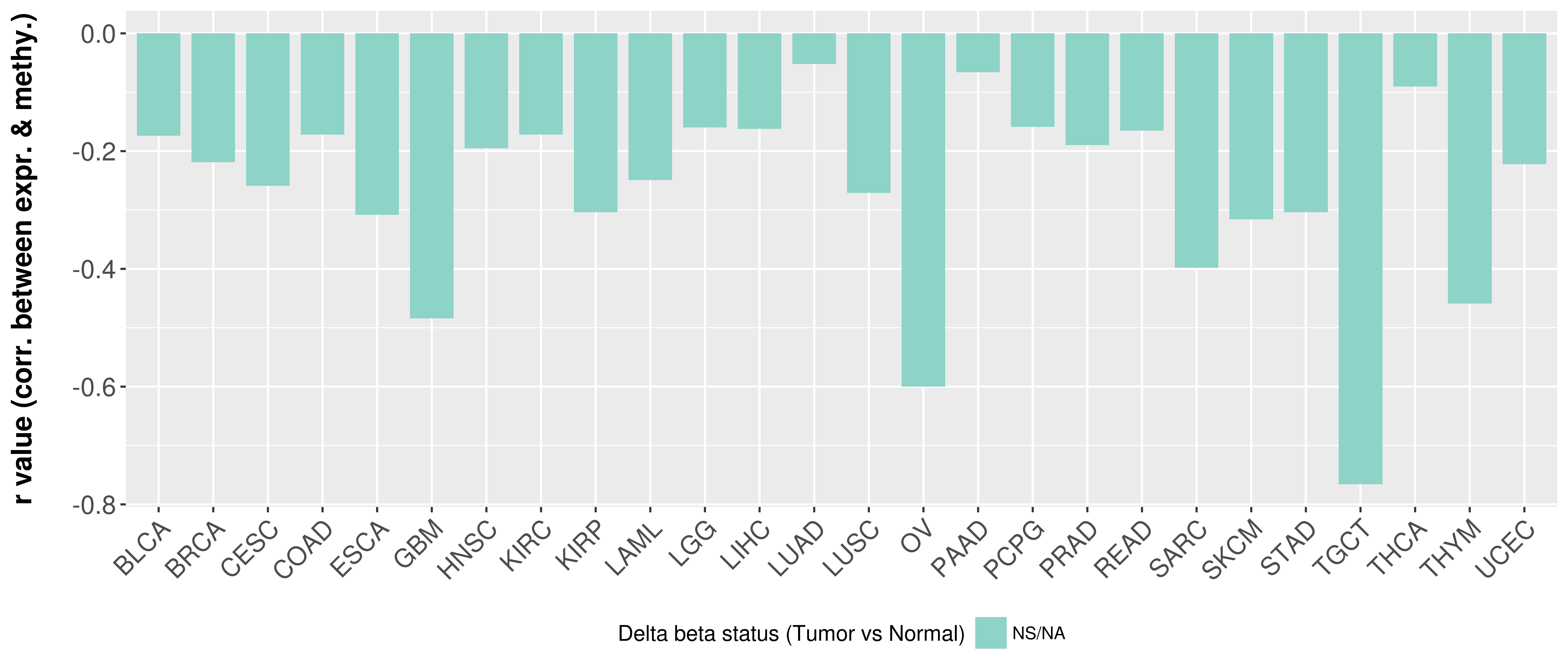
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
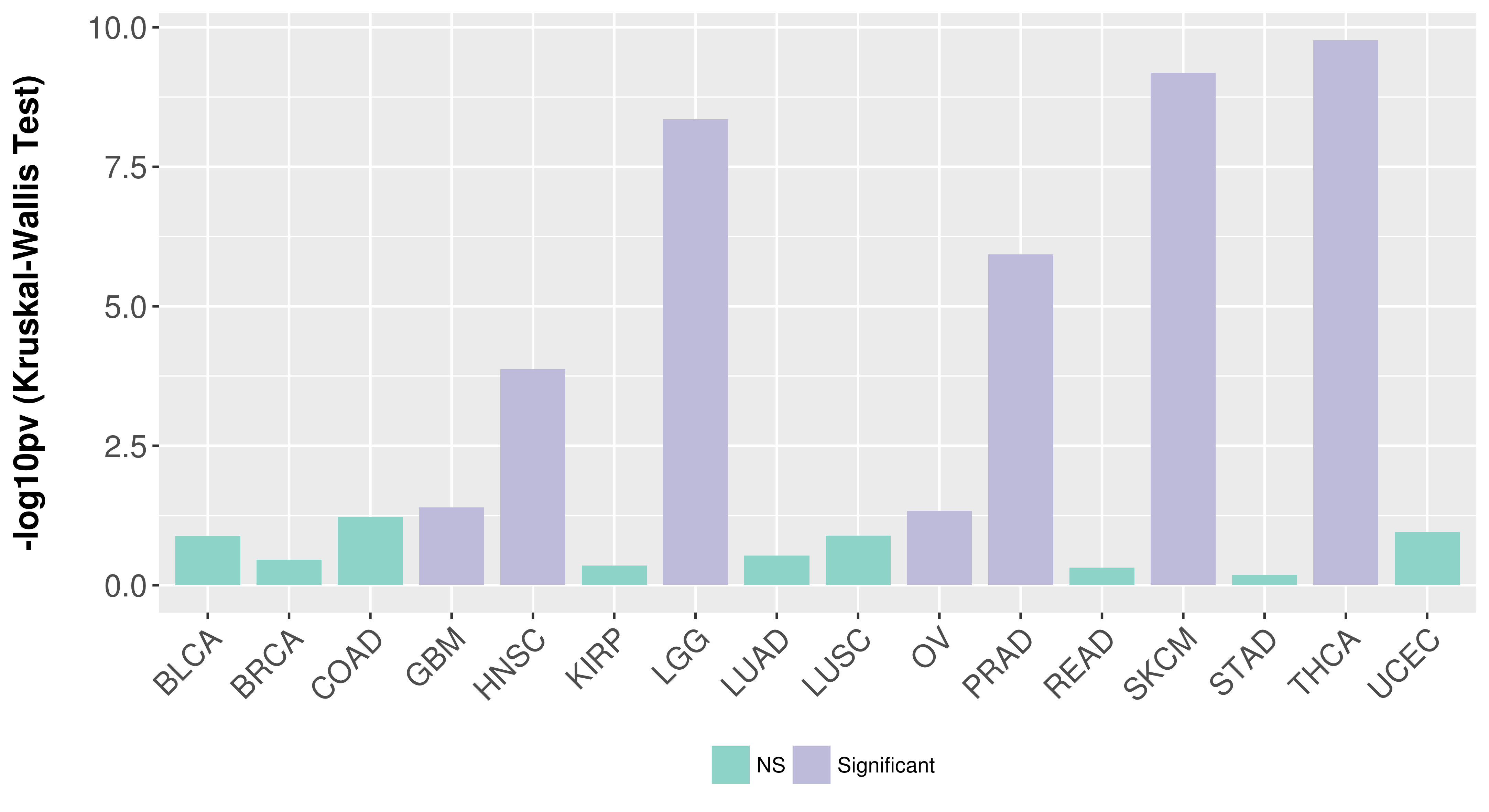
Association between expresson and subtype.
|
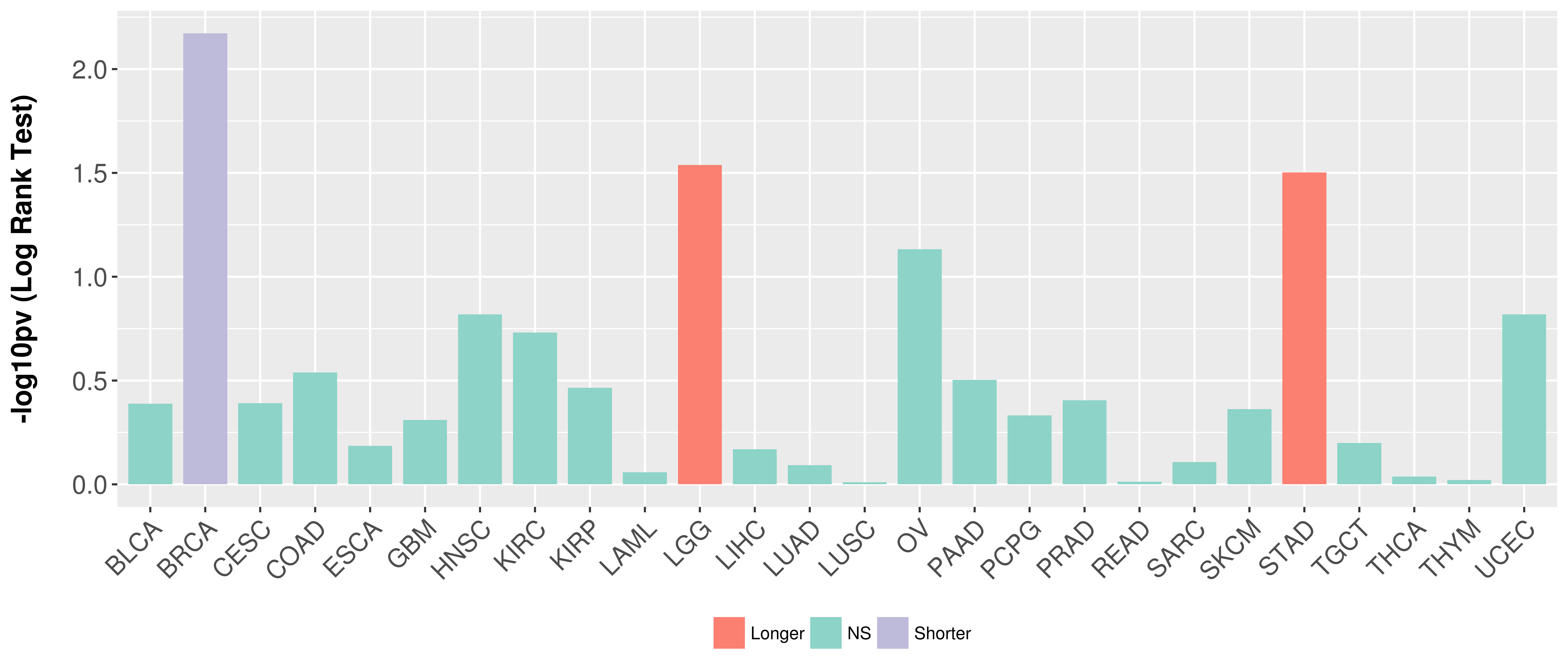
Overall survival analysis based on expression.
|
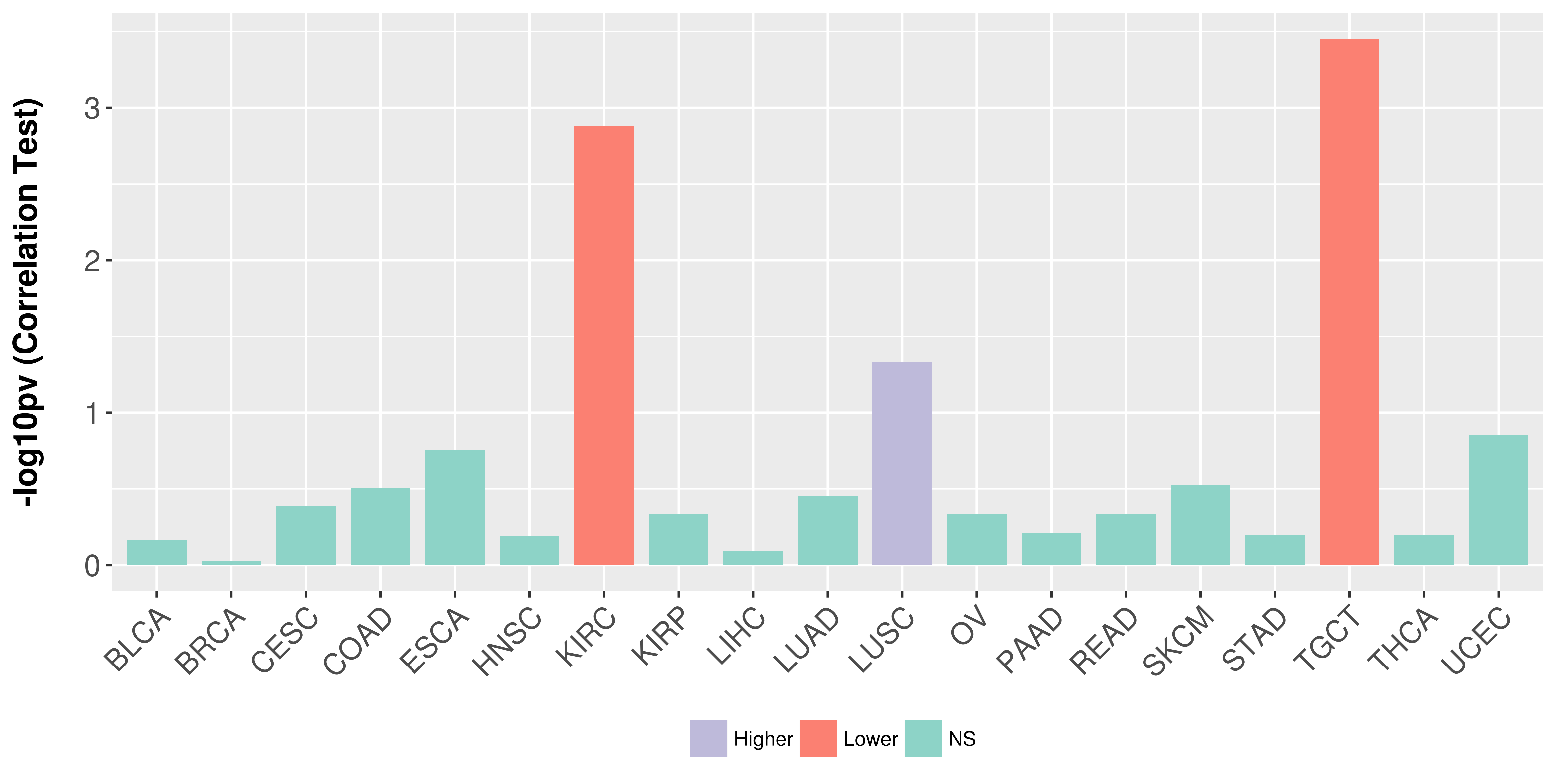
Association between expresson and stage.
|
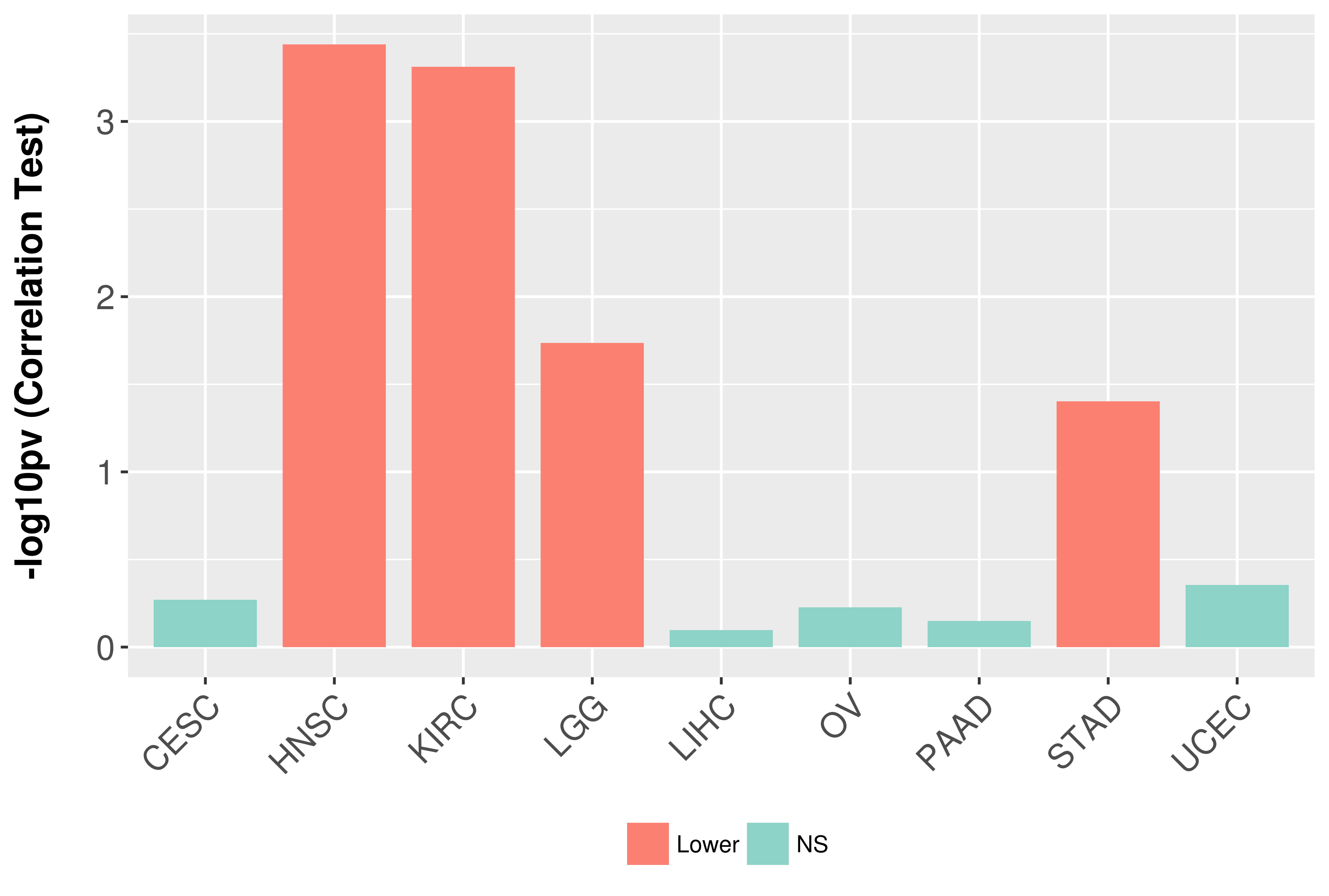
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ERCC6. |
| Summary | |
|---|---|
| Symbol | ERCC6 |
| Name | ERCC excision repair 6, chromatin remodeling factor |
| Aliases | RAD26; ARMD5; Cockayne syndrome B protein; CKN2; excision repair cross-complementing rodent repair deficienc ...... |
| Location | 10q11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for ERCC6. |