Browse HDAC4 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF12203 Glutamine rich N terminal domain of histone deacetylase 4 PF00850 Histone deacetylase domain |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Involved in muscle maturation via its interaction with the myocyte enhancer factors such as MEF2A, MEF2C and MEF2D. Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer. Deacetylates HSPA1A and HSPA1B at 'Lys-77' leading to their preferential binding to co-chaperone STUB1 (PubMed:27708256). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001501 skeletal system development GO:0001503 ossification GO:0001649 osteoblast differentiation GO:0002076 osteoblast development GO:0002521 leukocyte differentiation GO:0003012 muscle system process GO:0003013 circulatory system process GO:0003015 heart process GO:0003299 muscle hypertrophy in response to stress GO:0003300 cardiac muscle hypertrophy GO:0006090 pyruvate metabolic process GO:0006091 generation of precursor metabolites and energy GO:0006096 glycolytic process GO:0006109 regulation of carbohydrate metabolic process GO:0006110 regulation of glycolytic process GO:0006140 regulation of nucleotide metabolic process GO:0006165 nucleoside diphosphate phosphorylation GO:0006338 chromatin remodeling GO:0006476 protein deacetylation GO:0006732 coenzyme metabolic process GO:0006733 oxidoreduction coenzyme metabolic process GO:0006757 ATP generation from ADP GO:0006936 muscle contraction GO:0006937 regulation of muscle contraction GO:0006941 striated muscle contraction GO:0006942 regulation of striated muscle contraction GO:0007517 muscle organ development GO:0007519 skeletal muscle tissue development GO:0008015 blood circulation GO:0008016 regulation of heart contraction GO:0009116 nucleoside metabolic process GO:0009118 regulation of nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009123 nucleoside monophosphate metabolic process GO:0009126 purine nucleoside monophosphate metabolic process GO:0009132 nucleoside diphosphate metabolic process GO:0009135 purine nucleoside diphosphate metabolic process GO:0009141 nucleoside triphosphate metabolic process GO:0009144 purine nucleoside triphosphate metabolic process GO:0009150 purine ribonucleotide metabolic process GO:0009161 ribonucleoside monophosphate metabolic process GO:0009167 purine ribonucleoside monophosphate metabolic process GO:0009179 purine ribonucleoside diphosphate metabolic process GO:0009185 ribonucleoside diphosphate metabolic process GO:0009199 ribonucleoside triphosphate metabolic process GO:0009205 purine ribonucleoside triphosphate metabolic process GO:0009612 response to mechanical stimulus GO:0009894 regulation of catabolic process GO:0009895 negative regulation of catabolic process GO:0010591 regulation of lamellipodium assembly GO:0010592 positive regulation of lamellipodium assembly GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010677 negative regulation of cellular carbohydrate metabolic process GO:0010830 regulation of myotube differentiation GO:0010832 negative regulation of myotube differentiation GO:0010882 regulation of cardiac muscle contraction by calcium ion signaling GO:0014706 striated muscle tissue development GO:0014812 muscle cell migration GO:0014854 response to inactivity GO:0014870 response to muscle inactivity GO:0014874 response to stimulus involved in regulation of muscle adaptation GO:0014877 response to muscle inactivity involved in regulation of muscle adaptation GO:0014887 cardiac muscle adaptation GO:0014888 striated muscle adaptation GO:0014894 response to denervation involved in regulation of muscle adaptation GO:0014896 muscle hypertrophy GO:0014897 striated muscle hypertrophy GO:0014898 cardiac muscle hypertrophy in response to stress GO:0014902 myotube differentiation GO:0014904 myotube cell development GO:0014909 smooth muscle cell migration GO:0014910 regulation of smooth muscle cell migration GO:0014911 positive regulation of smooth muscle cell migration GO:0016052 carbohydrate catabolic process GO:0016202 regulation of striated muscle tissue development GO:0016570 histone modification GO:0016575 histone deacetylation GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0019362 pyridine nucleotide metabolic process GO:0019722 calcium-mediated signaling GO:0019932 second-messenger-mediated signaling GO:0030031 cell projection assembly GO:0030032 lamellipodium assembly GO:0030098 lymphocyte differentiation GO:0030183 B cell differentiation GO:0030278 regulation of ossification GO:0030279 negative regulation of ossification GO:0030335 positive regulation of cell migration GO:0031346 positive regulation of cell projection organization GO:0033002 muscle cell proliferation GO:0033233 regulation of protein sumoylation GO:0033235 positive regulation of protein sumoylation GO:0034612 response to tumor necrosis factor GO:0034983 peptidyl-lysine deacetylation GO:0035601 protein deacylation GO:0040017 positive regulation of locomotion GO:0040029 regulation of gene expression, epigenetic GO:0042113 B cell activation GO:0042278 purine nucleoside metabolic process GO:0042326 negative regulation of phosphorylation GO:0042493 response to drug GO:0042692 muscle cell differentiation GO:0043393 regulation of protein binding GO:0043433 negative regulation of sequence-specific DNA binding transcription factor activity GO:0043467 regulation of generation of precursor metabolites and energy GO:0043470 regulation of carbohydrate catabolic process GO:0043500 muscle adaptation GO:0043502 regulation of muscle adaptation GO:0043523 regulation of neuron apoptotic process GO:0043525 positive regulation of neuron apoptotic process GO:0044057 regulation of system process GO:0044089 positive regulation of cellular component biogenesis GO:0044262 cellular carbohydrate metabolic process GO:0044723 single-organism carbohydrate metabolic process GO:0044724 single-organism carbohydrate catabolic process GO:0045667 regulation of osteoblast differentiation GO:0045668 negative regulation of osteoblast differentiation GO:0045820 negative regulation of glycolytic process GO:0045912 negative regulation of carbohydrate metabolic process GO:0045978 negative regulation of nucleoside metabolic process GO:0045980 negative regulation of nucleotide metabolic process GO:0046031 ADP metabolic process GO:0046034 ATP metabolic process GO:0046128 purine ribonucleoside metabolic process GO:0046496 nicotinamide nucleotide metabolic process GO:0046939 nucleotide phosphorylation GO:0048634 regulation of muscle organ development GO:0048641 regulation of skeletal muscle tissue development GO:0048659 smooth muscle cell proliferation GO:0048660 regulation of smooth muscle cell proliferation GO:0048661 positive regulation of smooth muscle cell proliferation GO:0048741 skeletal muscle fiber development GO:0048742 regulation of skeletal muscle fiber development GO:0048747 muscle fiber development GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051098 regulation of binding GO:0051146 striated muscle cell differentiation GO:0051147 regulation of muscle cell differentiation GO:0051148 negative regulation of muscle cell differentiation GO:0051153 regulation of striated muscle cell differentiation GO:0051154 negative regulation of striated muscle cell differentiation GO:0051186 cofactor metabolic process GO:0051193 regulation of cofactor metabolic process GO:0051195 negative regulation of cofactor metabolic process GO:0051196 regulation of coenzyme metabolic process GO:0051198 negative regulation of coenzyme metabolic process GO:0051272 positive regulation of cellular component movement GO:0051402 neuron apoptotic process GO:0055001 muscle cell development GO:0055002 striated muscle cell development GO:0055117 regulation of cardiac muscle contraction GO:0060047 heart contraction GO:0060048 cardiac muscle contraction GO:0060491 regulation of cell projection assembly GO:0060537 muscle tissue development GO:0060538 skeletal muscle organ development GO:0070555 response to interleukin-1 GO:0070932 histone H3 deacetylation GO:0070933 histone H4 deacetylation GO:0070997 neuron death GO:0071107 response to parathyroid hormone GO:0071214 cellular response to abiotic stimulus GO:0071260 cellular response to mechanical stimulus GO:0071356 cellular response to tumor necrosis factor GO:0071374 cellular response to parathyroid hormone stimulus GO:0071496 cellular response to external stimulus GO:0072524 pyridine-containing compound metabolic process GO:0072593 reactive oxygen species metabolic process GO:0090257 regulation of muscle system process GO:0097581 lamellipodium organization GO:0098732 macromolecule deacylation GO:1900542 regulation of purine nucleotide metabolic process GO:1900543 negative regulation of purine nucleotide metabolic process GO:1901214 regulation of neuron death GO:1901216 positive regulation of neuron death GO:1901657 glycosyl compound metabolic process GO:1901861 regulation of muscle tissue development GO:1902743 regulation of lamellipodium organization GO:1902745 positive regulation of lamellipodium organization GO:1903320 regulation of protein modification by small protein conjugation or removal GO:1903322 positive regulation of protein modification by small protein conjugation or removal GO:1903409 reactive oxygen species biosynthetic process GO:1903426 regulation of reactive oxygen species biosynthetic process GO:1903428 positive regulation of reactive oxygen species biosynthetic process GO:1903522 regulation of blood circulation GO:1903578 regulation of ATP metabolic process GO:1903579 negative regulation of ATP metabolic process GO:2000147 positive regulation of cell motility GO:2000377 regulation of reactive oxygen species metabolic process GO:2000379 positive regulation of reactive oxygen species metabolic process |
| Molecular Function |
GO:0001025 RNA polymerase III transcription factor binding GO:0001047 core promoter binding GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0030955 potassium ion binding GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0031420 alkali metal ion binding GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0033558 protein deacetylase activity GO:0033613 activating transcription factor binding GO:0034979 NAD-dependent protein deacetylase activity GO:0042826 histone deacetylase binding GO:0070491 repressing transcription factor binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0015629 actin cytoskeleton GO:0017053 transcriptional repressor complex GO:0030016 myofibril GO:0030017 sarcomere GO:0030018 Z disc GO:0031594 neuromuscular junction GO:0031672 A band GO:0031674 I band GO:0042641 actomyosin GO:0043292 contractile fiber GO:0044449 contractile fiber part |
| KEGG | - |
| Reactome |
R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer |
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
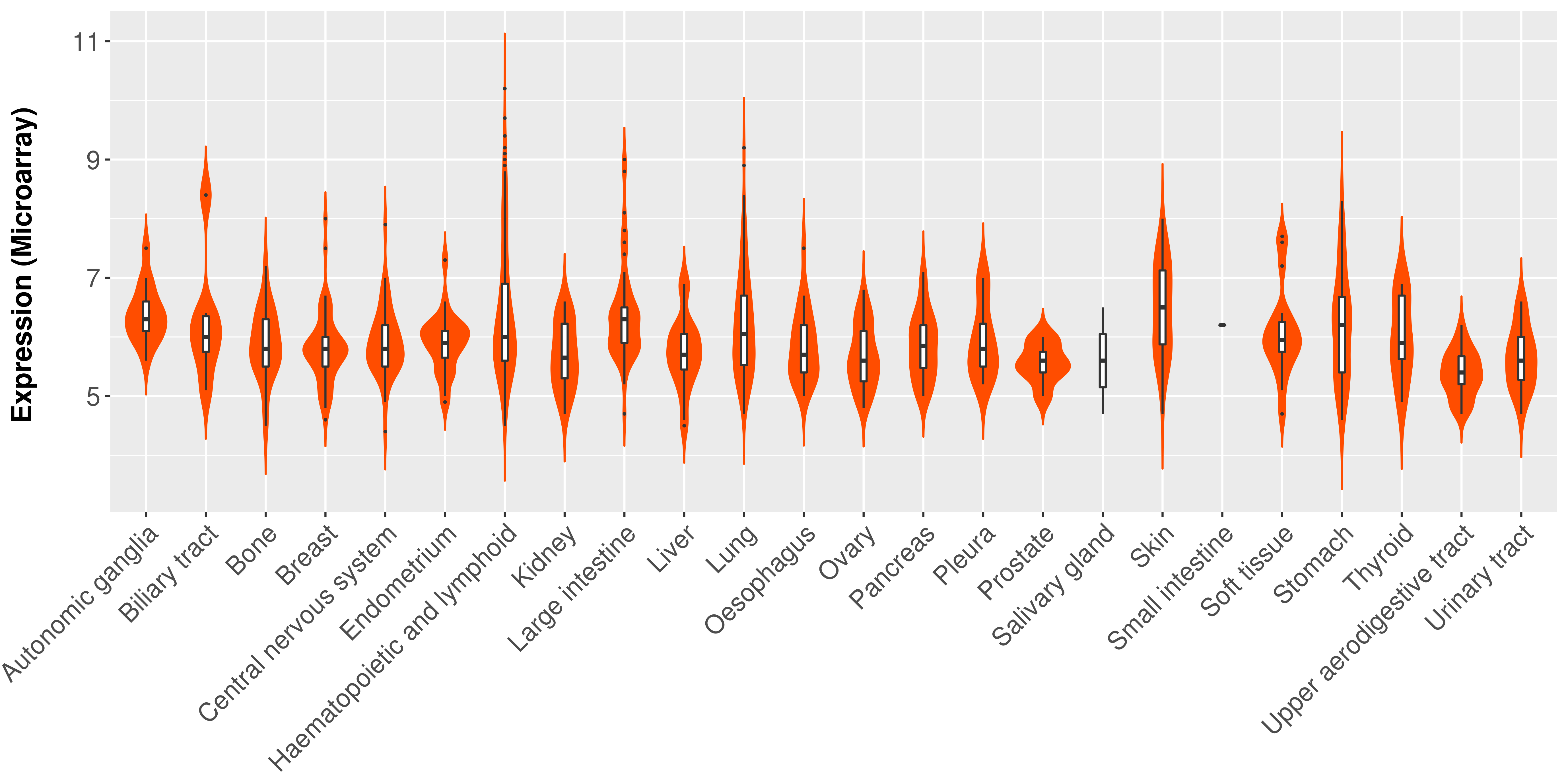
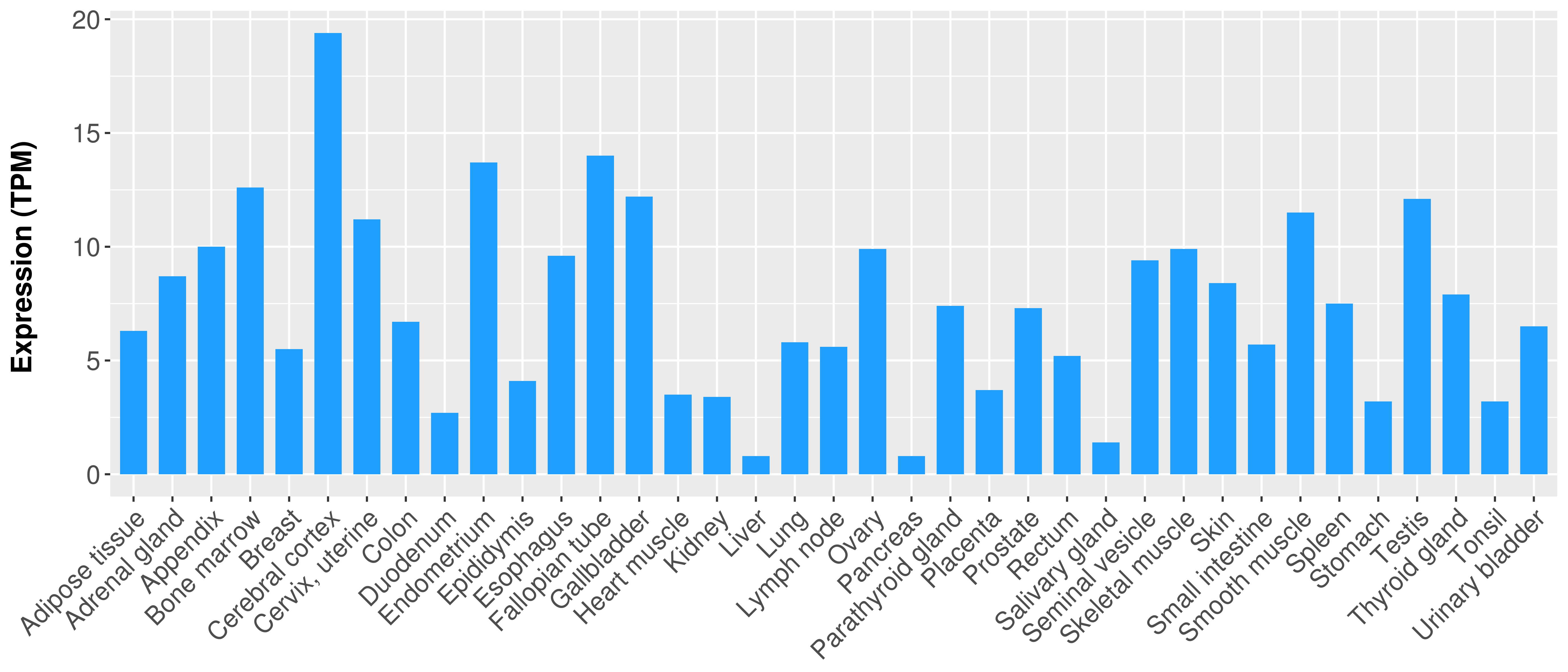
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
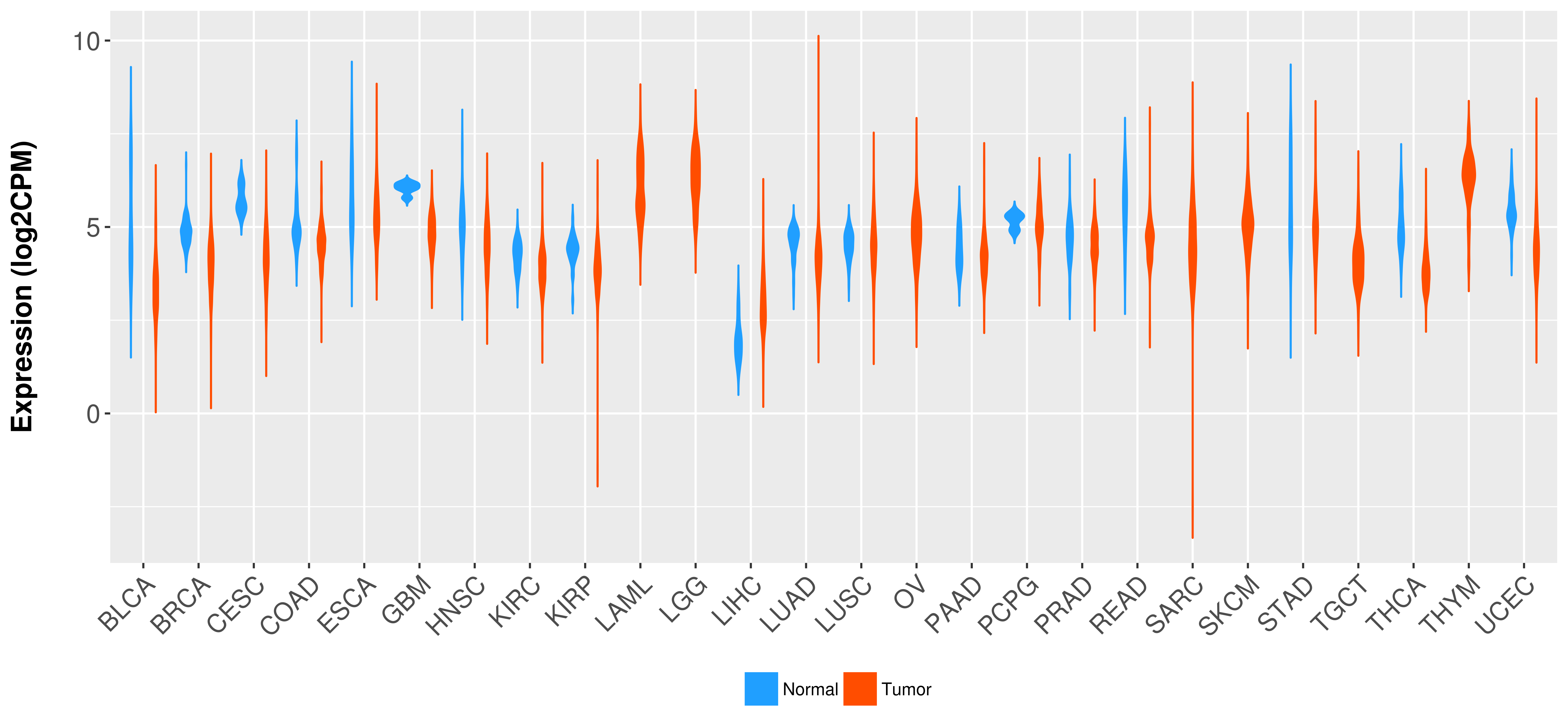
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
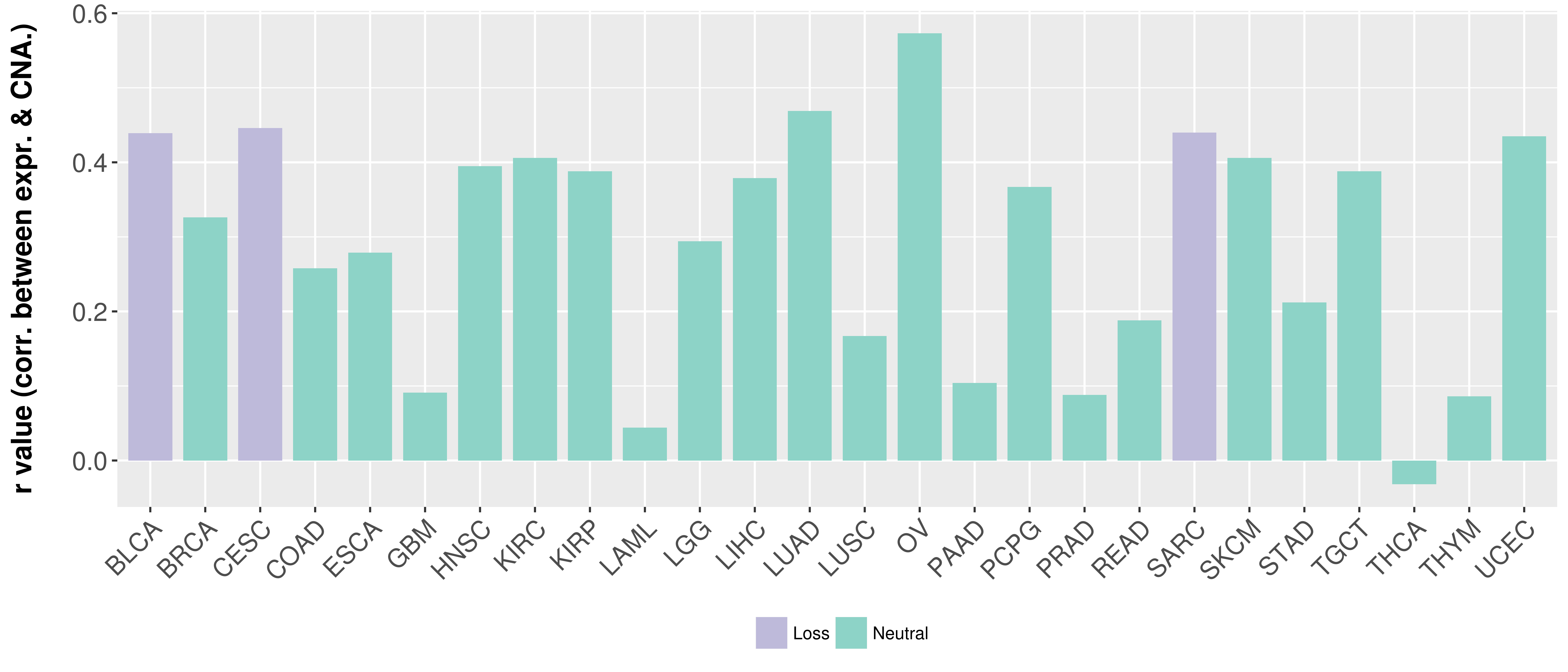
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
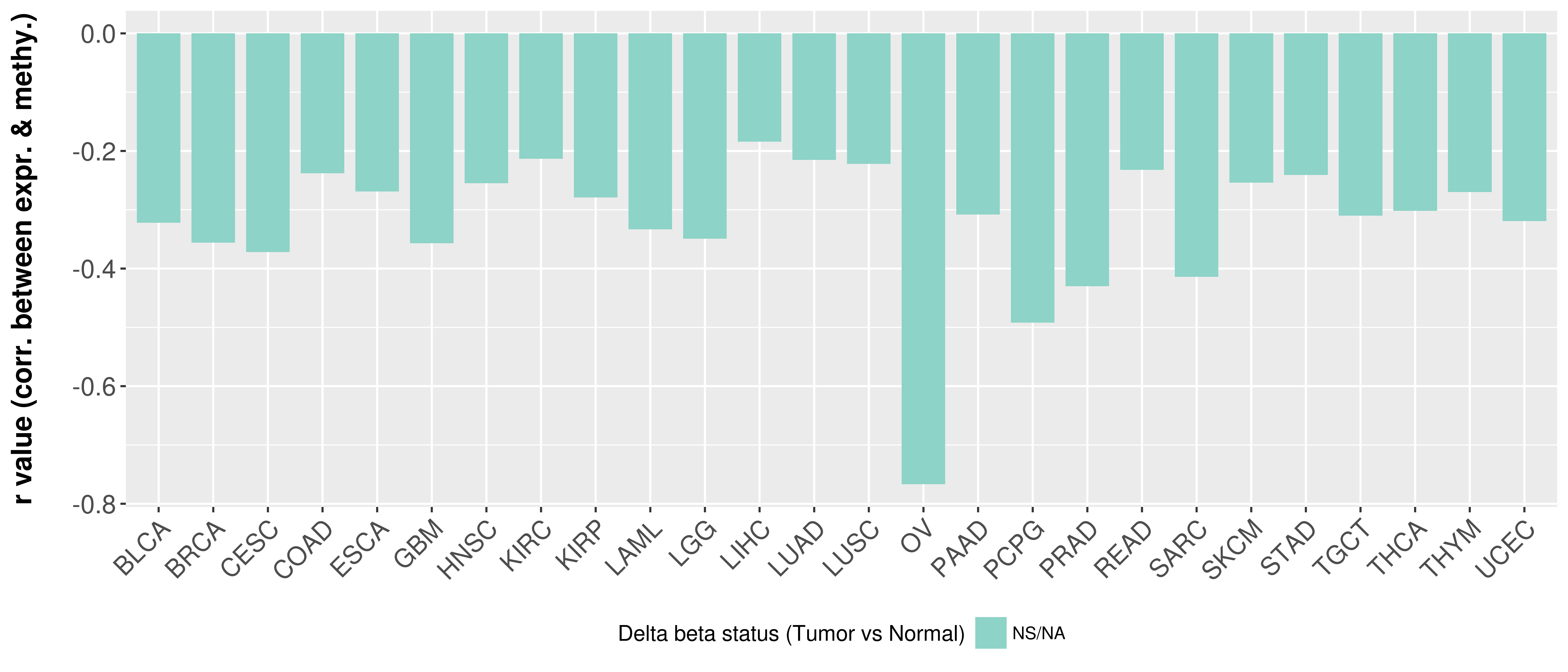
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
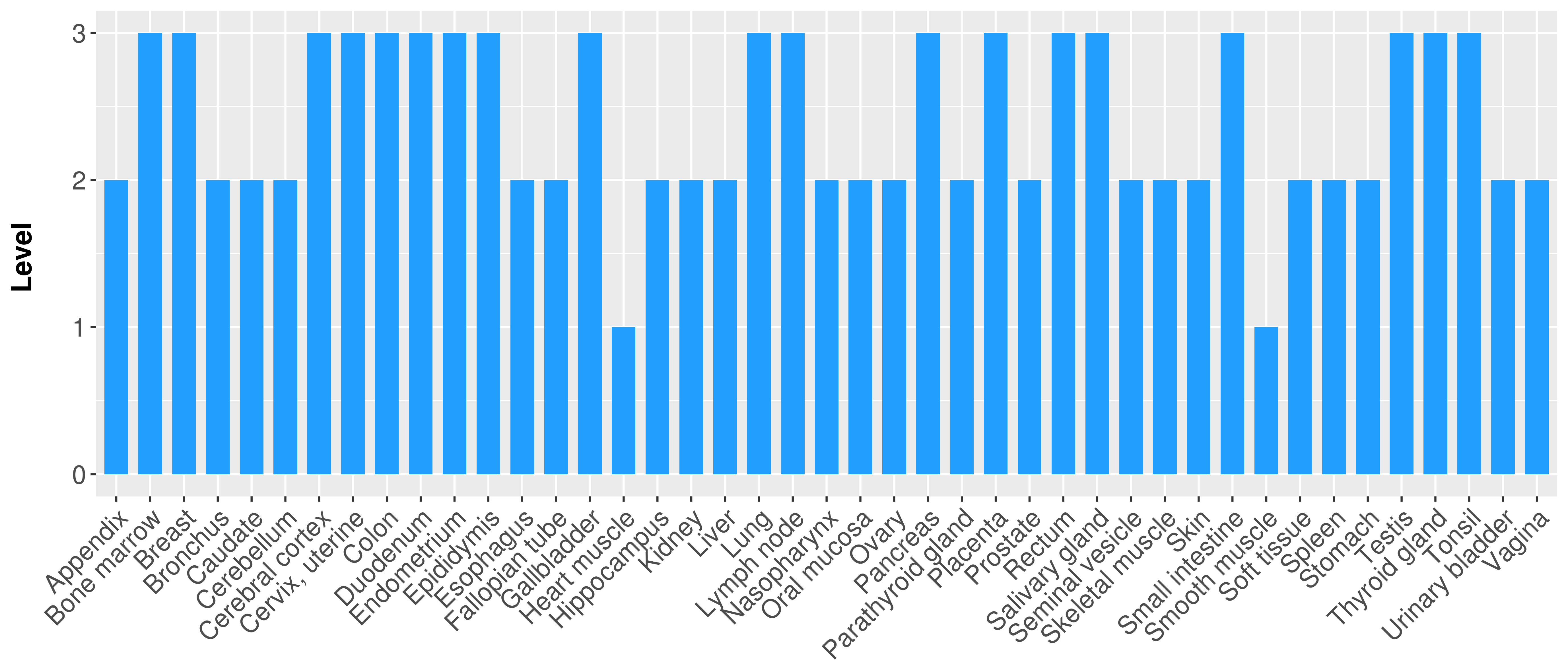
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
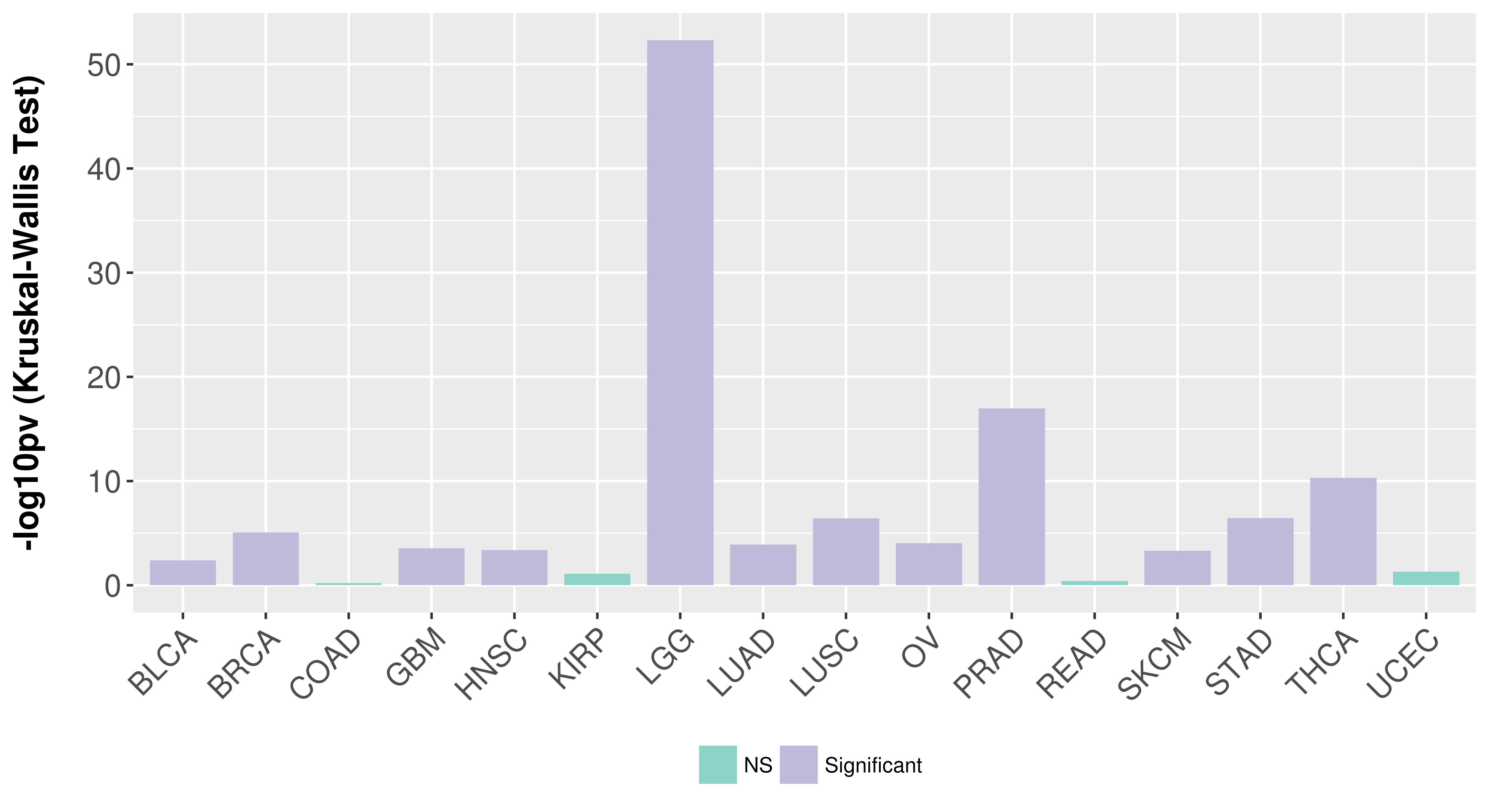
Association between expresson and subtype.
|
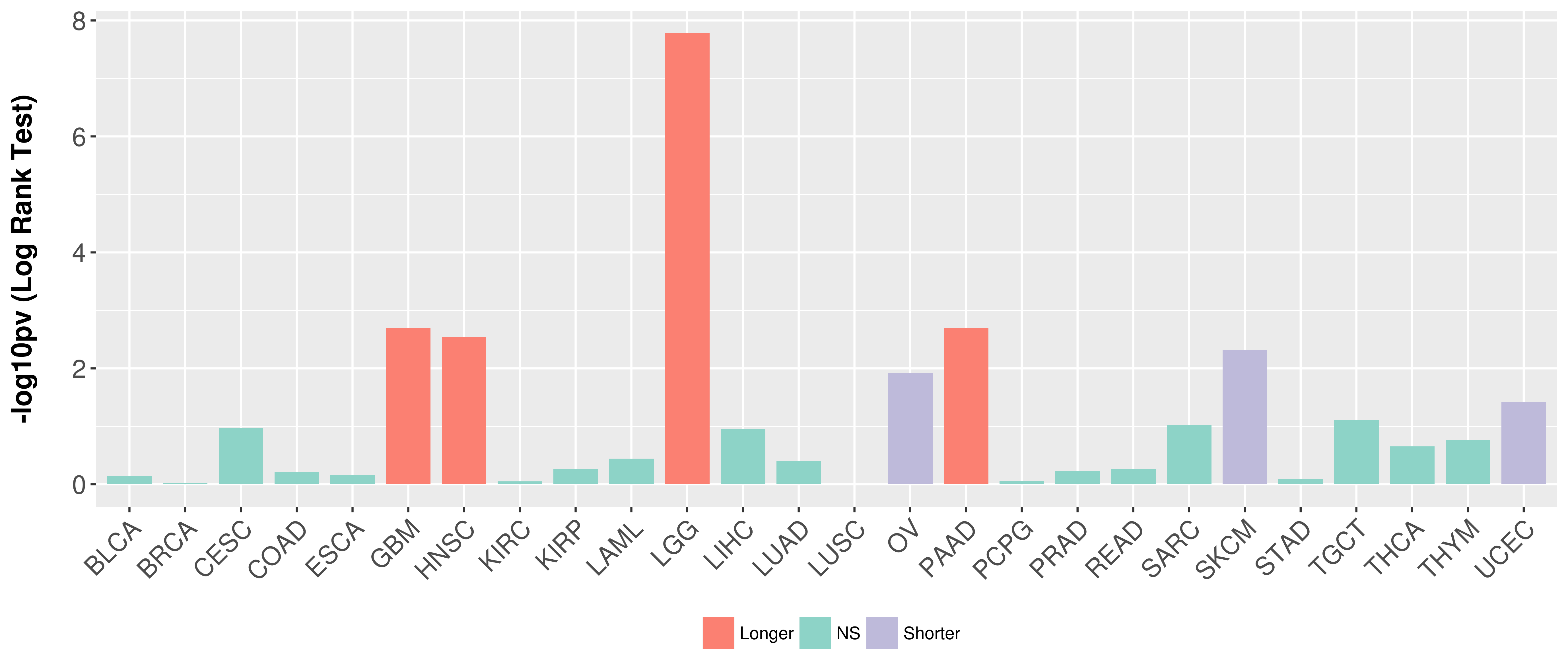
Overall survival analysis based on expression.
|
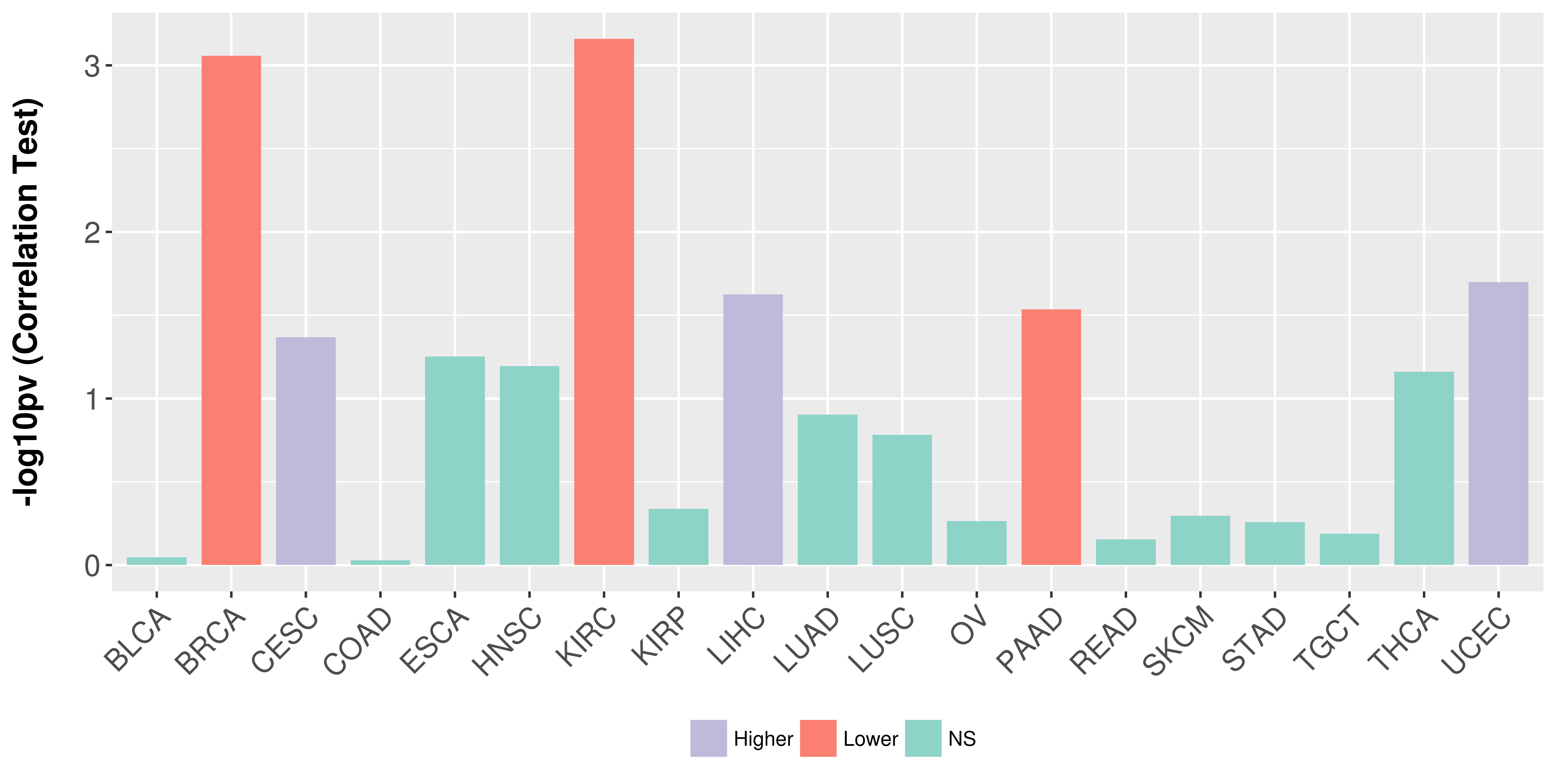
Association between expresson and stage.
|
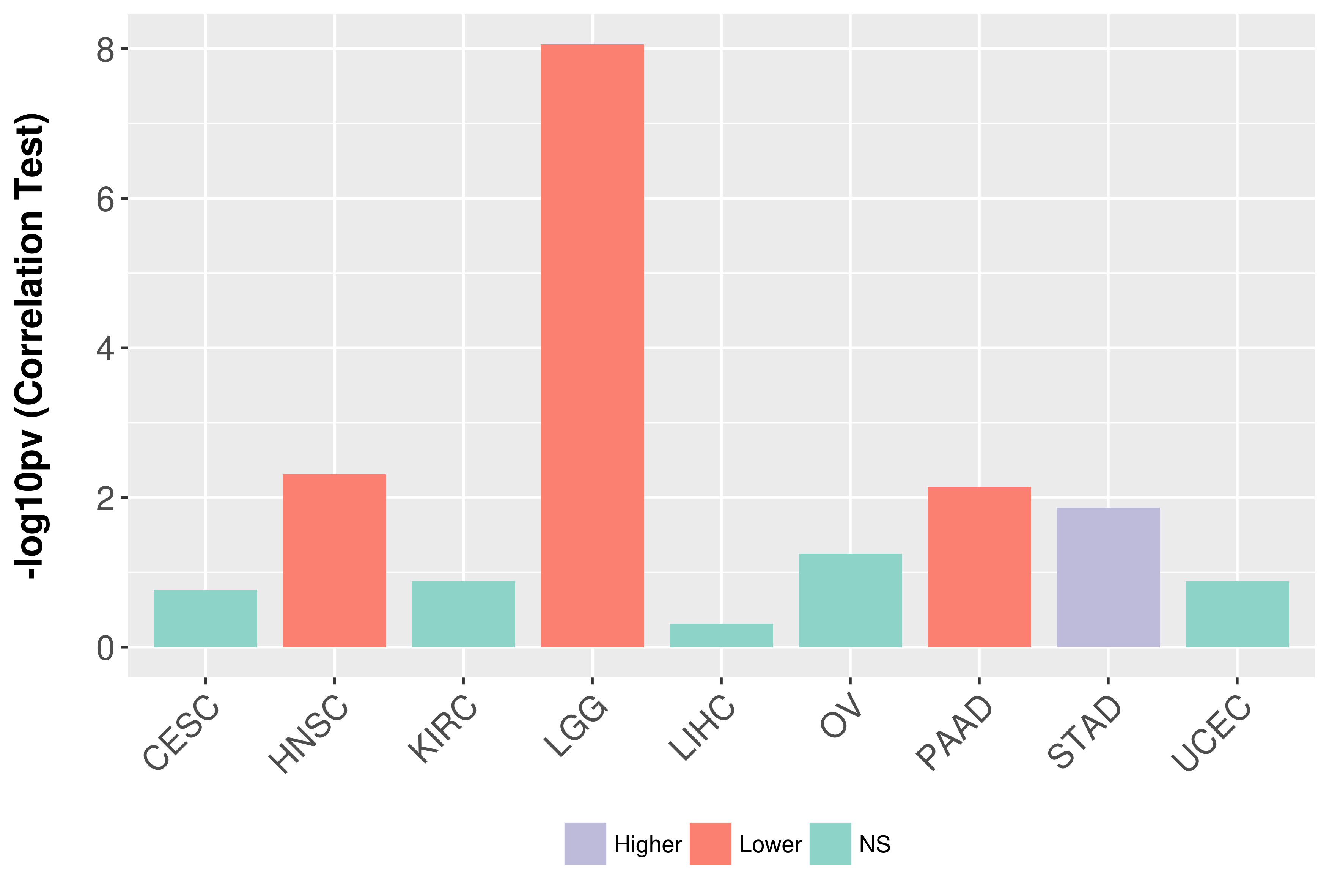
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
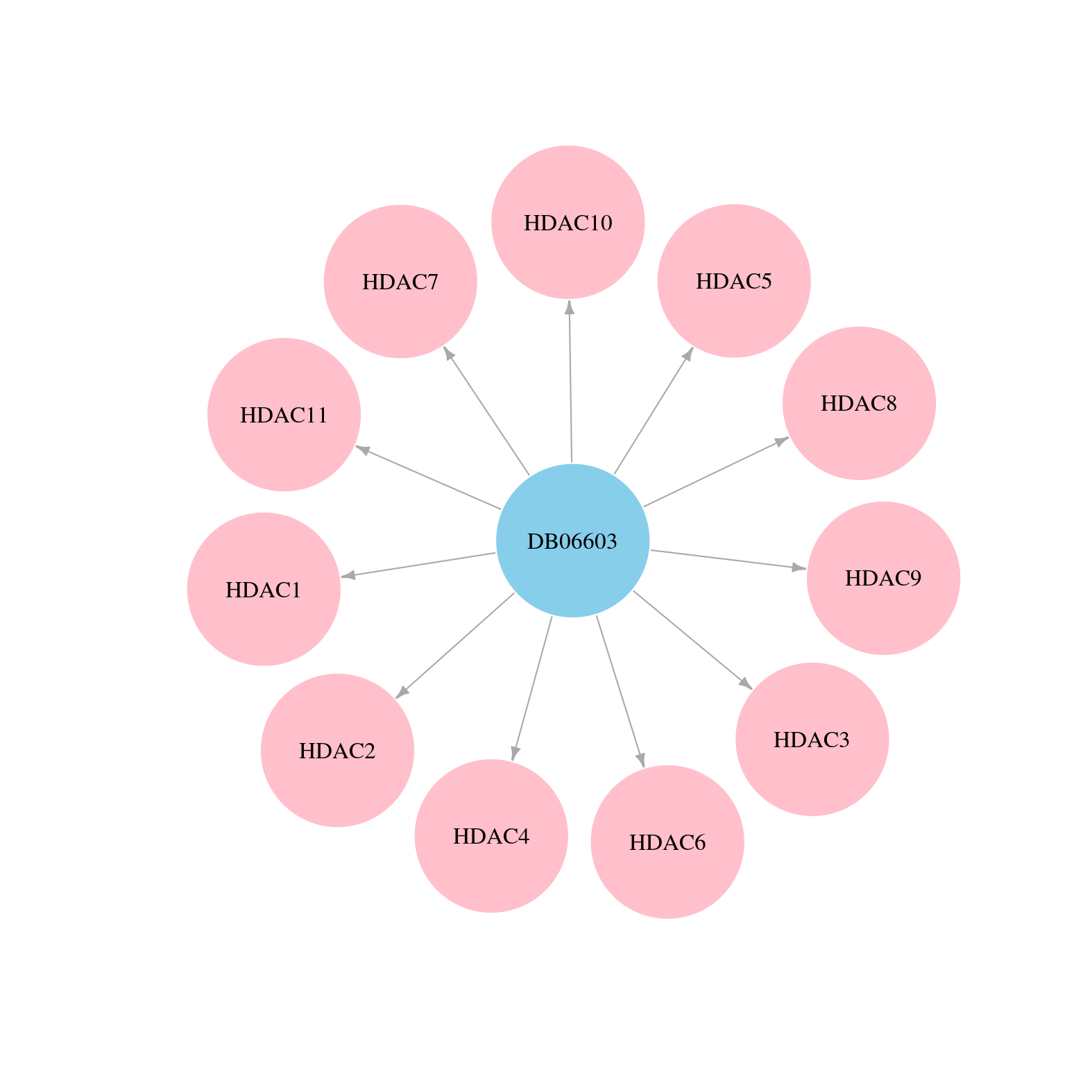
> Drugs from DrugBank database |
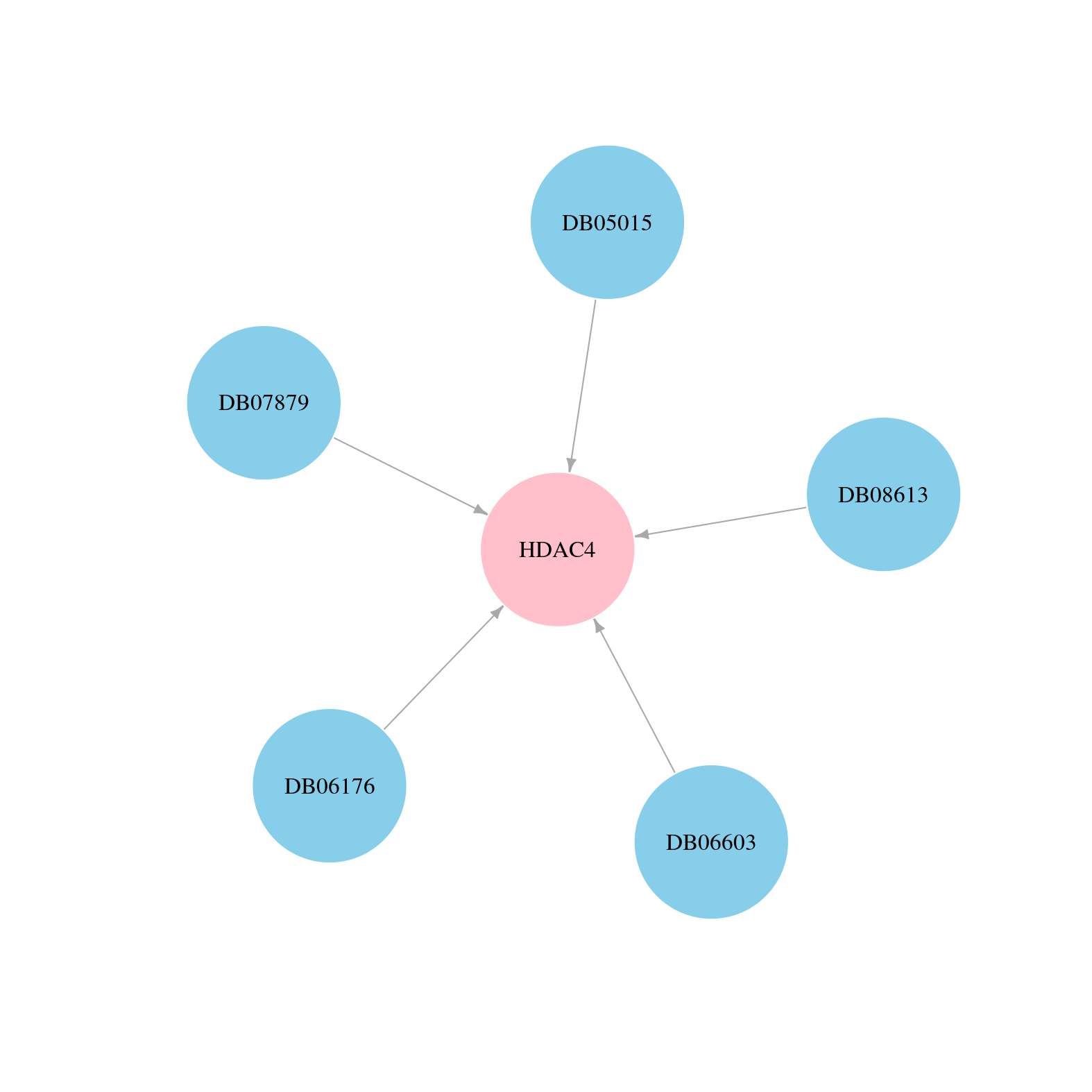
|
| Summary | |
|---|---|
| Symbol | HDAC4 |
| Name | histone deacetylase 4 |
| Aliases | KIAA0288; HDAC-A; HDACA; HD4; HA6116; HDAC-4; BDMR; brachydactyly-mental retardation syndrome; AHO3; histone ...... |
| Location | 2q37.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|