Browse HDAC6 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00850 Histone deacetylase domain PF02148 Zn-finger in ubiquitin-hydrolases and other protein |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes (By similarity). Plays a central role in microtubule-dependent cell motility via deacetylation of tubulin. Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer. ; FUNCTION: In addition to its protein deacetylase activity, plays a key role in the degradation of misfolded proteins: when misfolded proteins are too abundant to be degraded by the chaperone refolding system and the ubiquitin-proteasome, mediates the transport of misfolded proteins to a cytoplasmic juxtanuclear structure called aggresome. Probably acts as an adapter that recognizes polyubiquitinated misfolded proteins and target them to the aggresome, facilitating their clearance by autophagy. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000209 protein polyubiquitination GO:0000226 microtubule cytoskeleton organization GO:0000302 response to reactive oxygen species GO:0000422 mitophagy GO:0000423 macromitophagy GO:0001667 ameboidal-type cell migration GO:0006476 protein deacetylation GO:0006515 misfolded or incompletely synthesized protein catabolic process GO:0006914 autophagy GO:0006979 response to oxidative stress GO:0007018 microtubule-based movement GO:0007019 microtubule depolymerization GO:0007026 negative regulation of microtubule depolymerization GO:0007409 axonogenesis GO:0009636 response to toxic substance GO:0009755 hormone-mediated signaling pathway GO:0010035 response to inorganic substance GO:0010310 regulation of hydrogen peroxide metabolic process GO:0010421 hydrogen peroxide-mediated programmed cell death GO:0010469 regulation of receptor activity GO:0010506 regulation of autophagy GO:0010631 epithelial cell migration GO:0010632 regulation of epithelial cell migration GO:0010634 positive regulation of epithelial cell migration GO:0010639 negative regulation of organelle organization GO:0010727 negative regulation of hydrogen peroxide metabolic process GO:0010821 regulation of mitochondrion organization GO:0010869 regulation of receptor biosynthetic process GO:0010870 positive regulation of receptor biosynthetic process GO:0016049 cell growth GO:0016236 macroautophagy GO:0016358 dendrite development GO:0016570 histone modification GO:0016575 histone deacetylation GO:0018205 peptidyl-lysine modification GO:0030335 positive regulation of cell migration GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0031109 microtubule polymerization or depolymerization GO:0031110 regulation of microtubule polymerization or depolymerization GO:0031111 negative regulation of microtubule polymerization or depolymerization GO:0031114 regulation of microtubule depolymerization GO:0031334 positive regulation of protein complex assembly GO:0031647 regulation of protein stability GO:0032418 lysosome localization GO:0032800 receptor biosynthetic process GO:0032886 regulation of microtubule-based process GO:0032984 macromolecular complex disassembly GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0034599 cellular response to oxidative stress GO:0034614 cellular response to reactive oxygen species GO:0034983 peptidyl-lysine deacetylation GO:0035601 protein deacylation GO:0035966 response to topologically incorrect protein GO:0035967 cellular response to topologically incorrect protein GO:0036473 cell death in response to oxidative stress GO:0036474 cell death in response to hydrogen peroxide GO:0040017 positive regulation of locomotion GO:0040029 regulation of gene expression, epigenetic GO:0042542 response to hydrogen peroxide GO:0042743 hydrogen peroxide metabolic process GO:0043112 receptor metabolic process GO:0043162 ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway GO:0043241 protein complex disassembly GO:0043242 negative regulation of protein complex disassembly GO:0043244 regulation of protein complex disassembly GO:0043254 regulation of protein complex assembly GO:0043401 steroid hormone mediated signaling pathway GO:0043624 cellular protein complex disassembly GO:0044089 positive regulation of cellular component biogenesis GO:0045444 fat cell differentiation GO:0045598 regulation of fat cell differentiation GO:0045861 negative regulation of proteolysis GO:0048545 response to steroid hormone GO:0048588 developmental cell growth GO:0048667 cell morphogenesis involved in neuron differentiation GO:0048668 collateral sprouting GO:0048813 dendrite morphogenesis GO:0051131 chaperone-mediated protein complex assembly GO:0051261 protein depolymerization GO:0051272 positive regulation of cellular component movement GO:0051341 regulation of oxidoreductase activity GO:0051354 negative regulation of oxidoreductase activity GO:0051493 regulation of cytoskeleton organization GO:0051494 negative regulation of cytoskeleton organization GO:0051640 organelle localization GO:0051646 mitochondrion localization GO:0051788 response to misfolded protein GO:0060560 developmental growth involved in morphogenesis GO:0060632 regulation of microtubule-based movement GO:0060765 regulation of androgen receptor signaling pathway GO:0060996 dendritic spine development GO:0060997 dendritic spine morphogenesis GO:0061564 axon development GO:0061726 mitochondrion disassembly GO:0070301 cellular response to hydrogen peroxide GO:0070507 regulation of microtubule cytoskeleton organization GO:0070841 inclusion body assembly GO:0070842 aggresome assembly GO:0070843 misfolded protein transport GO:0070844 polyubiquitinated protein transport GO:0070845 polyubiquitinated misfolded protein transport GO:0070846 Hsp90 deacetylation GO:0070932 histone H3 deacetylation GO:0071218 cellular response to misfolded protein GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0072593 reactive oxygen species metabolic process GO:0090034 regulation of chaperone-mediated protein complex assembly GO:0090035 positive regulation of chaperone-mediated protein complex assembly GO:0090042 tubulin deacetylation GO:0090130 tissue migration GO:0090132 epithelium migration GO:0097061 dendritic spine organization GO:0097468 programmed cell death in response to reactive oxygen species GO:0098732 macromolecule deacylation GO:0098779 mitophagy in response to mitochondrial depolarization GO:0098780 response to mitochondrial depolarisation GO:1900407 regulation of cellular response to oxidative stress GO:1900409 positive regulation of cellular response to oxidative stress GO:1901031 regulation of response to reactive oxygen species GO:1901033 positive regulation of response to reactive oxygen species GO:1901298 regulation of hydrogen peroxide-mediated programmed cell death GO:1901300 positive regulation of hydrogen peroxide-mediated programmed cell death GO:1901879 regulation of protein depolymerization GO:1901880 negative regulation of protein depolymerization GO:1902882 regulation of response to oxidative stress GO:1902884 positive regulation of response to oxidative stress GO:1903008 organelle disassembly GO:1903146 regulation of mitophagy GO:1903201 regulation of oxidative stress-induced cell death GO:1903205 regulation of hydrogen peroxide-induced cell death GO:1903209 positive regulation of oxidative stress-induced cell death GO:1905206 positive regulation of hydrogen peroxide-induced cell death GO:2000147 positive regulation of cell motility GO:2000377 regulation of reactive oxygen species metabolic process GO:2000378 negative regulation of reactive oxygen species metabolic process |
| Molecular Function |
GO:0001047 core promoter binding GO:0003779 actin binding GO:0004407 histone deacetylase activity GO:0008013 beta-catenin binding GO:0008017 microtubule binding GO:0015631 tubulin binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0031072 heat shock protein binding GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0031593 polyubiquitin binding GO:0031625 ubiquitin protein ligase binding GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0032182 ubiquitin-like protein binding GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0042826 histone deacetylase binding GO:0042903 tubulin deacetylase activity GO:0043014 alpha-tubulin binding GO:0043130 ubiquitin binding GO:0044389 ubiquitin-like protein ligase binding GO:0048156 tau protein binding GO:0048487 beta-tubulin binding GO:0051787 misfolded protein binding GO:0051879 Hsp90 protein binding GO:0070840 dynein complex binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0005770 late endosome GO:0005771 multivesicular body GO:0005874 microtubule GO:0005875 microtubule associated complex GO:0005881 cytoplasmic microtubule GO:0005901 caveola GO:0016234 inclusion body GO:0016235 aggresome GO:0030286 dynein complex GO:0030424 axon GO:0030425 dendrite GO:0031252 cell leading edge GO:0043025 neuronal cell body GO:0043204 perikaryon GO:0044297 cell body GO:0044853 plasma membrane raft GO:0045121 membrane raft GO:0098589 membrane region GO:0098857 membrane microdomain |
| KEGG | - |
| Reactome |
R-HSA-3371556: Cellular response to heat stress R-HSA-2262752: Cellular responses to stress R-HSA-5617833: Cilium Assembly R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-3371511: HSF1 activation R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer |
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for HDAC6. |
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
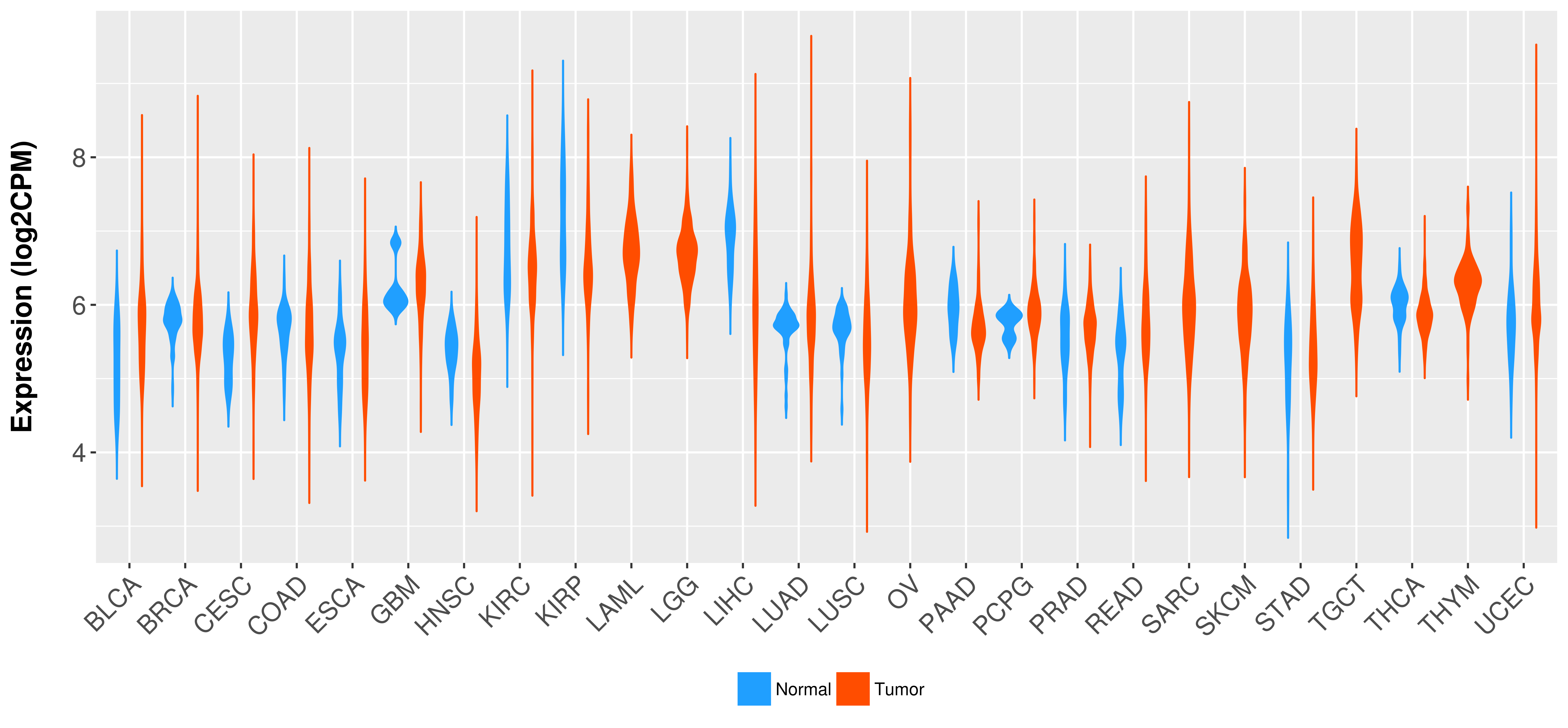
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
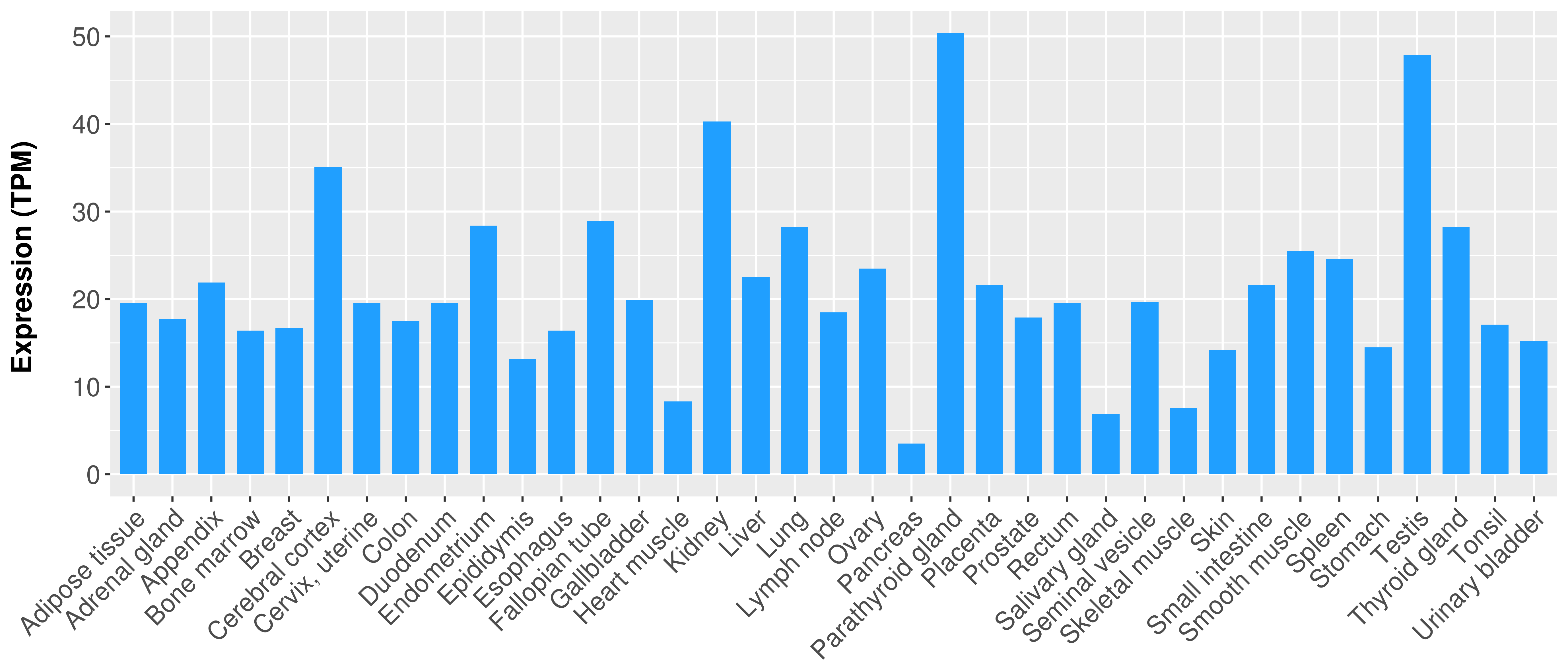
|
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
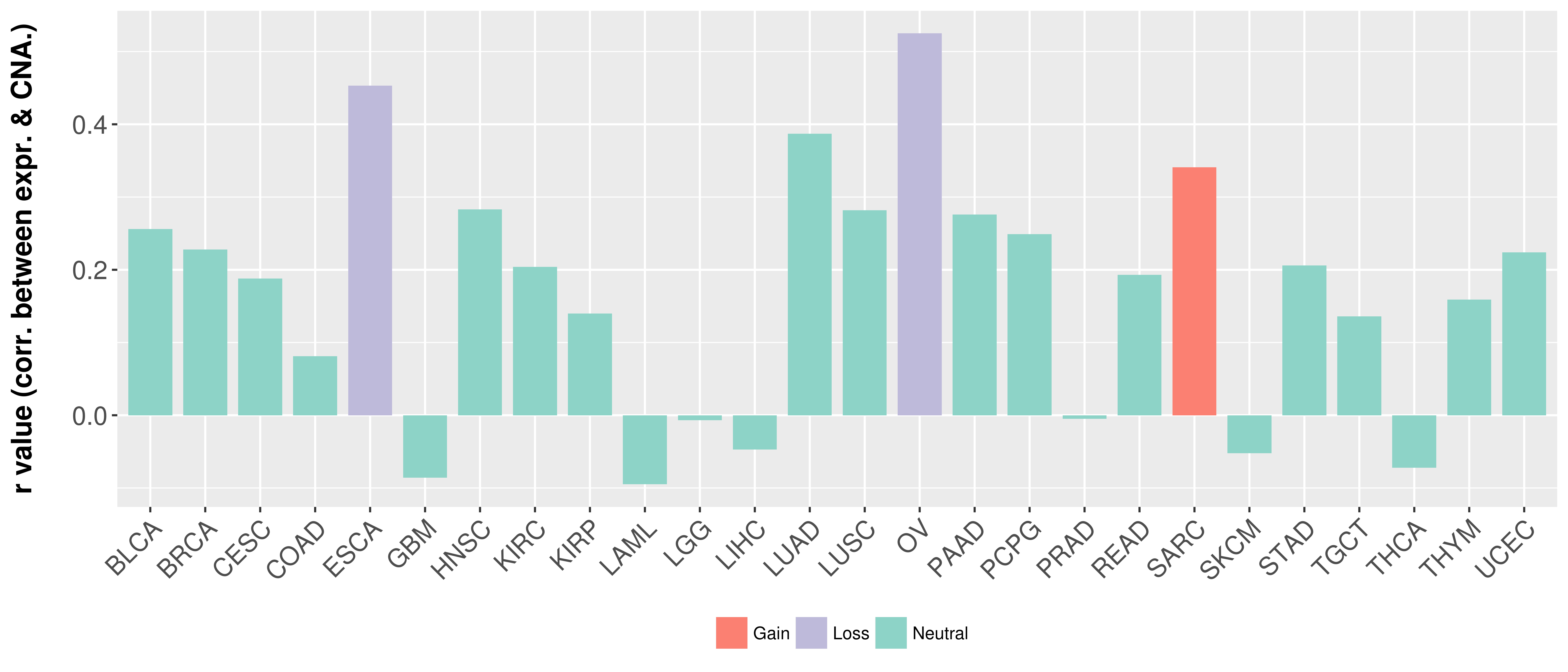
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
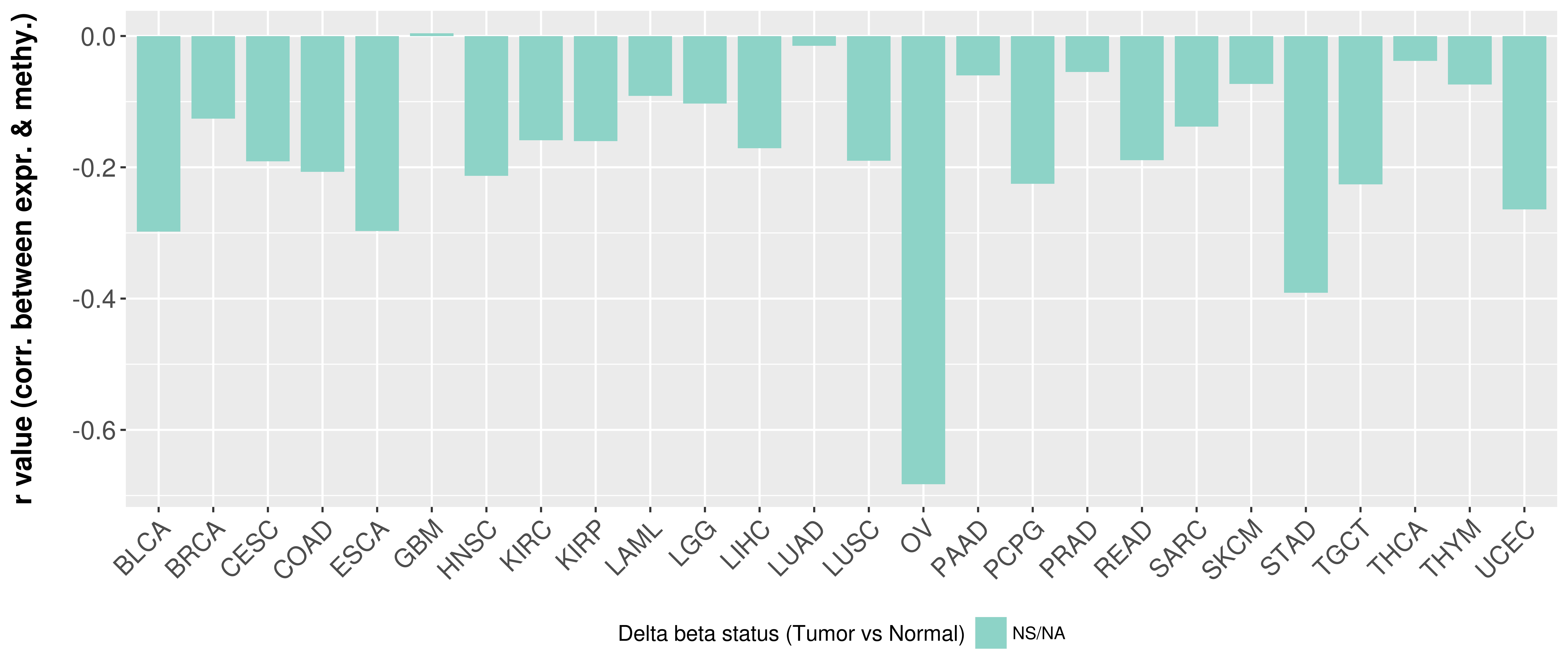
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
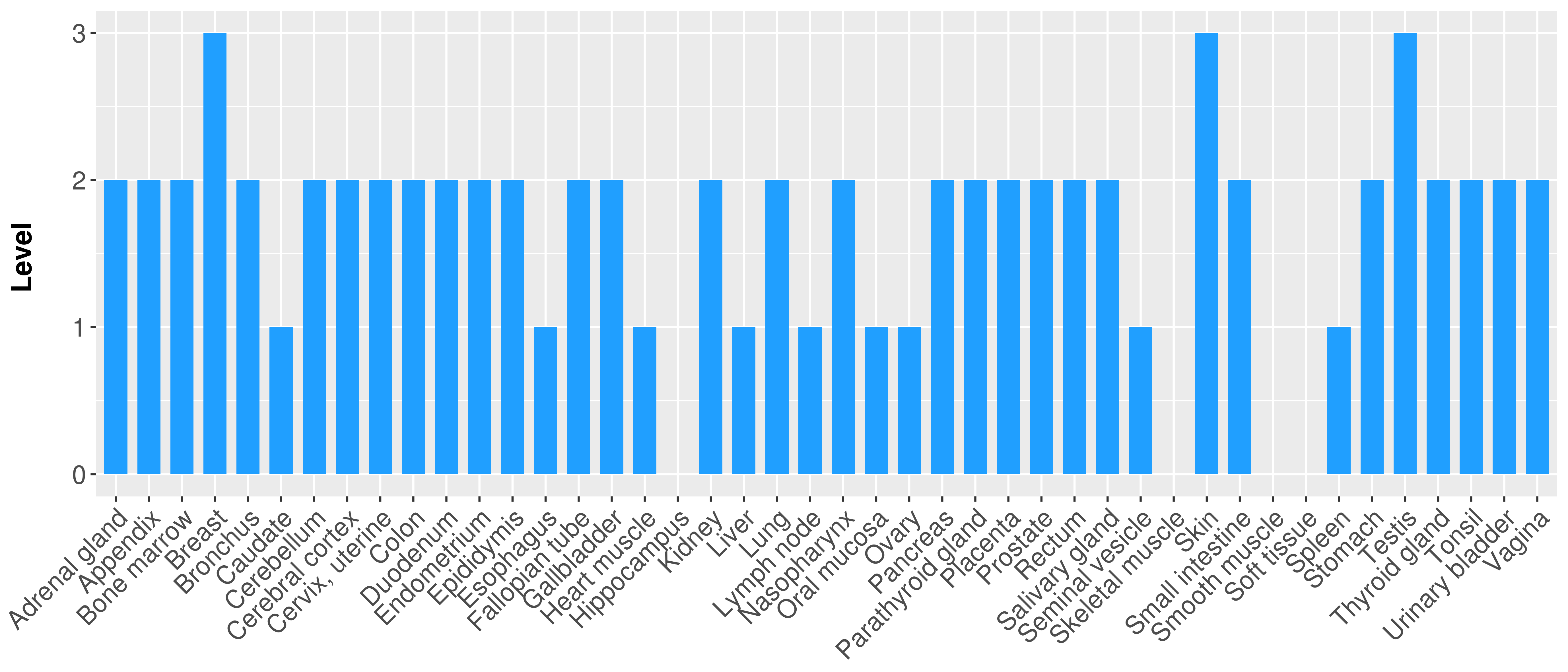
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
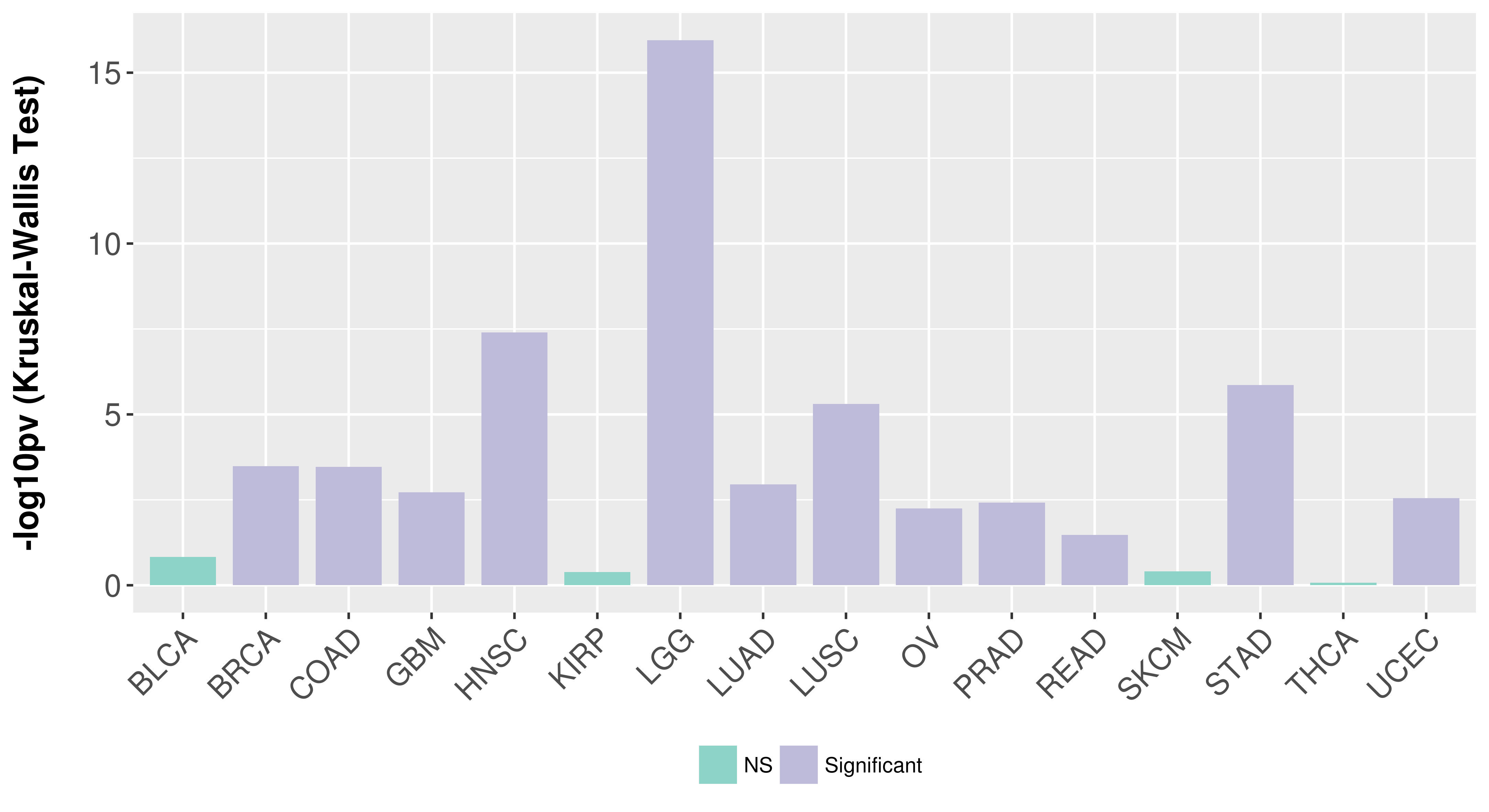
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
Association between expresson and subtype.
|
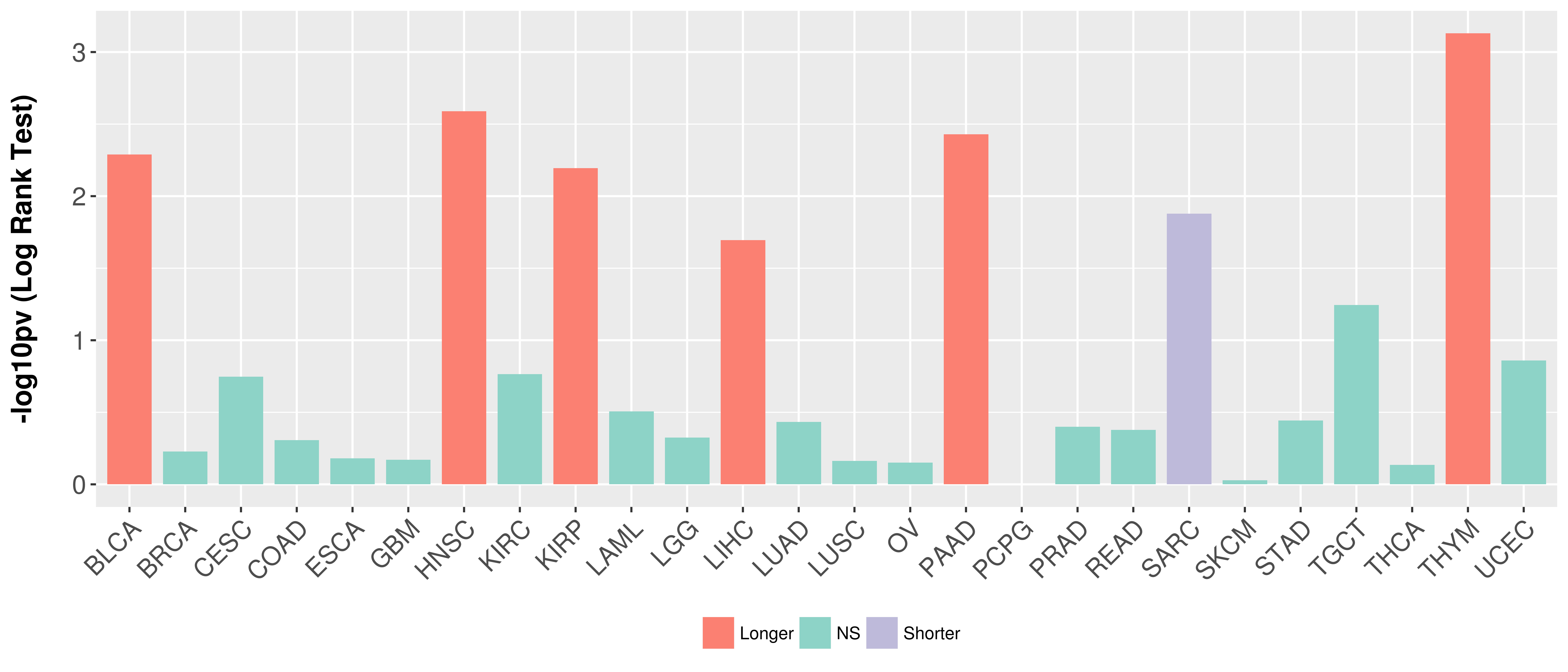
Overall survival analysis based on expression.
|
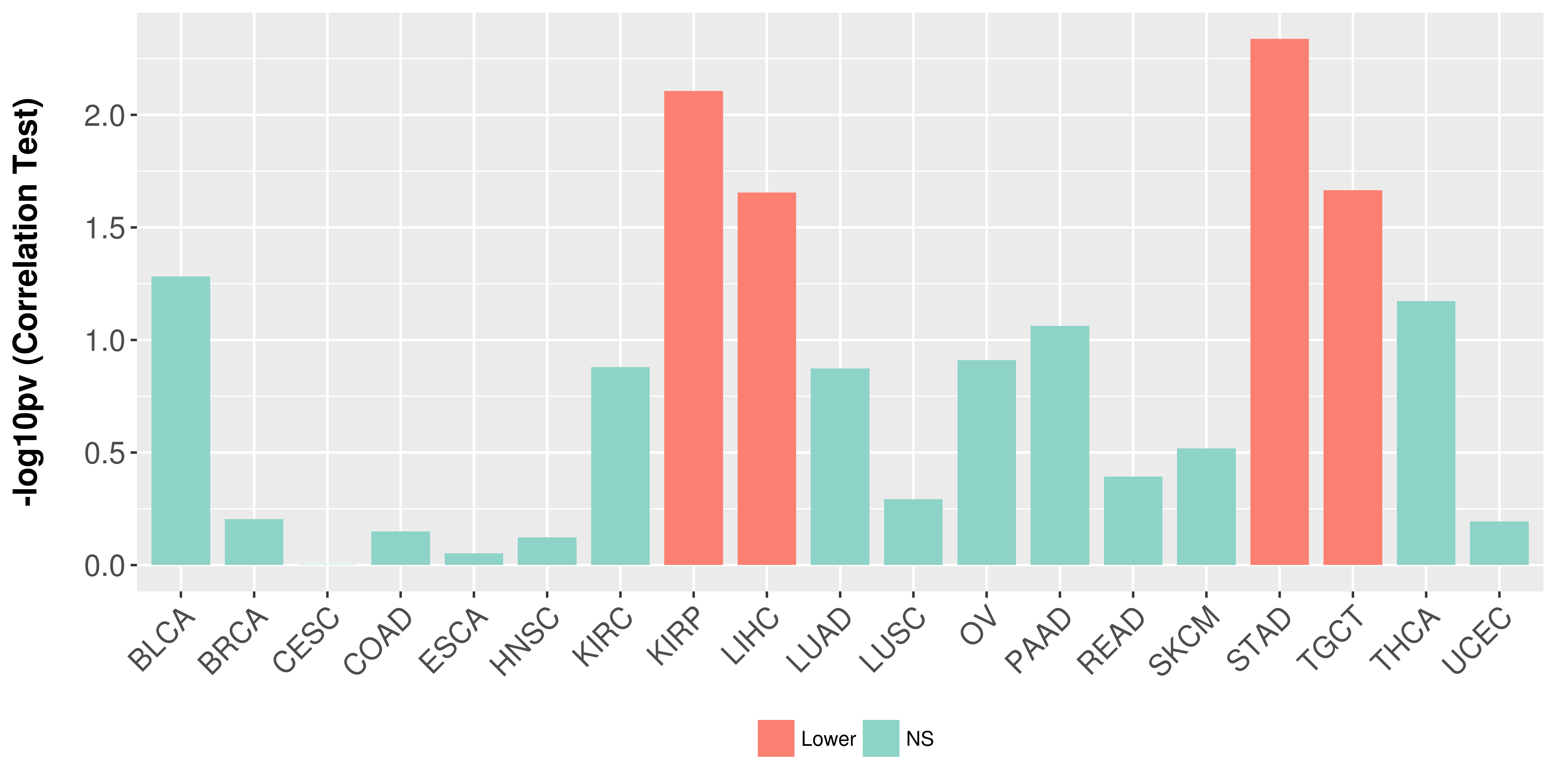
Association between expresson and stage.
|
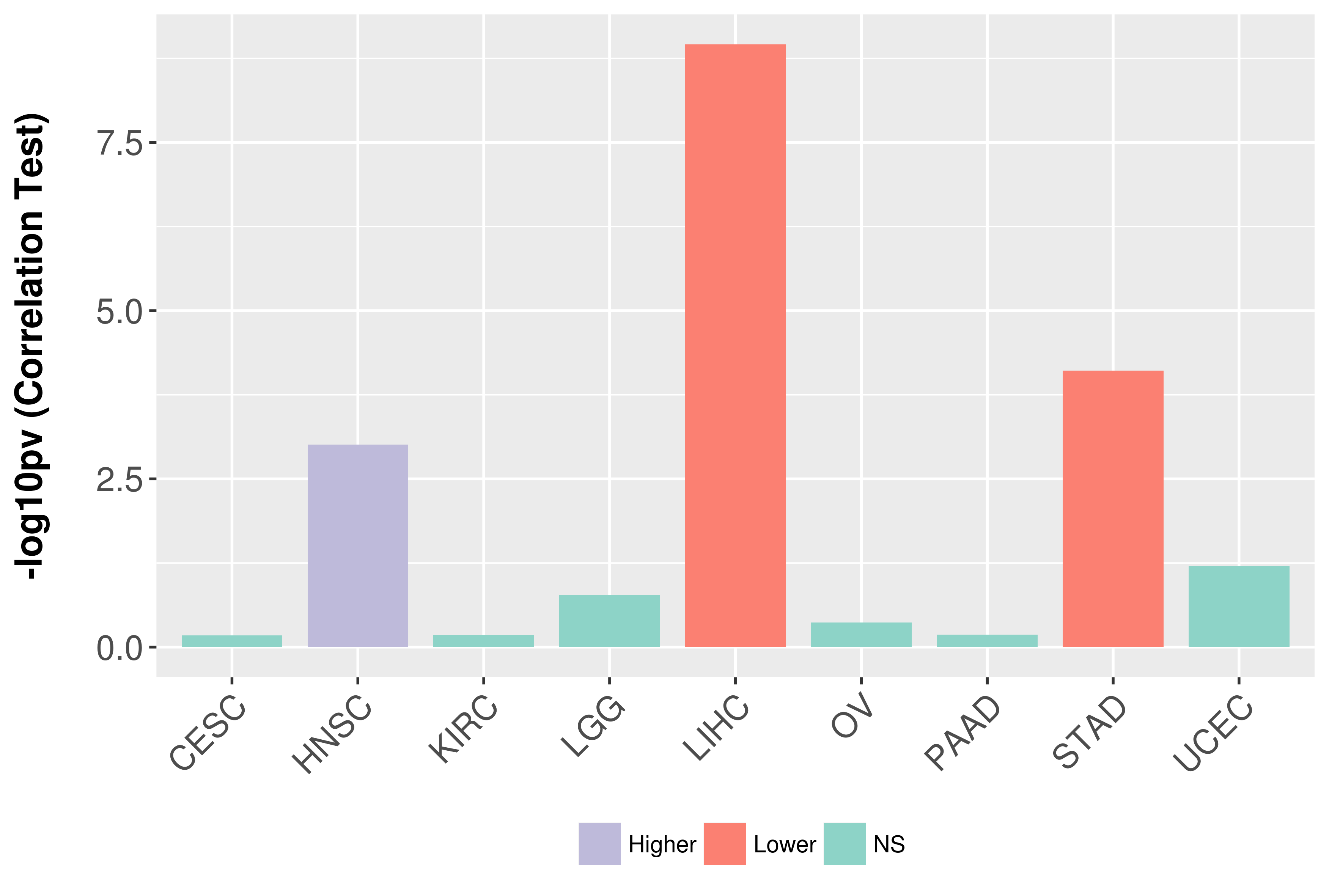
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
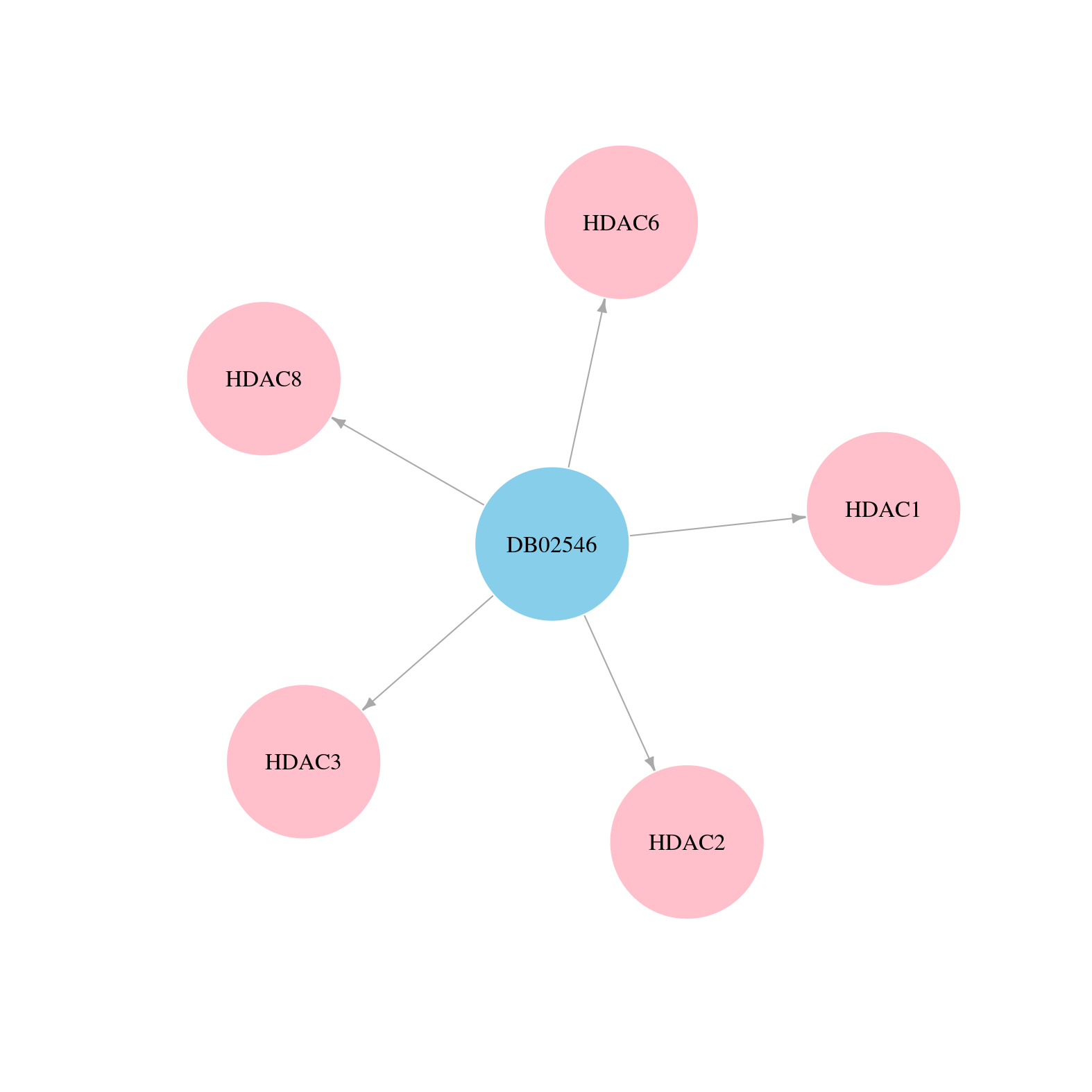
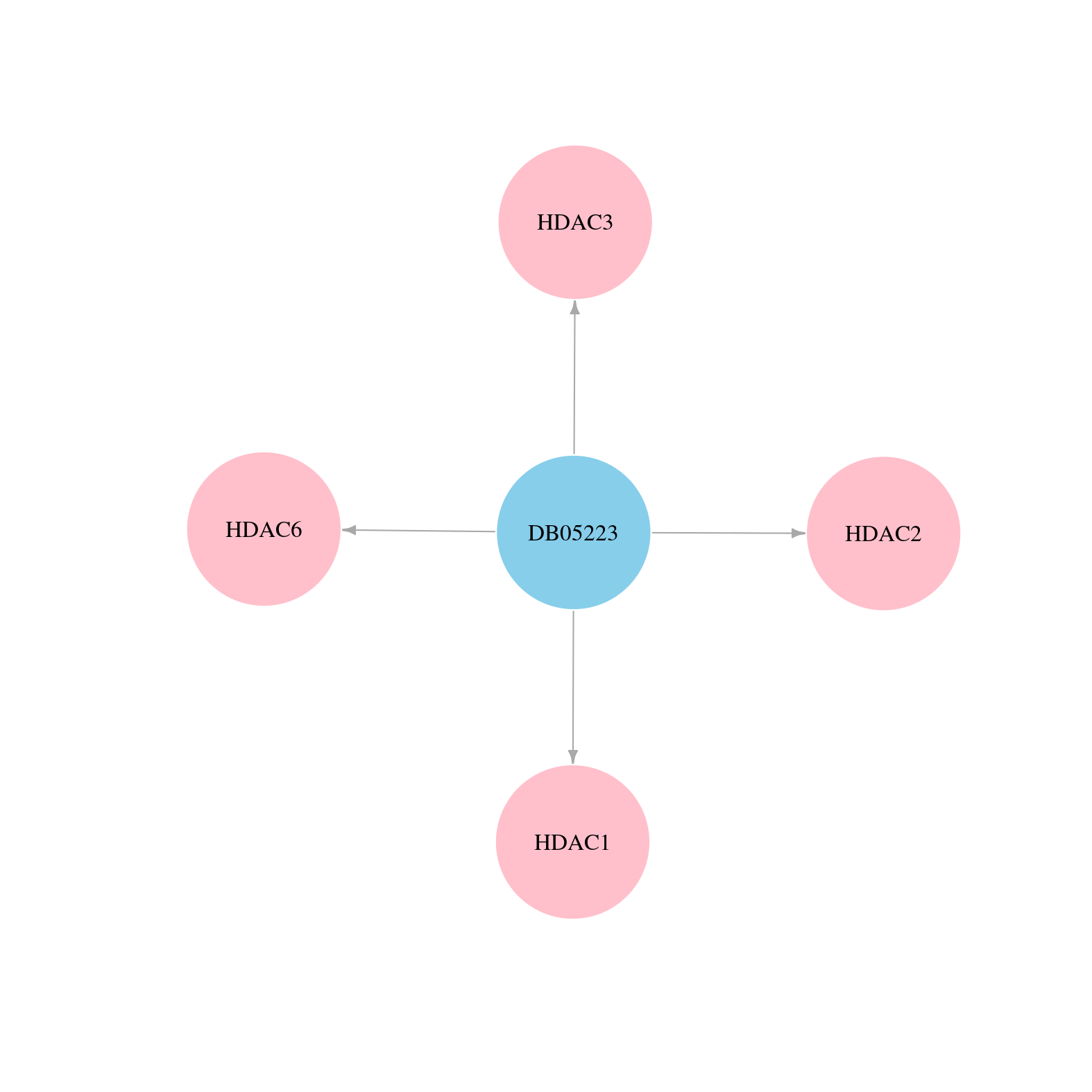
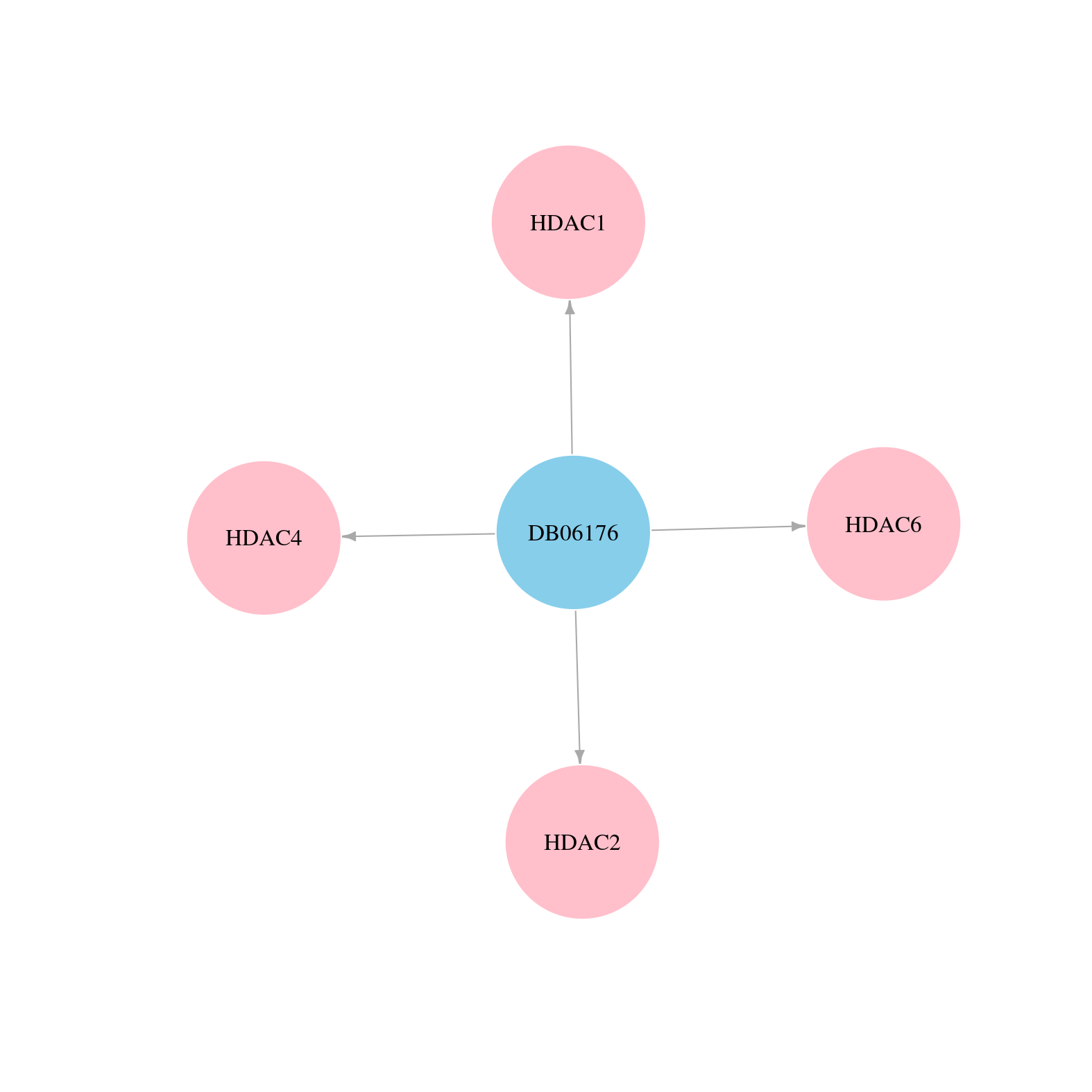
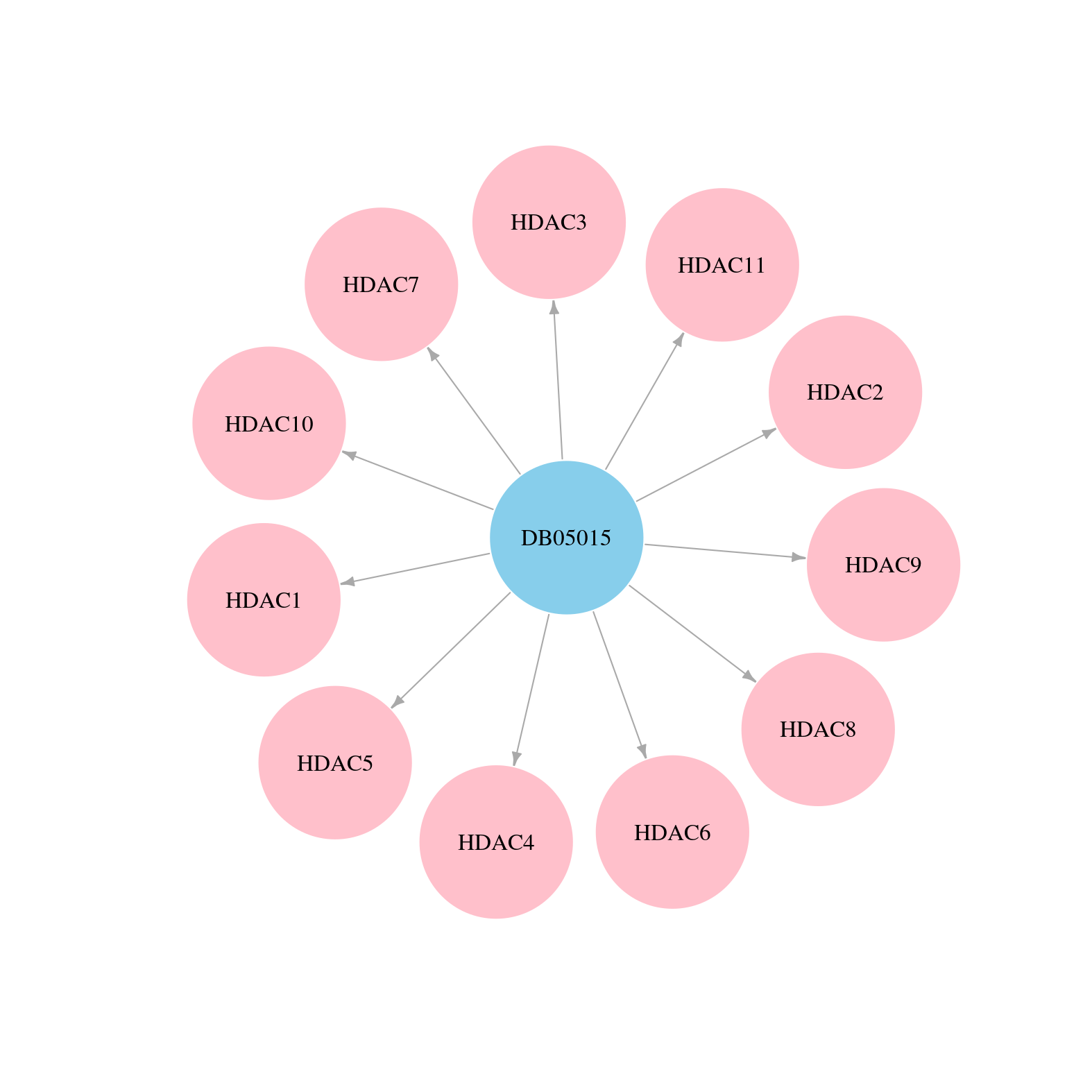
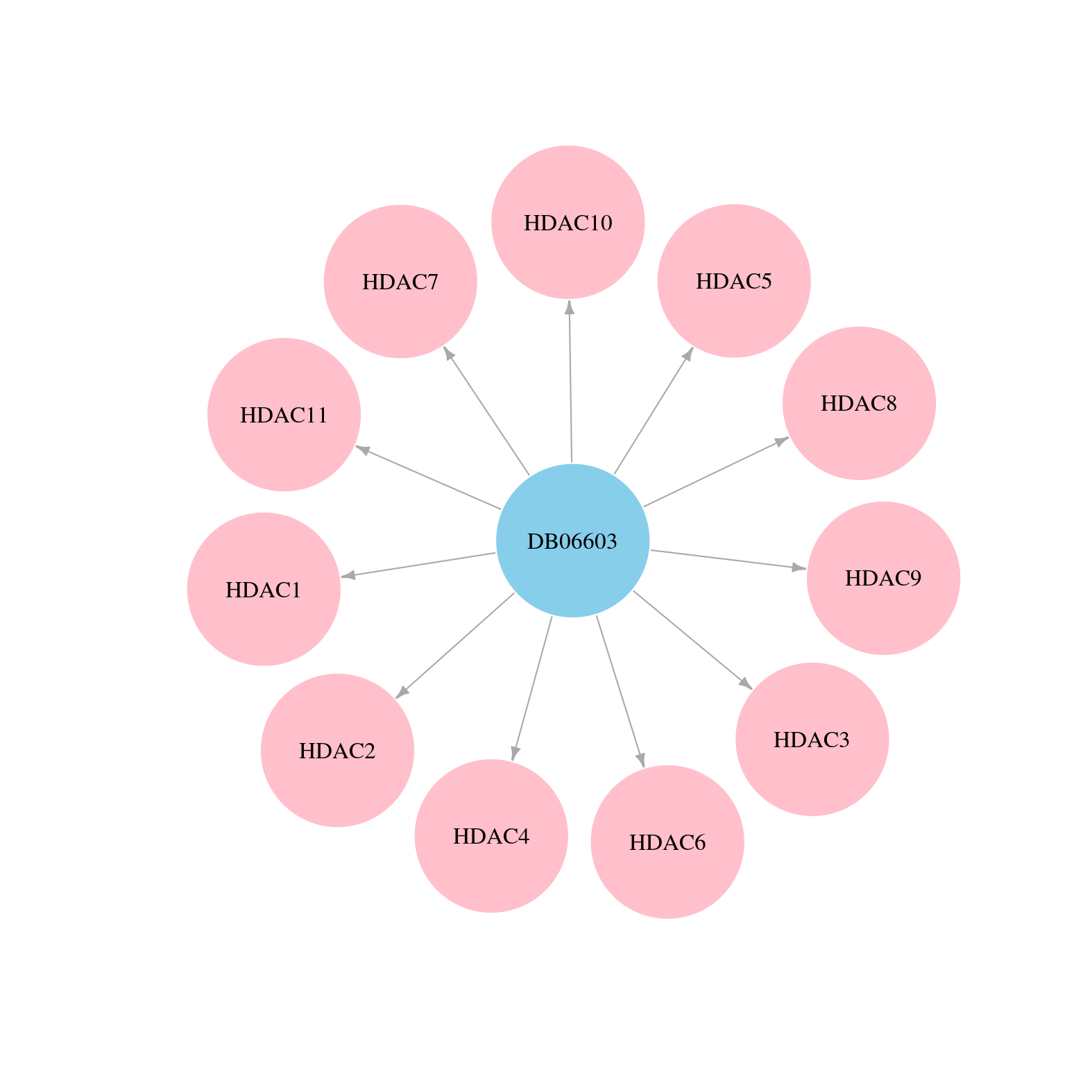
> Drugs from DrugBank database |
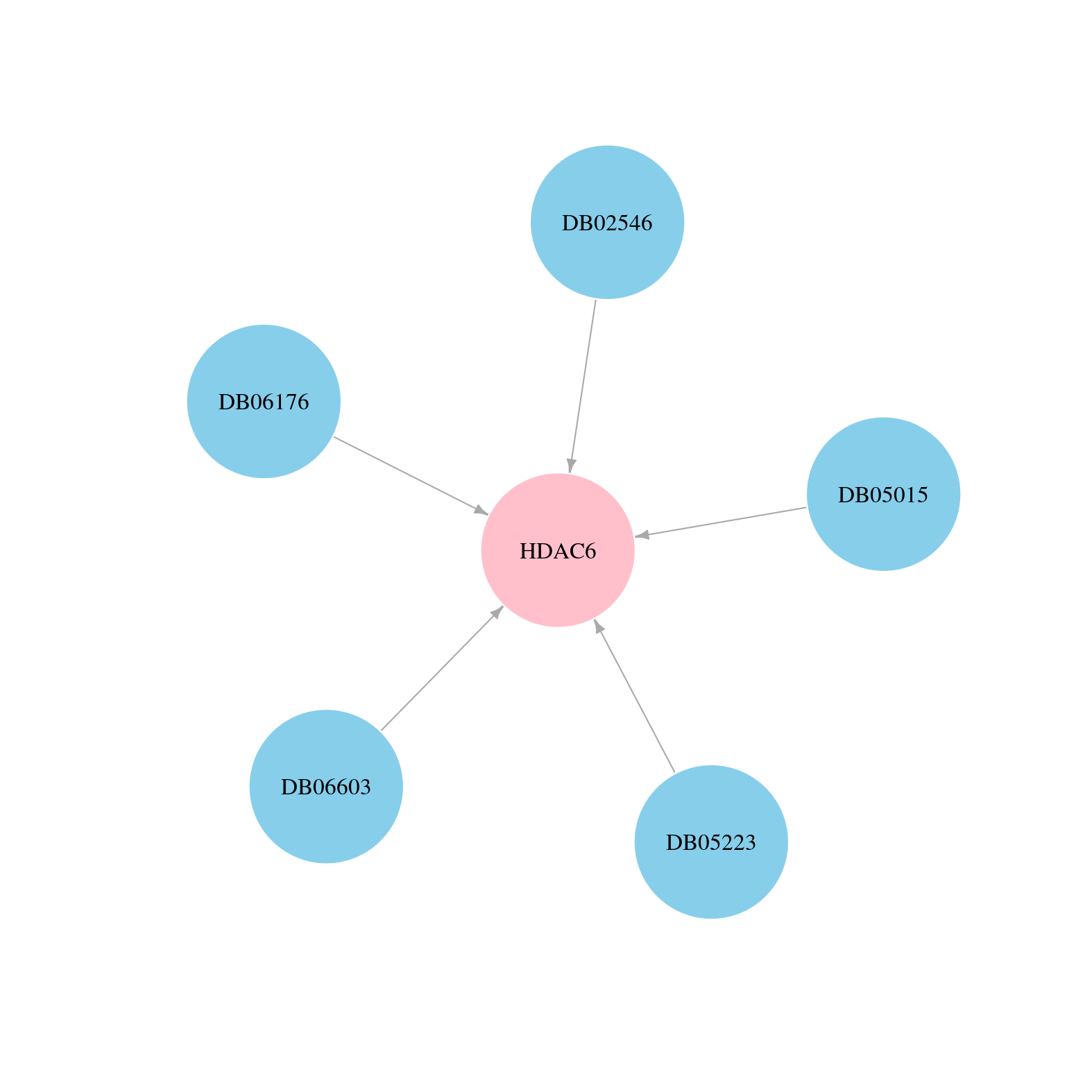
|
| Summary | |
|---|---|
| Symbol | HDAC6 |
| Name | histone deacetylase 6 |
| Aliases | KIAA0901; JM21; FLJ16239; PPP1R90; protein phosphatase 1, regulatory subunit 90; CPBHM |
| Location | Xp11.23 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for HDAC6. |