Browse KAT7 in pancancer
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01853 MOZ/SAS family PF01530 Zinc finger |
||||||||||
| Function |
Component of the HBO1 complex which has a histone H4-specific acetyltransferase activity, a reduced activity toward histone H3 and is responsible for the bulk of histone H4 acetylation in vivo. Through chromatin acetylation it may regulate DNA replication and act as a coactivator of TP53-dependent transcription. Acts as a coactivator of the licensing factor CDT1 (PubMed:18832067). Specifically represses AR-mediated transcription. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006260 DNA replication GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0034504 protein localization to nucleus GO:0043543 protein acylation GO:0043966 histone H3 acetylation GO:0043967 histone H4 acetylation GO:0043981 histone H4-K5 acetylation GO:0043982 histone H4-K8 acetylation GO:0043983 histone H4-K12 acetylation GO:1900180 regulation of protein localization to nucleus GO:1900182 positive regulation of protein localization to nucleus GO:1903829 positive regulation of cellular protein localization |
| Molecular Function |
GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0034212 peptide N-acetyltransferase activity GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0031248 protein acetyltransferase complex GO:1902493 acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214847: HATs acetylate histones |
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for KAT7. |
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
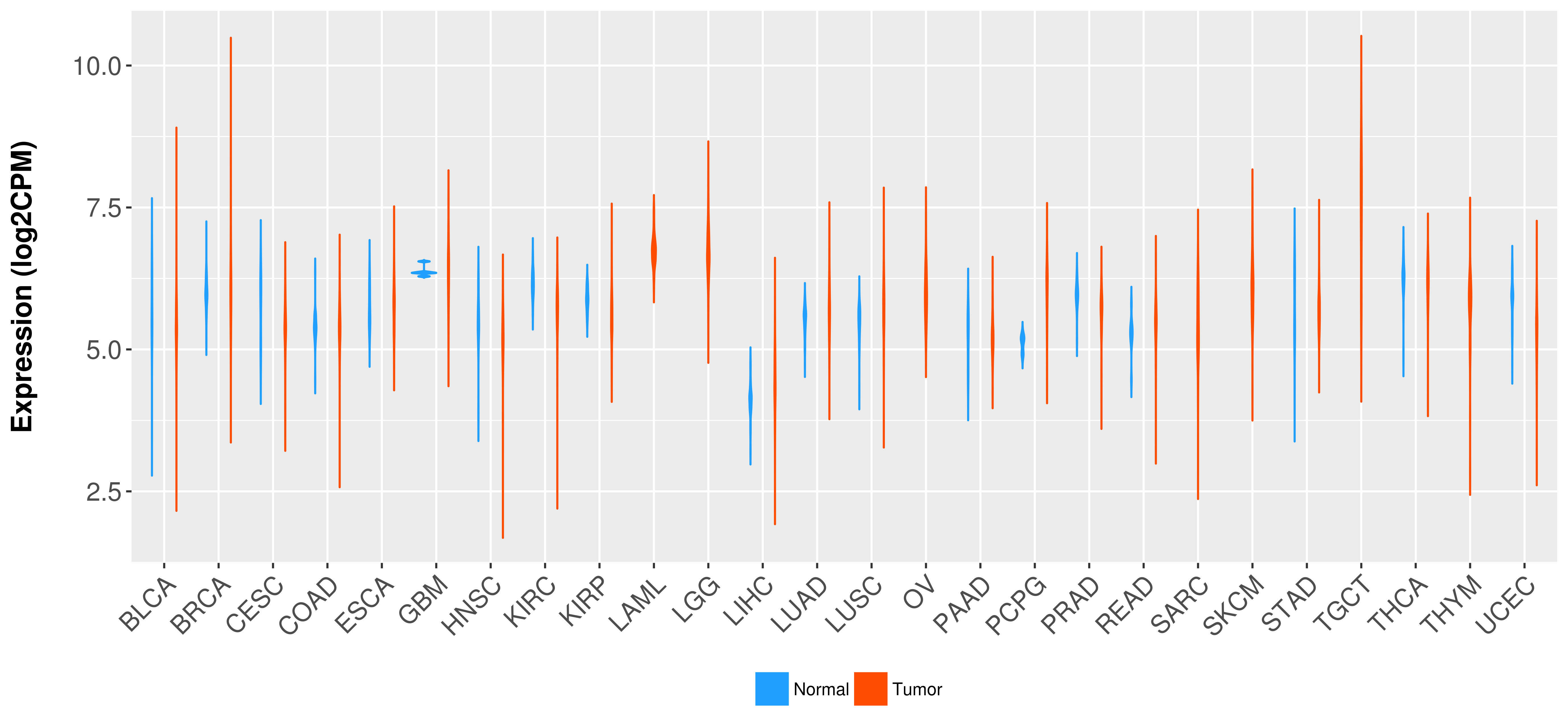
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
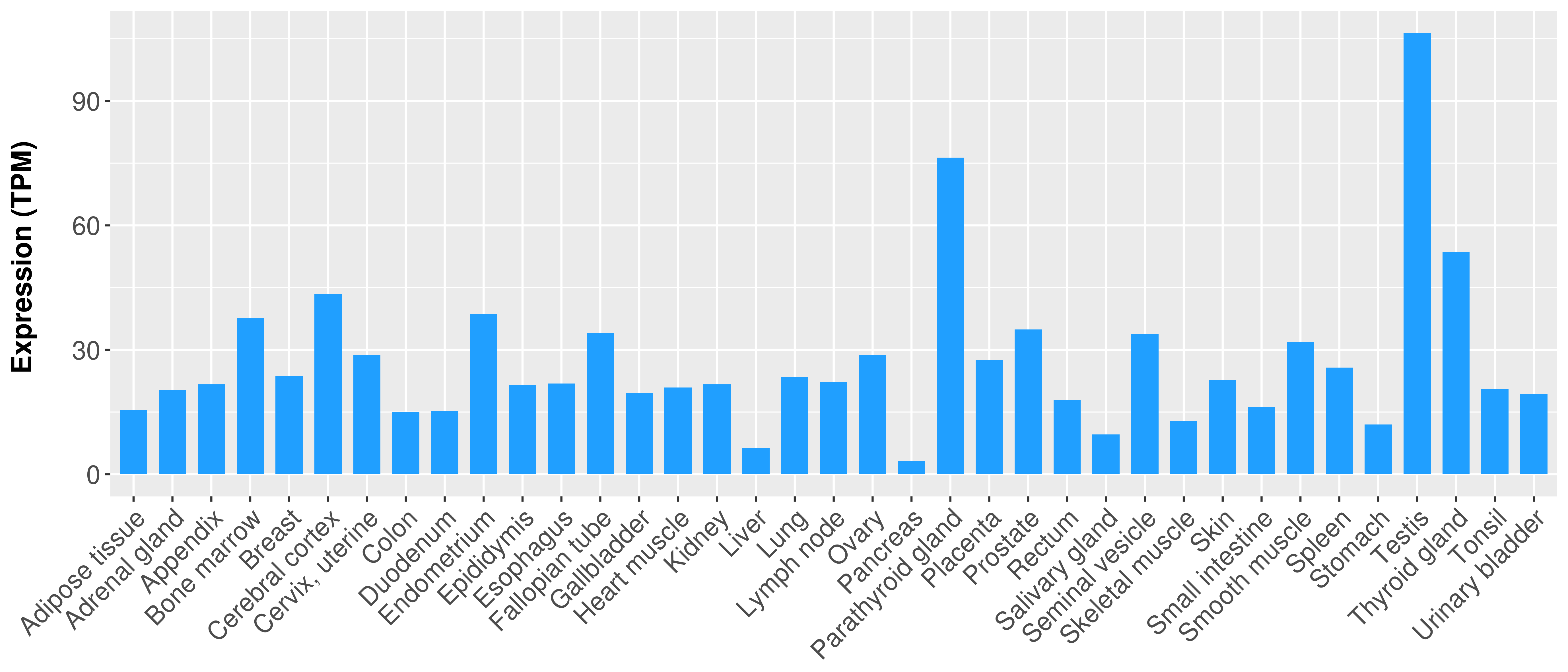
|
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
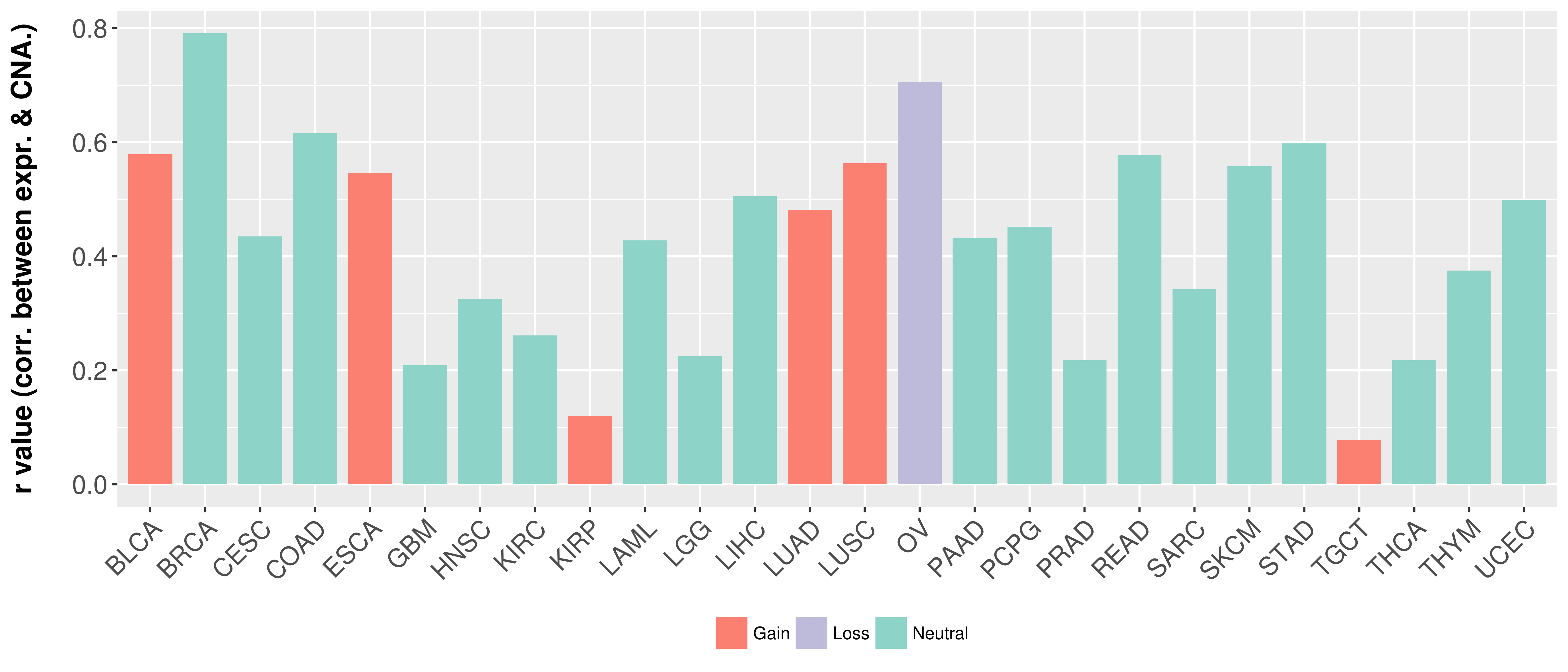
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
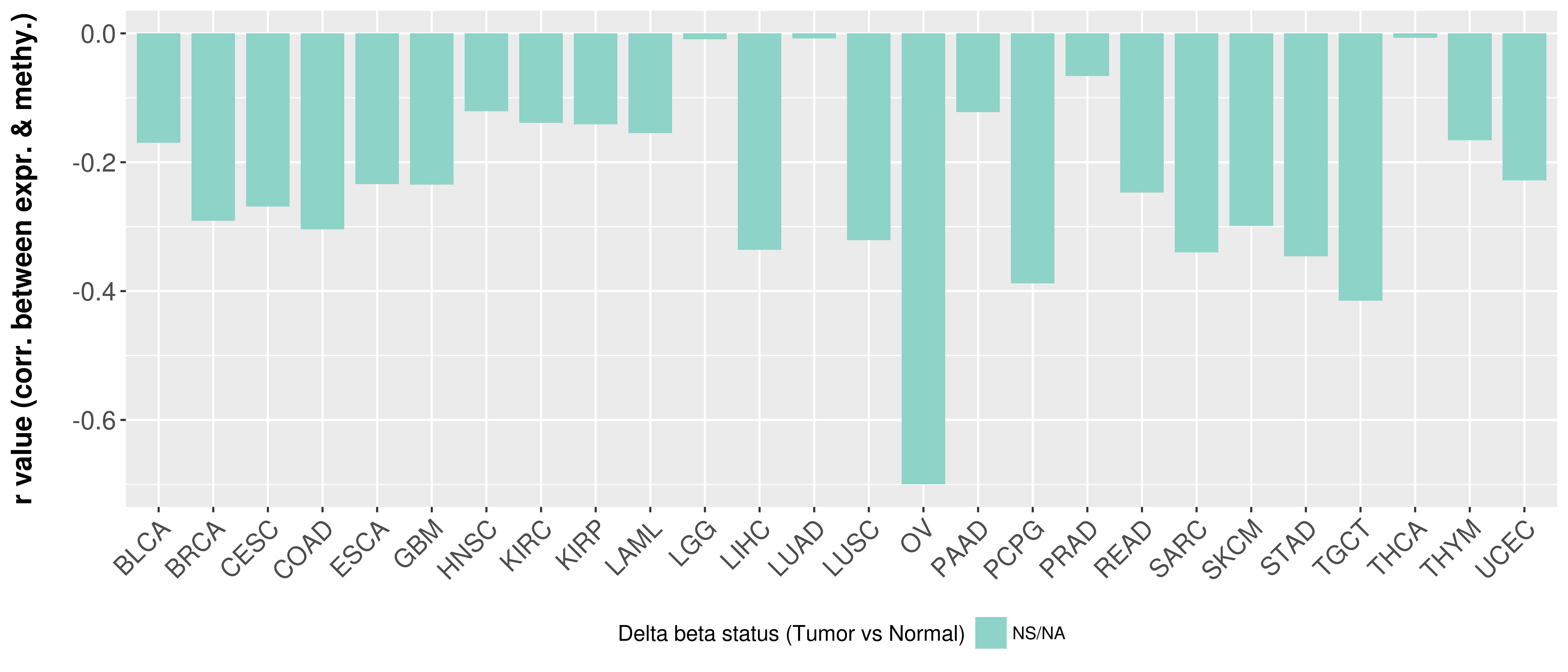
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
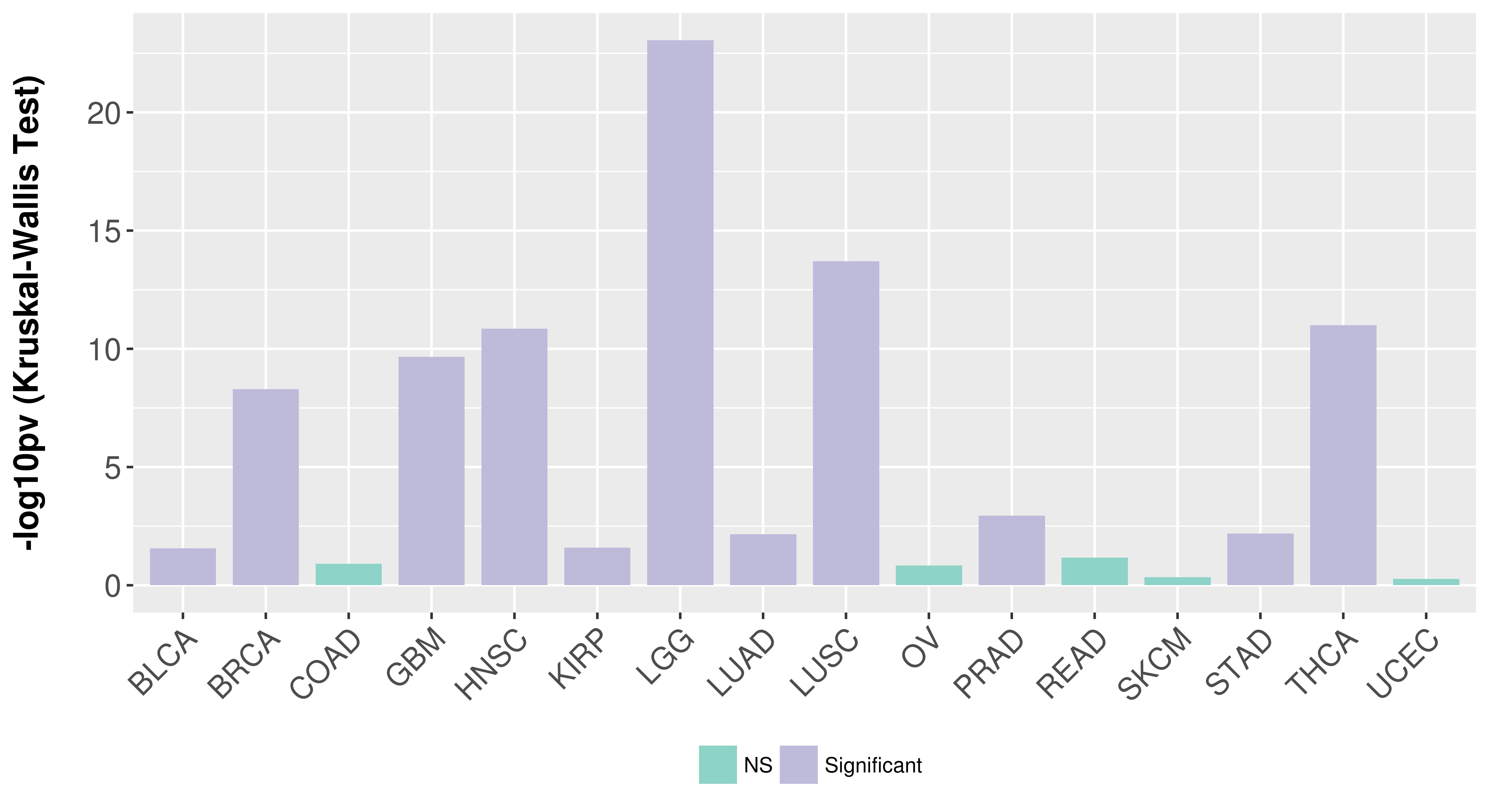
Association between expresson and subtype.
|
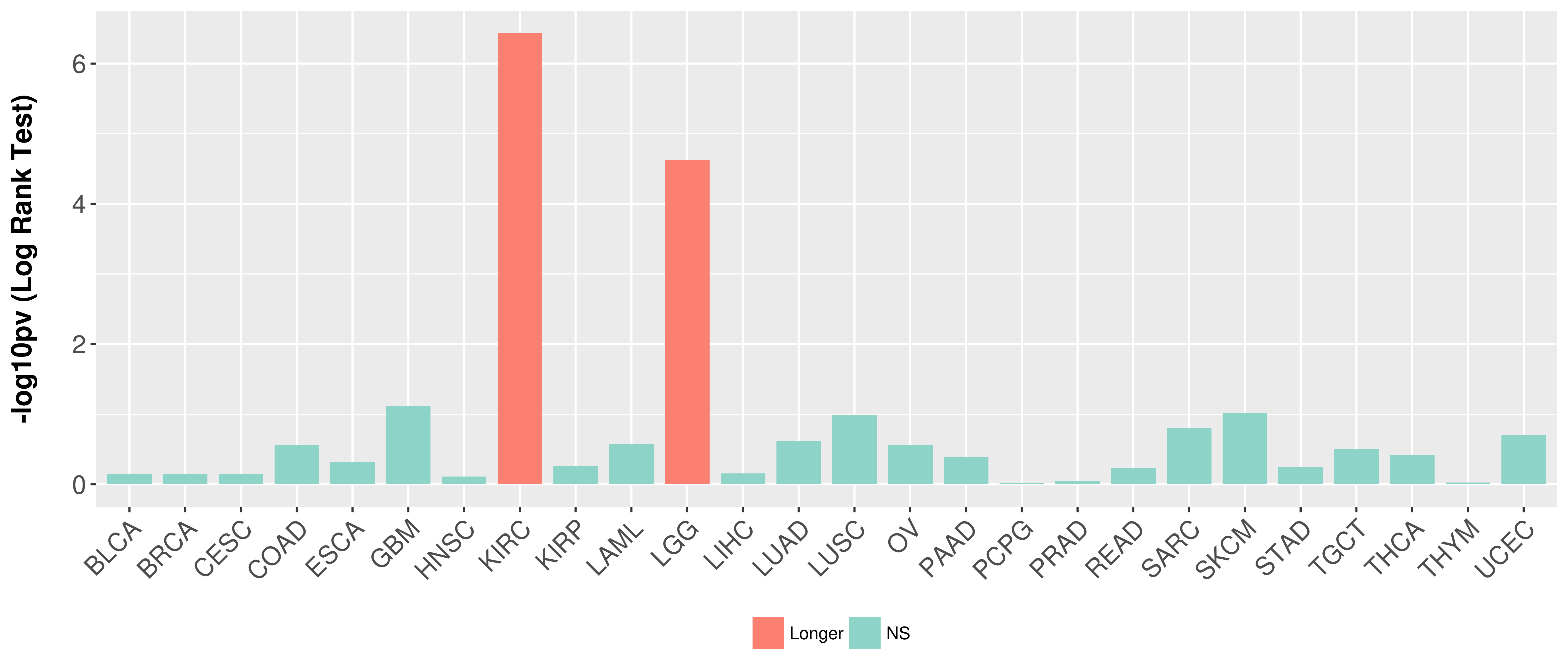
Overall survival analysis based on expression.
|
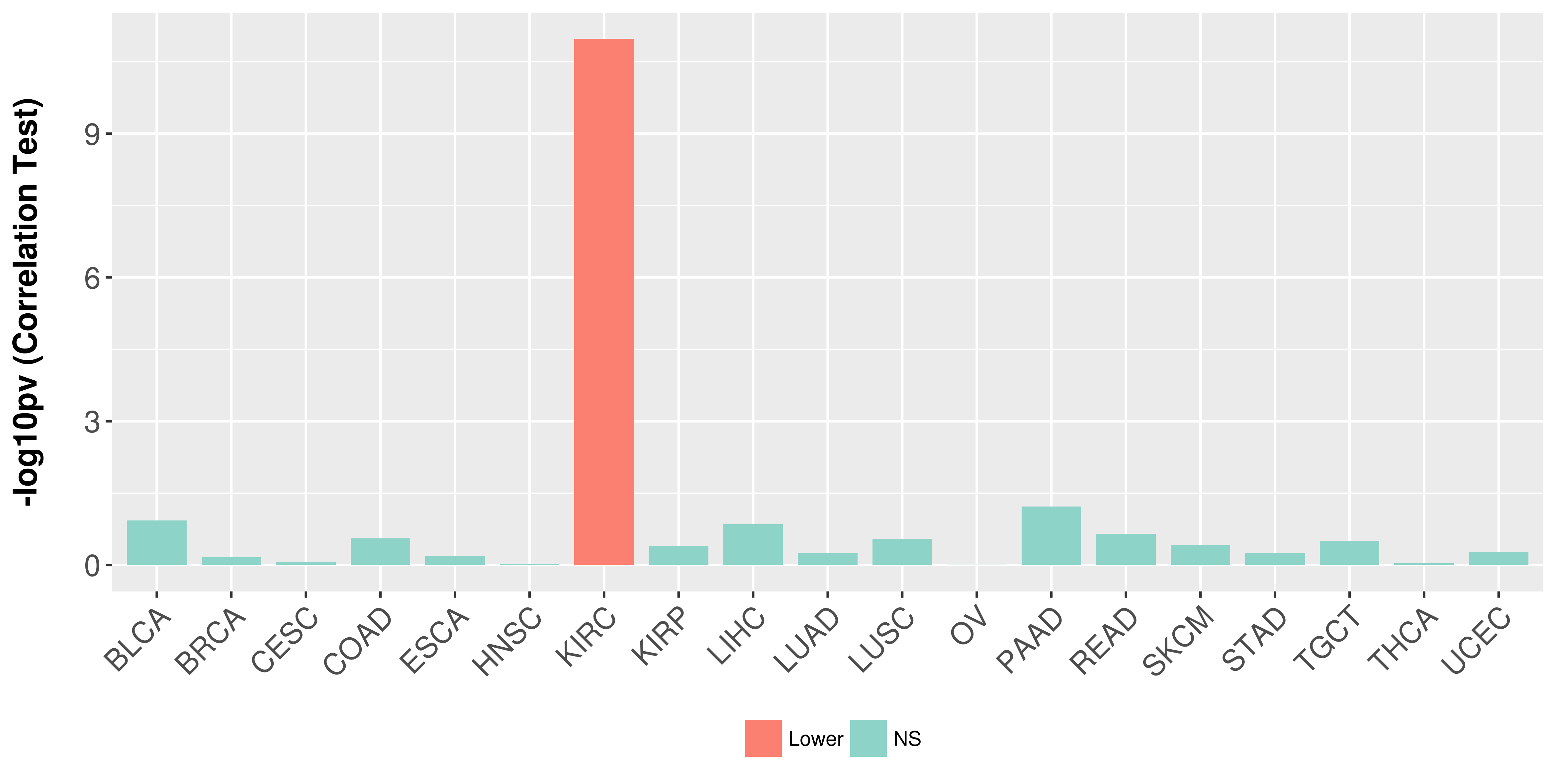
Association between expresson and stage.
|
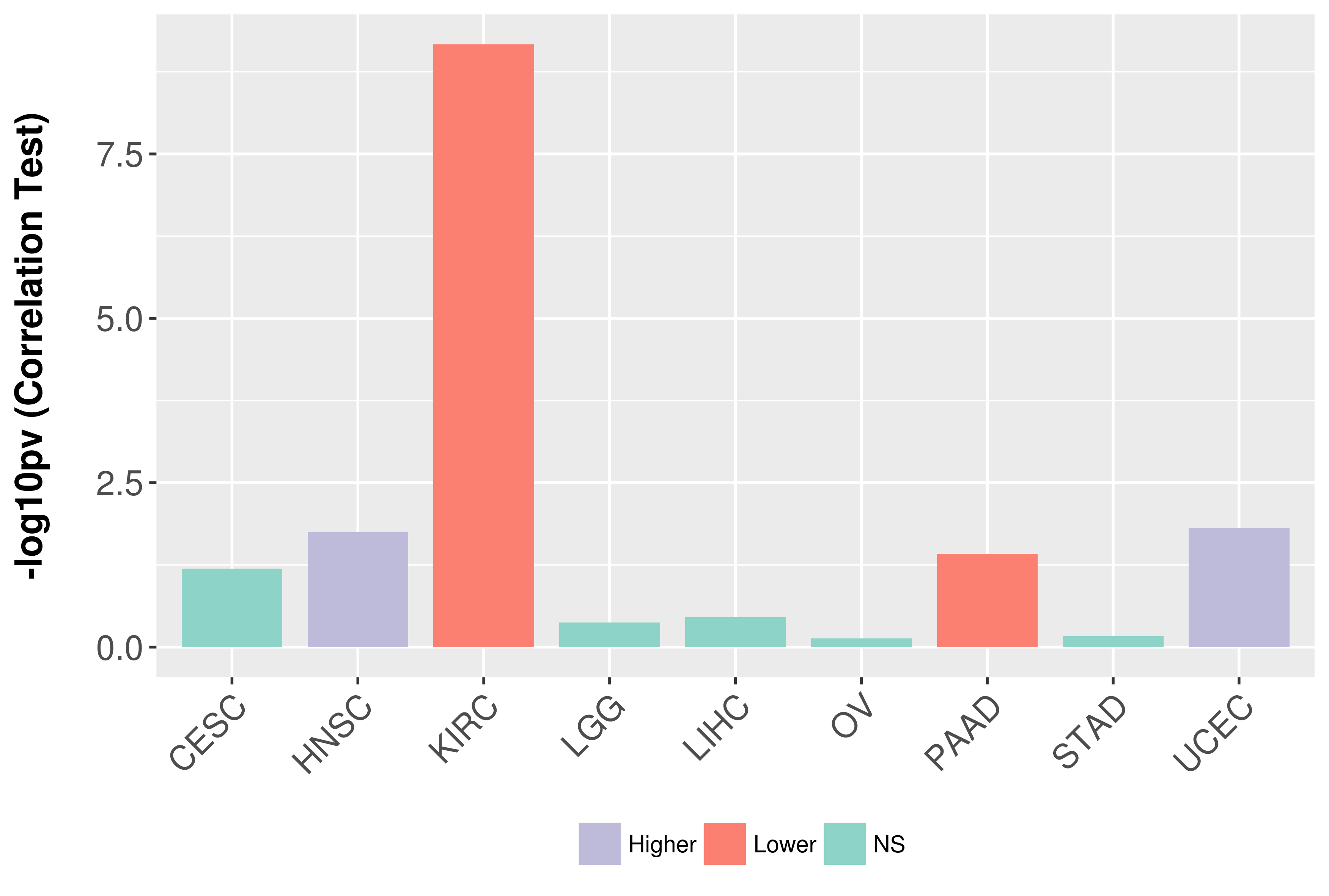
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for KAT7. |
| Summary | |
|---|---|
| Symbol | KAT7 |
| Name | lysine acetyltransferase 7 |
| Aliases | HBOA; HBO1; ZC2HC7; histone acetyltransferase binding to ORC1; MYST2; MYST histone acetyltransferase 2; MOZ, ...... |
| Location | 17q21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|