Browse SIRT1 in pancancer
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02146 Sir2 family |
||||||||||
| Function |
NAD-dependent protein deacetylase that links transcriptional regulation directly to intracellular energetics and participates in the coordination of several separated cellular functions such as cell cycle, response to DNA damage, metobolism, apoptosis and autophagy. Can modulate chromatin function through deacetylation of histones and can promote alterations in the methylation of histones and DNA, leading to transcriptional repression. Deacetylates a broad range of transcription factors and coregulators, thereby regulating target gene expression positively and negatively. Serves as a sensor of the cytosolic ratio of NAD(+)/NADH which is altered by glucose deprivation and metabolic changes associated with caloric restriction. Is essential in skeletal muscle cell differentiation and in response to low nutrients mediates the inhibitory effect on skeletal myoblast differentiation which also involves 5'-AMP-activated protein kinase (AMPK) and nicotinamide phosphoribosyltransferase (NAMPT). Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. The eNoSC complex is able to sense the energy status of cell: upon glucose starvation, elevation of NAD(+)/NADP(+) ratio activates SIRT1, leading to histone H3 deacetylation followed by dimethylation of H3 at 'Lys-9' (H3K9me2) by SUV39H1 and the formation of silent chromatin in the rDNA locus. Deacetylates 'Lys-266' of SUV39H1, leading to its activation. Inhibits skeletal muscle differentiation by deacetylating PCAF and MYOD1. Deacetylates H2A and 'Lys-26' of HIST1H1E. Deacetylates 'Lys-16' of histone H4 (in vitro). Involved in NR0B2/SHP corepression function through chromatin remodeling: Recruited to LRH1 target gene promoters by NR0B2/SHP thereby stimulating histone H3 and H4 deacetylation leading to transcriptional repression. Proposed to contribute to genomic integrity via positive regulation of telomere length; however, reports on localization to pericentromeric heterochromatin are conflicting. Proposed to play a role in constitutive heterochromatin (CH) formation and/or maintenance through regulation of the available pool of nuclear SUV39H1. Upon oxidative/metabolic stress decreases SUV39H1 degradation by inhibiting SUV39H1 polyubiquitination by MDM2. This increase in SUV39H1 levels enhances SUV39H1 turnover in CH, which in turn seems to accelerate renewal of the heterochromatin which correlates with greater genomic integrity during stress response. Deacetylates 'Lys-382' of p53/TP53 and impairs its ability to induce transcription-dependent proapoptotic program and modulate cell senescence. Deacetylates TAF1B and thereby represses rDNA transcription by the RNA polymerase I. Deacetylates MYC, promotes the association of MYC with MAX and decreases MYC stability leading to compromised transformational capability. Deacetylates FOXO3 in response to oxidative stress thereby increasing its ability to induce cell cycle arrest and resistance to oxidative stress but inhibiting FOXO3-mediated induction of apoptosis transcriptional activity; also leading to FOXO3 ubiquitination and protesomal degradation. Appears to have a similar effect on MLLT7/FOXO4 in regulation of transcriptional activity and apoptosis. Deacetylates DNMT1; thereby impairs DNMT1 methyltransferase-independent transcription repressor activity, modulates DNMT1 cell cycle regulatory function and DNMT1-mediated gene silencing. Deacetylates RELA/NF-kappa-B p65 thereby inhibiting its transactivating potential and augments apoptosis in response to TNF-alpha. Deacetylates HIF1A, KAT5/TIP60, RB1 and HIC1. Deacetylates FOXO1 resulting in its nuclear retention and enhancement of its transcriptional activity leading to increased gluconeogenesis in liver. Inhibits E2F1 transcriptional activity and apoptotic function, possibly by deacetylation. Involved in HES1- and HEY2-mediated transcriptional repression. In cooperation with MYCN seems to be involved in transcriptional repression of DUSP6/MAPK3 leading to MYCN stabilization by phosphorylation at 'Ser-62'. Deacetylates MEF2D. Required for antagonist-mediated transcription suppression of AR-dependent genes which may be linked to local deacetylation of histone H3. Represses HNF1A-mediated transcription. Required for the repression of ESRRG by CREBZF. Modulates AP-1 transcription factor activity. Deacetylates NR1H3 AND NR1H2 and deacetylation of NR1H3 at 'Lys-434' positively regulates transcription of NR1H3:RXR target genes, promotes NR1H3 proteosomal degradation and results in cholesterol efflux; a promoter clearing mechanism after reach round of transcription is proposed. Involved in lipid metabolism. Implicated in regulation of adipogenesis and fat mobilization in white adipocytes by repression of PPARG which probably involves association with NCOR1 and SMRT/NCOR2. Deacetylates ACSS2 leading to its activation, and HMGCS1. Involved in liver and muscle metabolism. Through deacteylation and activation of PPARGC1A is required to activate fatty acid oxidation in skeletel muscle under low-glucose conditions and is involved in glucose homeostasis. Involved in regulation of PPARA and fatty acid beta-oxidation in liver. Involved in positive regulation of insulin secretion in pancreatic beta cells in response to glucose; the function seems to imply transcriptional repression of UCP2. Proposed to deacetylate IRS2 thereby facilitating its insulin-induced tyrosine phosphorylation. Deacetylates SREBF1 isoform SREBP-1C thereby decreasing its stability and transactivation in lipogenic gene expression. Involved in DNA damage response by repressing genes which are involved in DNA repair, such as XPC and TP73, deacetylating XRCC6/Ku70, and faciliting recruitment of additional factors to sites of damaged DNA, such as SIRT1-deacetylated NBN can recruit ATM to initiate DNA repair and SIRT1-deacetylated XPA interacts with RPA2. Also involved in DNA repair of DNA double-strand breaks by homologous recombination and specifically single-strand annealing independently of XRCC6/Ku70 and NBN. Transcriptional suppression of XPC probably involves an E2F4:RBL2 suppressor complex and protein kinase B (AKT) signaling. Transcriptional suppression of TP73 probably involves E2F4 and PCAF. Deacetylates WRN thereby regulating its helicase and exonuclease activities and regulates WRN nuclear translocation in response to DNA damage. Deacetylates APEX1 at 'Lys-6' and 'Lys-7' and stimulates cellular AP endonuclease activity by promoting the association of APEX1 to XRCC1. Increases p53/TP53-mediated transcription-independent apoptosis by blocking nuclear translocation of cytoplasmic p53/TP53 and probably redirecting it to mitochondria. Deacetylates XRCC6/Ku70 at 'Lys-539' and 'Lys-542' causing it to sequester BAX away from mitochondria thereby inhibiting stress-induced apoptosis. Is involved in autophagy, presumably by deacetylating ATG5, ATG7 and MAP1LC3B/ATG8. Deacetylates AKT1 which leads to enhanced binding of AKT1 and PDK1 to PIP3 and promotes their activation. Proposed to play role in regulation of STK11/LBK1-dependent AMPK signaling pathways implicated in cellular senescence which seems to involve the regulation of the acetylation status of STK11/LBK1. Can deacetylate STK11/LBK1 and thereby increase its activity, cytoplasmic localization and association with STRAD; however, the relevance of such activity in normal cells is unclear. In endothelial cells is shown to inhibit STK11/LBK1 activity and to promote its degradation. Deacetylates SMAD7 at 'Lys-64' and 'Lys-70' thereby promoting its degradation. Deacetylates CIITA and augments its MHC class II transactivation and contributes to its stability. Deacteylates MECOM/EVI1. Deacetylates PML at 'Lys-487' and this deacetylation promotes PML control of PER2 nuclear localization. During the neurogenic transition, repress selective NOTCH1-target genes through histone deacetylation in a BCL6-dependent manner and leading to neuronal differentiation. Regulates the circadian expression of several core clock genes, including ARNTL/BMAL1, RORC, PER2 and CRY1 and plays a critical role in maintaining a controlled rhythmicity in histone acetylation, thereby contributing to circadian chromatin remodeling. Deacetylates ARNTL/BMAL1 and histones at the circadian gene promoters in order to facilitate repression by inhibitory components of the circadian oscillator. Deacetylates PER2, facilitating its ubiquitination and degradation by the proteosome. Protects cardiomyocytes against palmitate-induced apoptosis (PubMed:11672523, PubMed:12006491, PubMed:14976264, PubMed:14980222, PubMed:15126506, PubMed:15152190, PubMed:15205477, PubMed:15469825, PubMed:15692560, PubMed:16079181, PubMed:16166628, PubMed:16892051, PubMed:16998810, PubMed:17283066, PubMed:17334224, PubMed:17505061, PubMed:17612497, PubMed:17620057, PubMed:17936707, PubMed:18203716, PubMed:18296641, PubMed:18662546, PubMed:18687677, PubMed:19188449, PubMed:19220062, PubMed:19364925, PubMed:19690166, PubMed:19934257, PubMed:20097625, PubMed:20100829, PubMed:20203304, PubMed:20375098, PubMed:20620956, PubMed:20670893, PubMed:20817729, PubMed:21149730, PubMed:21245319, PubMed:21471201, PubMed:21504832, PubMed:21555002, PubMed:21698133, PubMed:21701047, PubMed:21775285, PubMed:21807113, PubMed:21841822, PubMed:21890893, PubMed:21909281, PubMed:21947282, PubMed:22274616). Deacetylates XBP1 isoform 2; deacetylation decreases protein stability of XBP1 isoform 2 and inhibits its transcriptional activity (PubMed:20955178). Involved in the CCAR2-mediated regulation of PCK1 and NR1D1 (PubMed:24415752). Deacetylates CTNB1 at 'Lys-49' (PubMed:24824780). In POMC (pro-opiomelanocortin) neurons, required for leptin-induced activation of PI3K signaling (By similarity). ; FUNCTION: Isoform 2: Isoform 2 is shown to deacetylate 'Lys-382' of p53/TP53, however with lower activity than isoform 1. In combination, the two isoforms exert an additive effect. Isoform 2 regulates p53/TP53 expression and cellular stress response and is in turn repressed by p53/TP53 presenting a SIRT1 isoform-dependent auto-regulatory loop. ; FUNCTION: (Microbial infection) In case of HIV-1 infection, interacts with and deacetylates the viral Tat protein. The viral Tat protein inhibits SIRT1 deacetylation activity toward RELA/NF-kappa-B p65, thereby potentiates its transcriptional activity and SIRT1 is proposed to contribute to T-cell hyperactivation during infection. ; FUNCTION: SirtT1 75 kDa fragment: catalytically inactive 75SirT1 may be involved in regulation of apoptosis. May be involved in protecting chondrocytes from apoptotic death by associating with cytochrome C and interfering with apoptosome assembly. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000012 single strand break repair GO:0000060 protein import into nucleus, translocation GO:0000183 chromatin silencing at rDNA GO:0000302 response to reactive oxygen species GO:0000720 pyrimidine dimer repair by nucleotide-excision repair GO:0000731 DNA synthesis involved in DNA repair GO:0001516 prostaglandin biosynthetic process GO:0001525 angiogenesis GO:0001542 ovulation from ovarian follicle GO:0001558 regulation of cell growth GO:0001666 response to hypoxia GO:0001678 cellular glucose homeostasis GO:0001933 negative regulation of protein phosphorylation GO:0001935 endothelial cell proliferation GO:0001936 regulation of endothelial cell proliferation GO:0001938 positive regulation of endothelial cell proliferation GO:0002250 adaptive immune response GO:0002367 cytokine production involved in immune response GO:0002440 production of molecular mediator of immune response GO:0002521 leukocyte differentiation GO:0002573 myeloid leukocyte differentiation GO:0002819 regulation of adaptive immune response GO:0002821 positive regulation of adaptive immune response GO:0005996 monosaccharide metabolic process GO:0006006 glucose metabolic process GO:0006109 regulation of carbohydrate metabolic process GO:0006260 DNA replication GO:0006282 regulation of DNA repair GO:0006289 nucleotide-excision repair GO:0006290 pyrimidine dimer repair GO:0006342 chromatin silencing GO:0006343 establishment of chromatin silencing GO:0006344 maintenance of chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006364 rRNA processing GO:0006469 negative regulation of protein kinase activity GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006476 protein deacetylation GO:0006479 protein methylation GO:0006606 protein import into nucleus GO:0006631 fatty acid metabolic process GO:0006633 fatty acid biosynthetic process GO:0006636 unsaturated fatty acid biosynthetic process GO:0006638 neutral lipid metabolic process GO:0006639 acylglycerol metabolic process GO:0006641 triglyceride metabolic process GO:0006642 triglyceride mobilization GO:0006690 icosanoid metabolic process GO:0006692 prostanoid metabolic process GO:0006693 prostaglandin metabolic process GO:0006694 steroid biosynthetic process GO:0006699 bile acid biosynthetic process GO:0006869 lipid transport GO:0006913 nucleocytoplasmic transport GO:0006914 autophagy GO:0006925 inflammatory cell apoptotic process GO:0006979 response to oxidative stress GO:0007178 transmembrane receptor protein serine/threonine kinase signaling pathway GO:0007179 transforming growth factor beta receptor signaling pathway GO:0007249 I-kappaB kinase/NF-kappaB signaling GO:0007283 spermatogenesis GO:0007292 female gamete generation GO:0007346 regulation of mitotic cell cycle GO:0007517 muscle organ development GO:0007548 sex differentiation GO:0007568 aging GO:0007569 cell aging GO:0007623 circadian rhythm GO:0008202 steroid metabolic process GO:0008206 bile acid metabolic process GO:0008213 protein alkylation GO:0008286 insulin receptor signaling pathway GO:0008406 gonad development GO:0008585 female gonad development GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0008631 intrinsic apoptotic signaling pathway in response to oxidative stress GO:0009266 response to temperature stimulus GO:0009267 cellular response to starvation GO:0009314 response to radiation GO:0009408 response to heat GO:0009411 response to UV GO:0009416 response to light stimulus GO:0009755 hormone-mediated signaling pathway GO:0009991 response to extracellular stimulus GO:0010035 response to inorganic substance GO:0010212 response to ionizing radiation GO:0010498 proteasomal protein catabolic process GO:0010506 regulation of autophagy GO:0010508 positive regulation of autophagy GO:0010565 regulation of cellular ketone metabolic process GO:0010657 muscle cell apoptotic process GO:0010660 regulation of muscle cell apoptotic process GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010874 regulation of cholesterol efflux GO:0010875 positive regulation of cholesterol efflux GO:0010876 lipid localization GO:0010883 regulation of lipid storage GO:0010906 regulation of glucose metabolic process GO:0010934 macrophage cytokine production GO:0010950 positive regulation of endopeptidase activity GO:0010952 positive regulation of peptidase activity GO:0014065 phosphatidylinositol 3-kinase signaling GO:0014066 regulation of phosphatidylinositol 3-kinase signaling GO:0014068 positive regulation of phosphatidylinositol 3-kinase signaling GO:0015850 organic hydroxy compound transport GO:0015918 sterol transport GO:0016049 cell growth GO:0016053 organic acid biosynthetic process GO:0016072 rRNA metabolic process GO:0016236 macroautophagy GO:0016239 positive regulation of macroautophagy GO:0016241 regulation of macroautophagy GO:0016458 gene silencing GO:0016570 histone modification GO:0016571 histone methylation GO:0016573 histone acetylation GO:0016575 histone deacetylation GO:0017015 regulation of transforming growth factor beta receptor signaling pathway GO:0017038 protein import GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019216 regulation of lipid metabolic process GO:0019217 regulation of fatty acid metabolic process GO:0019218 regulation of steroid metabolic process GO:0019318 hexose metabolic process GO:0019915 lipid storage GO:0022602 ovulation cycle process GO:0022613 ribonucleoprotein complex biogenesis GO:0030099 myeloid cell differentiation GO:0030225 macrophage differentiation GO:0030301 cholesterol transport GO:0030308 negative regulation of cell growth GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0030512 negative regulation of transforming growth factor beta receptor signaling pathway GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0030728 ovulation GO:0031056 regulation of histone modification GO:0031057 negative regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031060 regulation of histone methylation GO:0031062 positive regulation of histone methylation GO:0031392 regulation of prostaglandin biosynthetic process GO:0031393 negative regulation of prostaglandin biosynthetic process GO:0031647 regulation of protein stability GO:0031648 protein destabilization GO:0031667 response to nutrient levels GO:0031668 cellular response to extracellular stimulus GO:0031669 cellular response to nutrient levels GO:0031929 TOR signaling GO:0031935 regulation of chromatin silencing GO:0031937 positive regulation of chromatin silencing GO:0032006 regulation of TOR signaling GO:0032007 negative regulation of TOR signaling GO:0032069 regulation of nuclease activity GO:0032070 regulation of deoxyribonuclease activity GO:0032071 regulation of endodeoxyribonuclease activity GO:0032088 negative regulation of NF-kappaB transcription factor activity GO:0032259 methylation GO:0032368 regulation of lipid transport GO:0032370 positive regulation of lipid transport GO:0032371 regulation of sterol transport GO:0032373 positive regulation of sterol transport GO:0032374 regulation of cholesterol transport GO:0032376 positive regulation of cholesterol transport GO:0032386 regulation of intracellular transport GO:0032868 response to insulin GO:0032869 cellular response to insulin stimulus GO:0032922 circadian regulation of gene expression GO:0033028 myeloid cell apoptotic process GO:0033032 regulation of myeloid cell apoptotic process GO:0033034 positive regulation of myeloid cell apoptotic process GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0033144 negative regulation of intracellular steroid hormone receptor signaling pathway GO:0033157 regulation of intracellular protein transport GO:0033158 regulation of protein import into nucleus, translocation GO:0033210 leptin-mediated signaling pathway GO:0033344 cholesterol efflux GO:0033500 carbohydrate homeostasis GO:0033559 unsaturated fatty acid metabolic process GO:0033574 response to testosterone GO:0033673 negative regulation of kinase activity GO:0033674 positive regulation of kinase activity GO:0034390 smooth muscle cell apoptotic process GO:0034391 regulation of smooth muscle cell apoptotic process GO:0034470 ncRNA processing GO:0034504 protein localization to nucleus GO:0034599 cellular response to oxidative stress GO:0034605 cellular response to heat GO:0034612 response to tumor necrosis factor GO:0034614 cellular response to reactive oxygen species GO:0034644 cellular response to UV GO:0034968 histone lysine methylation GO:0034976 response to endoplasmic reticulum stress GO:0034983 peptidyl-lysine deacetylation GO:0035065 regulation of histone acetylation GO:0035067 negative regulation of histone acetylation GO:0035356 cellular triglyceride homeostasis GO:0035357 peroxisome proliferator activated receptor signaling pathway GO:0035358 regulation of peroxisome proliferator activated receptor signaling pathway GO:0035601 protein deacylation GO:0036293 response to decreased oxygen levels GO:0036294 cellular response to decreased oxygen levels GO:0036473 cell death in response to oxidative stress GO:0040029 regulation of gene expression, epigenetic GO:0042180 cellular ketone metabolic process GO:0042254 ribosome biogenesis GO:0042304 regulation of fatty acid biosynthetic process GO:0042306 regulation of protein import into nucleus GO:0042326 negative regulation of phosphorylation GO:0042542 response to hydrogen peroxide GO:0042593 glucose homeostasis GO:0042594 response to starvation GO:0042595 behavioral response to starvation GO:0042632 cholesterol homeostasis GO:0042698 ovulation cycle GO:0042770 signal transduction in response to DNA damage GO:0042771 intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:0043122 regulation of I-kappaB kinase/NF-kappaB signaling GO:0043124 negative regulation of I-kappaB kinase/NF-kappaB signaling GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043280 positive regulation of cysteine-type endopeptidase activity involved in apoptotic process GO:0043281 regulation of cysteine-type endopeptidase activity involved in apoptotic process GO:0043401 steroid hormone mediated signaling pathway GO:0043414 macromolecule methylation GO:0043433 negative regulation of sequence-specific DNA binding transcription factor activity GO:0043434 response to peptide hormone GO:0043491 protein kinase B signaling GO:0043516 regulation of DNA damage response, signal transduction by p53 class mediator GO:0043518 negative regulation of DNA damage response, signal transduction by p53 class mediator GO:0043543 protein acylation GO:0043966 histone H3 acetylation GO:0043967 histone H4 acetylation GO:0043984 histone H4-K16 acetylation GO:0044154 histone H3-K14 acetylation GO:0044262 cellular carbohydrate metabolic process GO:0044283 small molecule biosynthetic process GO:0044320 cellular response to leptin stimulus GO:0044321 response to leptin GO:0044723 single-organism carbohydrate metabolic process GO:0044744 protein targeting to nucleus GO:0045137 development of primary sexual characteristics GO:0045342 MHC class II biosynthetic process GO:0045346 regulation of MHC class II biosynthetic process GO:0045348 positive regulation of MHC class II biosynthetic process GO:0045444 fat cell differentiation GO:0045598 regulation of fat cell differentiation GO:0045599 negative regulation of fat cell differentiation GO:0045717 negative regulation of fatty acid biosynthetic process GO:0045739 positive regulation of DNA repair GO:0045765 regulation of angiogenesis GO:0045766 positive regulation of angiogenesis GO:0045814 negative regulation of gene expression, epigenetic GO:0045833 negative regulation of lipid metabolic process GO:0045860 positive regulation of protein kinase activity GO:0045862 positive regulation of proteolysis GO:0045922 negative regulation of fatty acid metabolic process GO:0045926 negative regulation of growth GO:0046394 carboxylic acid biosynthetic process GO:0046456 icosanoid biosynthetic process GO:0046457 prostanoid biosynthetic process GO:0046486 glycerolipid metabolic process GO:0046545 development of primary female sexual characteristics GO:0046626 regulation of insulin receptor signaling pathway GO:0046628 positive regulation of insulin receptor signaling pathway GO:0046660 female sex differentiation GO:0046822 regulation of nucleocytoplasmic transport GO:0046890 regulation of lipid biosynthetic process GO:0048015 phosphatidylinositol-mediated signaling GO:0048017 inositol lipid-mediated signaling GO:0048232 male gamete generation GO:0048511 rhythmic process GO:0048514 blood vessel morphogenesis GO:0048545 response to steroid hormone GO:0048608 reproductive structure development GO:0050673 epithelial cell proliferation GO:0050678 regulation of epithelial cell proliferation GO:0050679 positive regulation of epithelial cell proliferation GO:0050810 regulation of steroid biosynthetic process GO:0050872 white fat cell differentiation GO:0050873 brown fat cell differentiation GO:0051052 regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051055 negative regulation of lipid biosynthetic process GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051095 regulation of helicase activity GO:0051097 negative regulation of helicase activity GO:0051169 nuclear transport GO:0051170 nuclear import GO:0051235 maintenance of location GO:0051346 negative regulation of hydrolase activity GO:0051348 negative regulation of transferase activity GO:0051567 histone H3-K9 methylation GO:0051570 regulation of histone H3-K9 methylation GO:0051574 positive regulation of histone H3-K9 methylation GO:0051896 regulation of protein kinase B signaling GO:0051898 negative regulation of protein kinase B signaling GO:0052547 regulation of peptidase activity GO:0052548 regulation of endopeptidase activity GO:0055088 lipid homeostasis GO:0055089 fatty acid homeostasis GO:0055090 acylglycerol homeostasis GO:0055092 sterol homeostasis GO:0060612 adipose tissue development GO:0060765 regulation of androgen receptor signaling pathway GO:0060766 negative regulation of androgen receptor signaling pathway GO:0060968 regulation of gene silencing GO:0061082 myeloid leukocyte cytokine production GO:0061448 connective tissue development GO:0061458 reproductive system development GO:0061647 histone H3-K9 modification GO:0070059 intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress GO:0070301 cellular response to hydrogen peroxide GO:0070328 triglyceride homeostasis GO:0070482 response to oxygen levels GO:0070857 regulation of bile acid biosynthetic process GO:0070914 UV-damage excision repair GO:0070932 histone H3 deacetylation GO:0070997 neuron death GO:0071214 cellular response to abiotic stimulus GO:0071356 cellular response to tumor necrosis factor GO:0071375 cellular response to peptide hormone stimulus GO:0071383 cellular response to steroid hormone stimulus GO:0071394 cellular response to testosterone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071417 cellular response to organonitrogen compound GO:0071440 regulation of histone H3-K14 acetylation GO:0071441 negative regulation of histone H3-K14 acetylation GO:0071453 cellular response to oxygen levels GO:0071456 cellular response to hypoxia GO:0071478 cellular response to radiation GO:0071479 cellular response to ionizing radiation GO:0071482 cellular response to light stimulus GO:0071496 cellular response to external stimulus GO:0071559 response to transforming growth factor beta GO:0071560 cellular response to transforming growth factor beta stimulus GO:0071887 leukocyte apoptotic process GO:0071888 macrophage apoptotic process GO:0071897 DNA biosynthetic process GO:0071900 regulation of protein serine/threonine kinase activity GO:0071901 negative regulation of protein serine/threonine kinase activity GO:0071902 positive regulation of protein serine/threonine kinase activity GO:0072330 monocarboxylic acid biosynthetic process GO:0072331 signal transduction by p53 class mediator GO:0072332 intrinsic apoptotic signaling pathway by p53 class mediator GO:0090092 regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090101 negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090239 regulation of histone H4 acetylation GO:0090241 negative regulation of histone H4 acetylation GO:0090287 regulation of cellular response to growth factor stimulus GO:0090288 negative regulation of cellular response to growth factor stimulus GO:0090335 regulation of brown fat cell differentiation GO:0090342 regulation of cell aging GO:0090343 positive regulation of cell aging GO:0090344 negative regulation of cell aging GO:0090398 cellular senescence GO:0090400 stress-induced premature senescence GO:0097193 intrinsic apoptotic signaling pathway GO:0098732 macromolecule deacylation GO:1900034 regulation of cellular response to heat GO:1900076 regulation of cellular response to insulin stimulus GO:1900078 positive regulation of cellular response to insulin stimulus GO:1900180 regulation of protein localization to nucleus GO:1900407 regulation of cellular response to oxidative stress GO:1900408 negative regulation of cellular response to oxidative stress GO:1901214 regulation of neuron death GO:1901215 negative regulation of neuron death GO:1901342 regulation of vasculature development GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1901654 response to ketone GO:1901655 cellular response to ketone GO:1901796 regulation of signal transduction by p53 class mediator GO:1901797 negative regulation of signal transduction by p53 class mediator GO:1901983 regulation of protein acetylation GO:1901984 negative regulation of protein acetylation GO:1902165 regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902166 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator GO:1902175 regulation of oxidative stress-induced intrinsic apoptotic signaling pathway GO:1902176 negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway GO:1902229 regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902230 negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage GO:1902235 regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway GO:1902237 positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway GO:1902253 regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902254 negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator GO:1902275 regulation of chromatin organization GO:1902532 negative regulation of intracellular signal transduction GO:1902593 single-organism nuclear import GO:1902882 regulation of response to oxidative stress GO:1902883 negative regulation of response to oxidative stress GO:1903201 regulation of oxidative stress-induced cell death GO:1903202 negative regulation of oxidative stress-induced cell death GO:1903533 regulation of protein targeting GO:1903844 regulation of cellular response to transforming growth factor beta stimulus GO:1903845 negative regulation of cellular response to transforming growth factor beta stimulus GO:1904018 positive regulation of vasculature development GO:1904177 regulation of adipose tissue development GO:1904179 positive regulation of adipose tissue development GO:1904251 regulation of bile acid metabolic process GO:1904589 regulation of protein import GO:1905268 negative regulation of chromatin organization GO:1905269 positive regulation of chromatin organization GO:1990619 histone H3-K9 deacetylation GO:2000106 regulation of leukocyte apoptotic process GO:2000108 positive regulation of leukocyte apoptotic process GO:2000109 regulation of macrophage apoptotic process GO:2000111 positive regulation of macrophage apoptotic process GO:2000116 regulation of cysteine-type endopeptidase activity GO:2000479 regulation of cAMP-dependent protein kinase activity GO:2000480 negative regulation of cAMP-dependent protein kinase activity GO:2000481 positive regulation of cAMP-dependent protein kinase activity GO:2000618 regulation of histone H4-K16 acetylation GO:2000619 negative regulation of histone H4-K16 acetylation GO:2000654 regulation of cellular response to testosterone stimulus GO:2000655 negative regulation of cellular response to testosterone stimulus GO:2000756 regulation of peptidyl-lysine acetylation GO:2000757 negative regulation of peptidyl-lysine acetylation GO:2000772 regulation of cellular senescence GO:2000773 negative regulation of cellular senescence GO:2000774 positive regulation of cellular senescence GO:2001020 regulation of response to DNA damage stimulus GO:2001021 negative regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus GO:2001056 positive regulation of cysteine-type endopeptidase activity GO:2001233 regulation of apoptotic signaling pathway GO:2001234 negative regulation of apoptotic signaling pathway GO:2001235 positive regulation of apoptotic signaling pathway GO:2001242 regulation of intrinsic apoptotic signaling pathway GO:2001243 negative regulation of intrinsic apoptotic signaling pathway GO:2001244 positive regulation of intrinsic apoptotic signaling pathway GO:2001279 regulation of unsaturated fatty acid biosynthetic process |
| Molecular Function |
GO:0001046 core promoter sequence-specific DNA binding GO:0001047 core promoter binding GO:0002039 p53 binding GO:0003714 transcription corepressor activity GO:0004407 histone deacetylase activity GO:0008022 protein C-terminus binding GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0019215 intermediate filament binding GO:0032129 histone deacetylase activity (H3-K9 specific) GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0035257 nuclear hormone receptor binding GO:0042393 histone binding GO:0043398 HLH domain binding GO:0043425 bHLH transcription factor binding GO:0046969 NAD-dependent histone deacetylase activity (H3-K9 specific) GO:0048037 cofactor binding GO:0050662 coenzyme binding GO:0051019 mitogen-activated protein kinase binding GO:0051287 NAD binding GO:0051427 hormone receptor binding GO:0070403 NAD+ binding GO:1990254 keratin filament binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000791 euchromatin GO:0000792 heterochromatin GO:0005635 nuclear envelope GO:0005637 nuclear inner membrane GO:0005677 chromatin silencing complex GO:0005719 nuclear euchromatin GO:0005720 nuclear heterochromatin GO:0016604 nuclear body GO:0016605 PML body GO:0017053 transcriptional repressor complex GO:0031519 PcG protein complex GO:0031965 nuclear membrane GO:0033553 rDNA heterochromatin GO:0034708 methyltransferase complex GO:0035097 histone methyltransferase complex GO:0035098 ESC/E(Z) complex GO:0044454 nuclear chromosome part GO:0090568 nuclear transcriptional repressor complex |
| KEGG |
hsa04068 FoxO signaling pathway hsa04152 AMPK signaling pathway hsa04922 Glucagon signaling pathway |
| Reactome |
R-HSA-3371556: Cellular response to heat stress R-HSA-2262752: Cellular responses to stress R-HSA-400253: Circadian Clock R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-5250941: Negative epigenetic regulation of rRNA expression R-HSA-1368082: RORA activates gene expression R-HSA-3371453: Regulation of HSF1-mediated heat shock response R-HSA-427359: SIRT1 negatively regulates rRNA Expression |
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
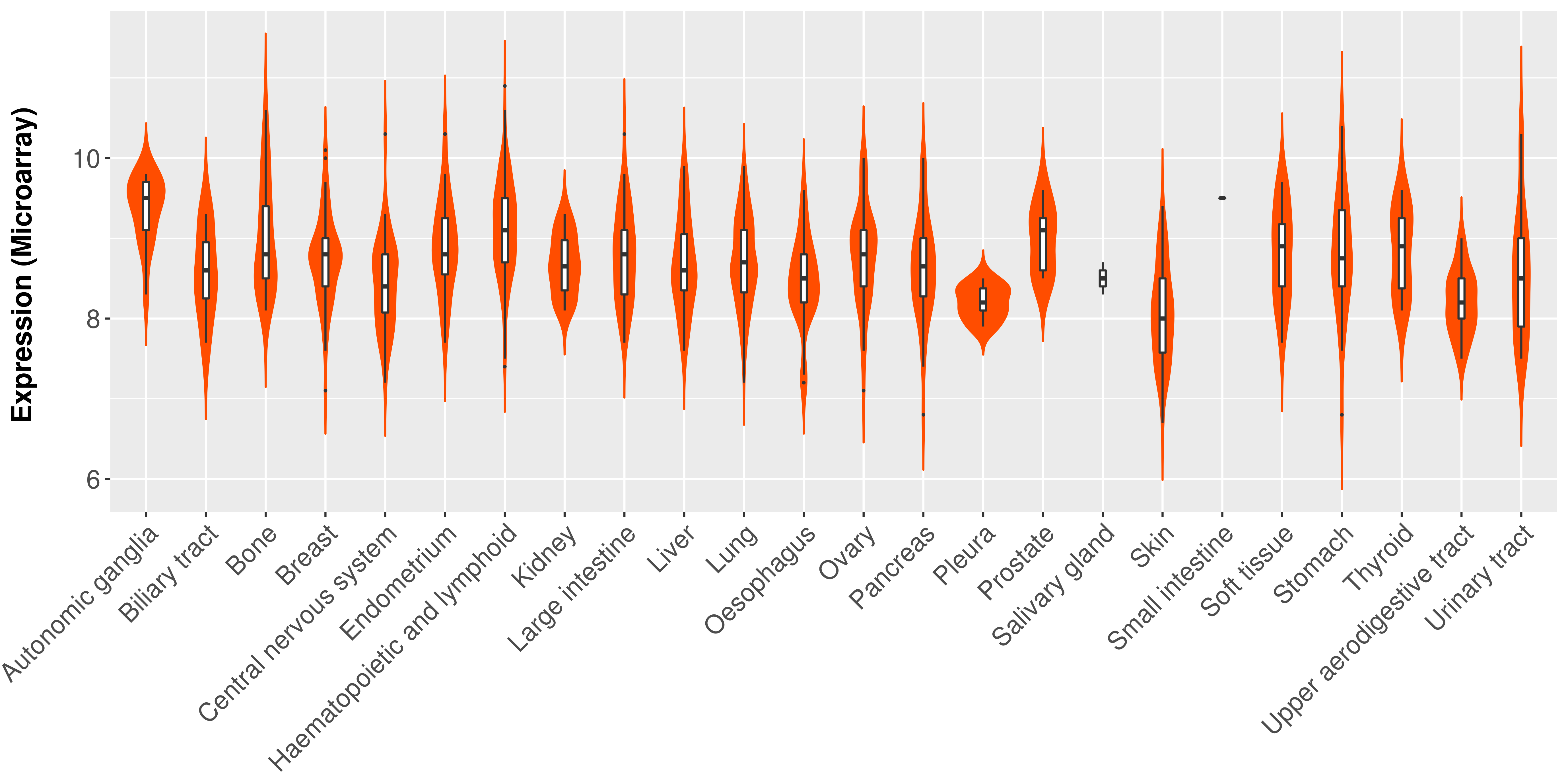
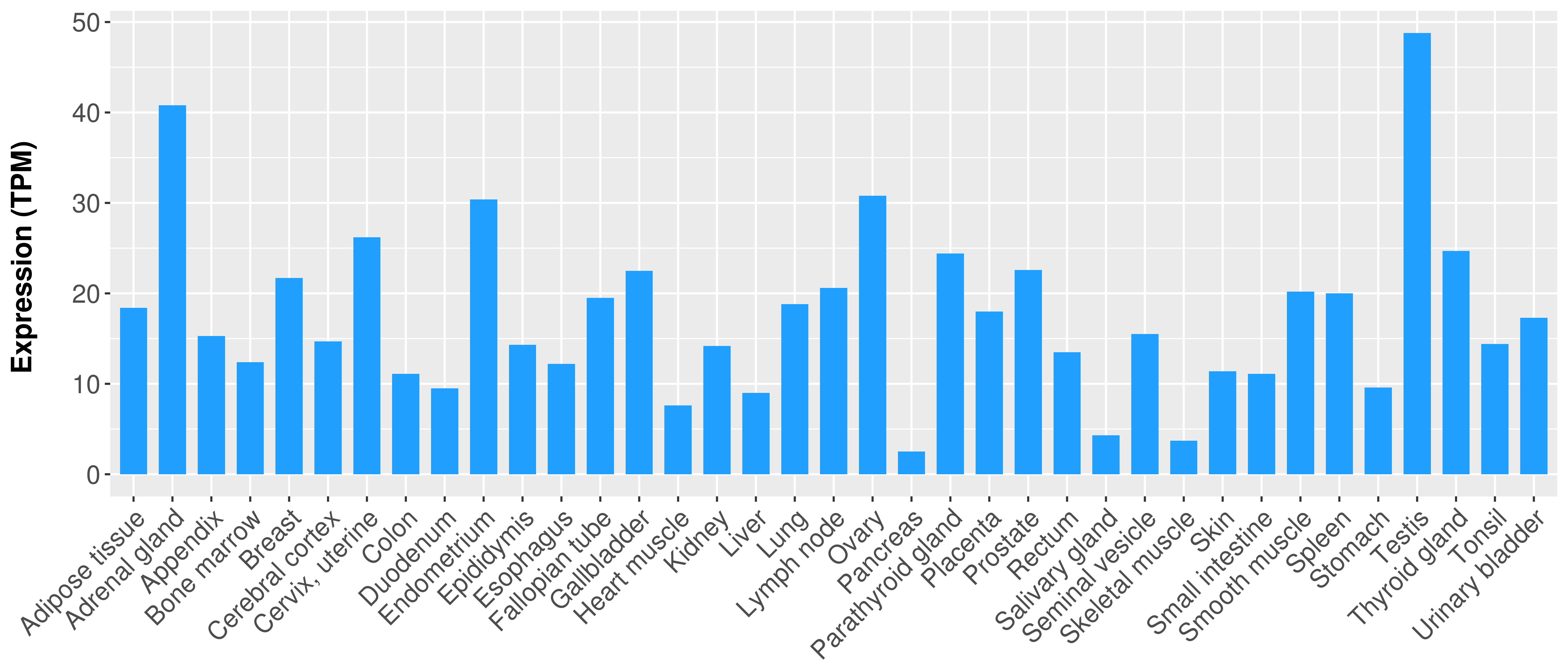
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
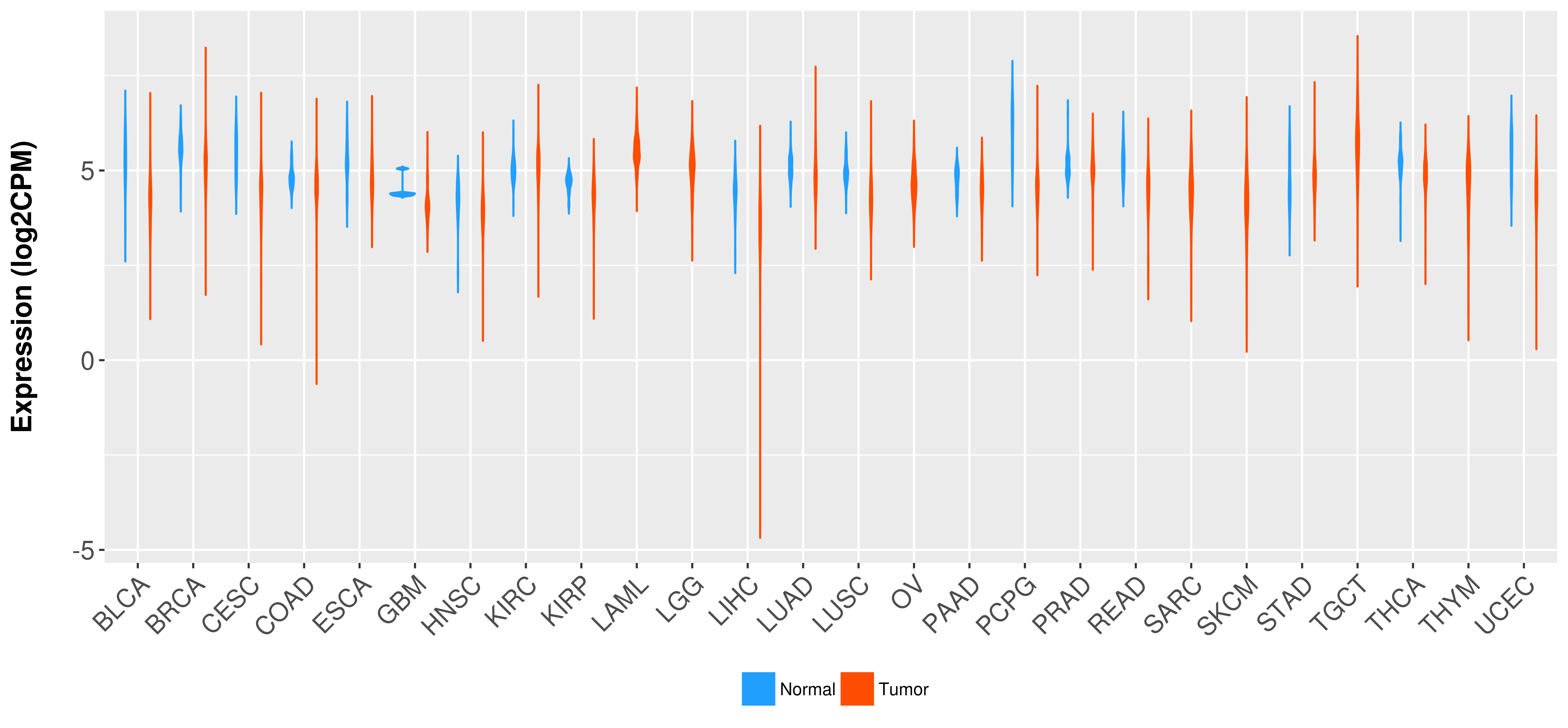
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
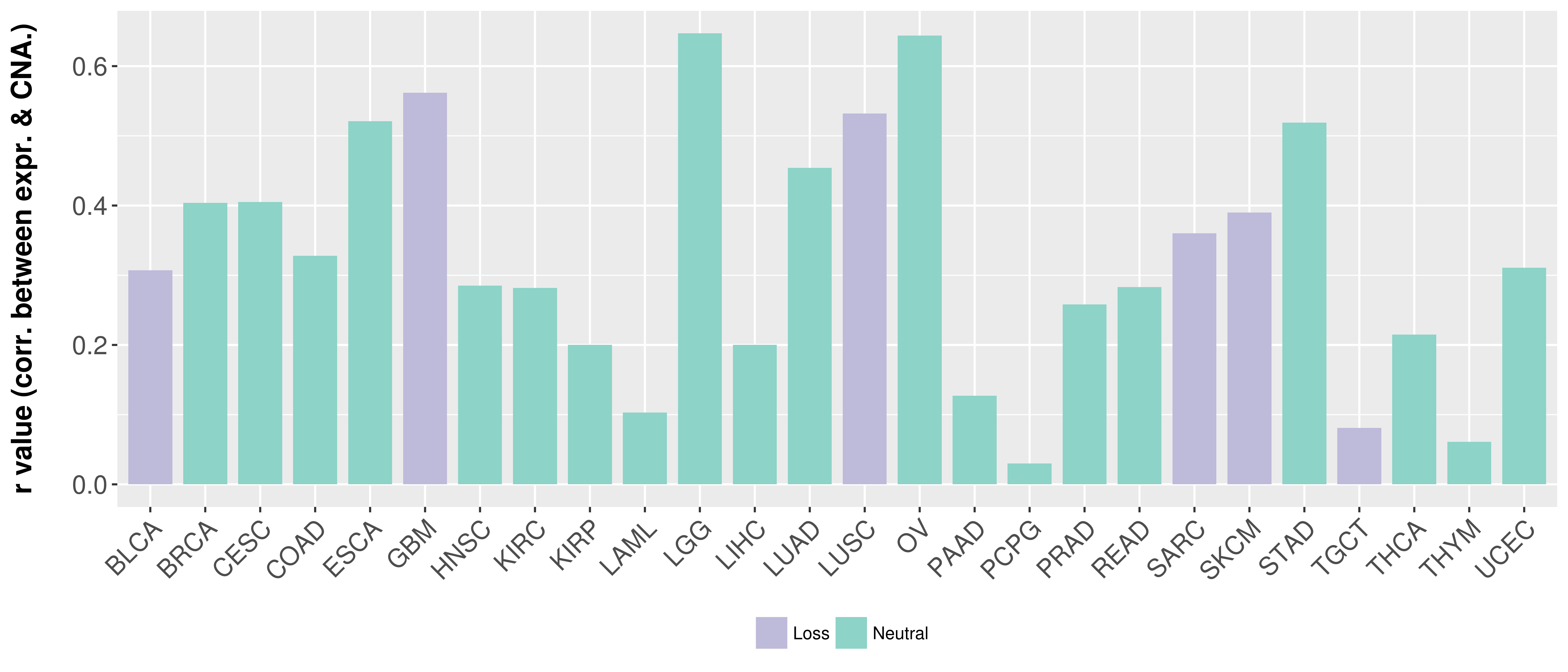
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
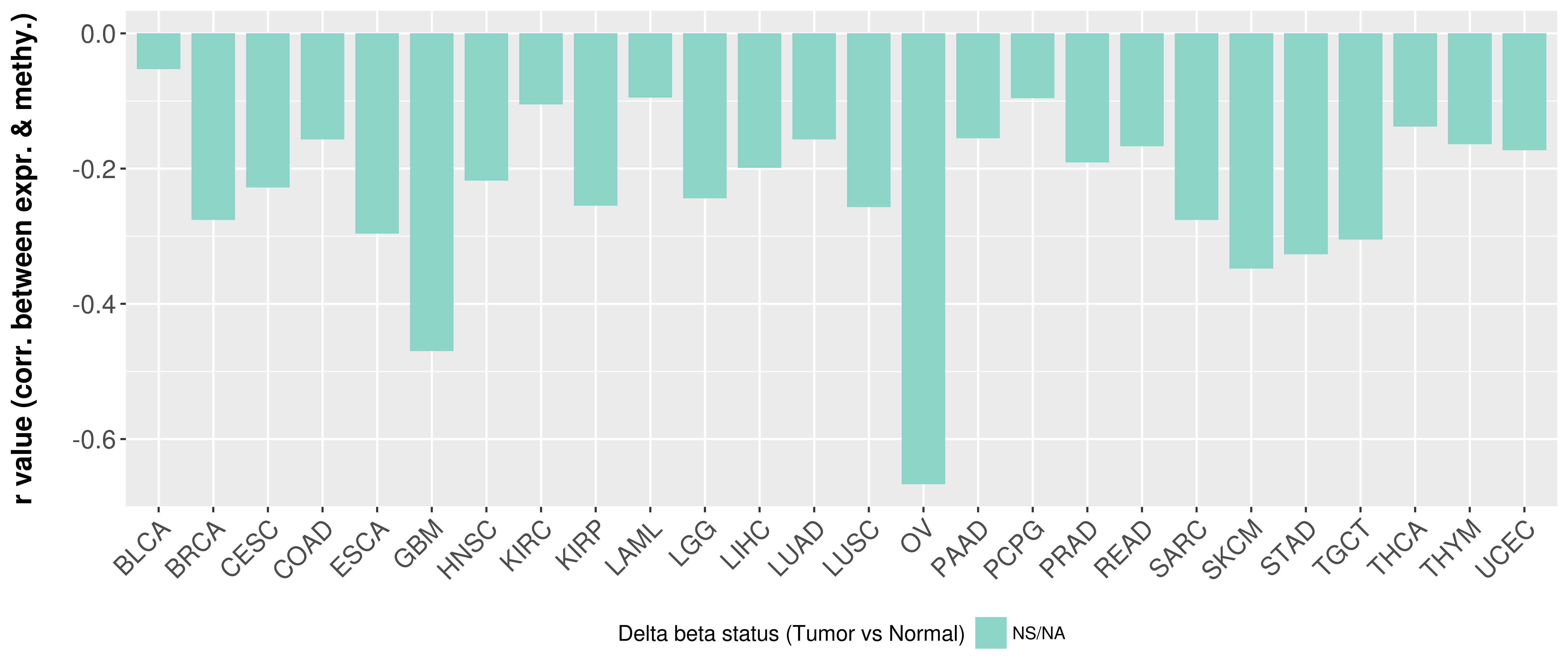
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
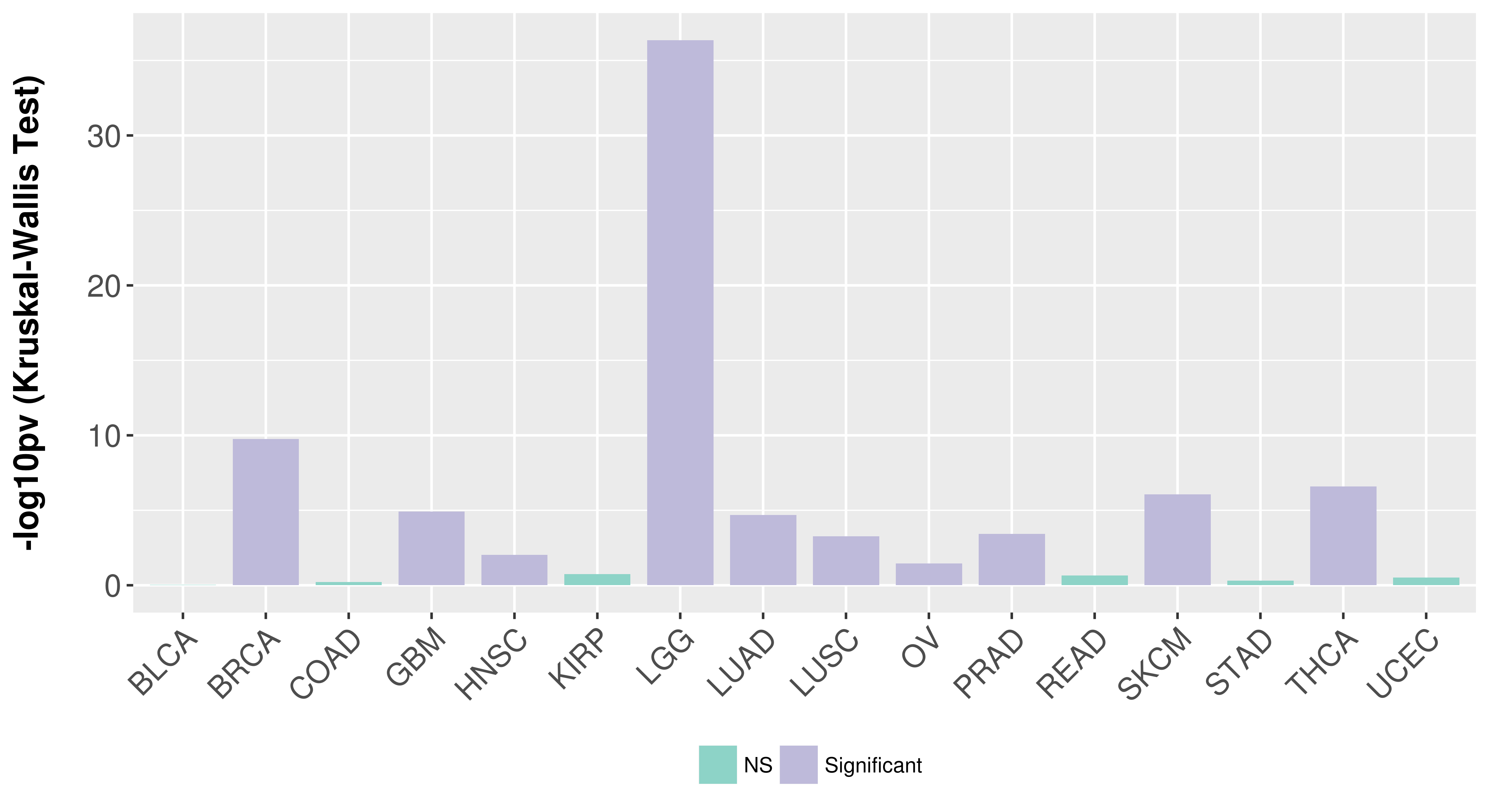
Association between expresson and subtype.
|
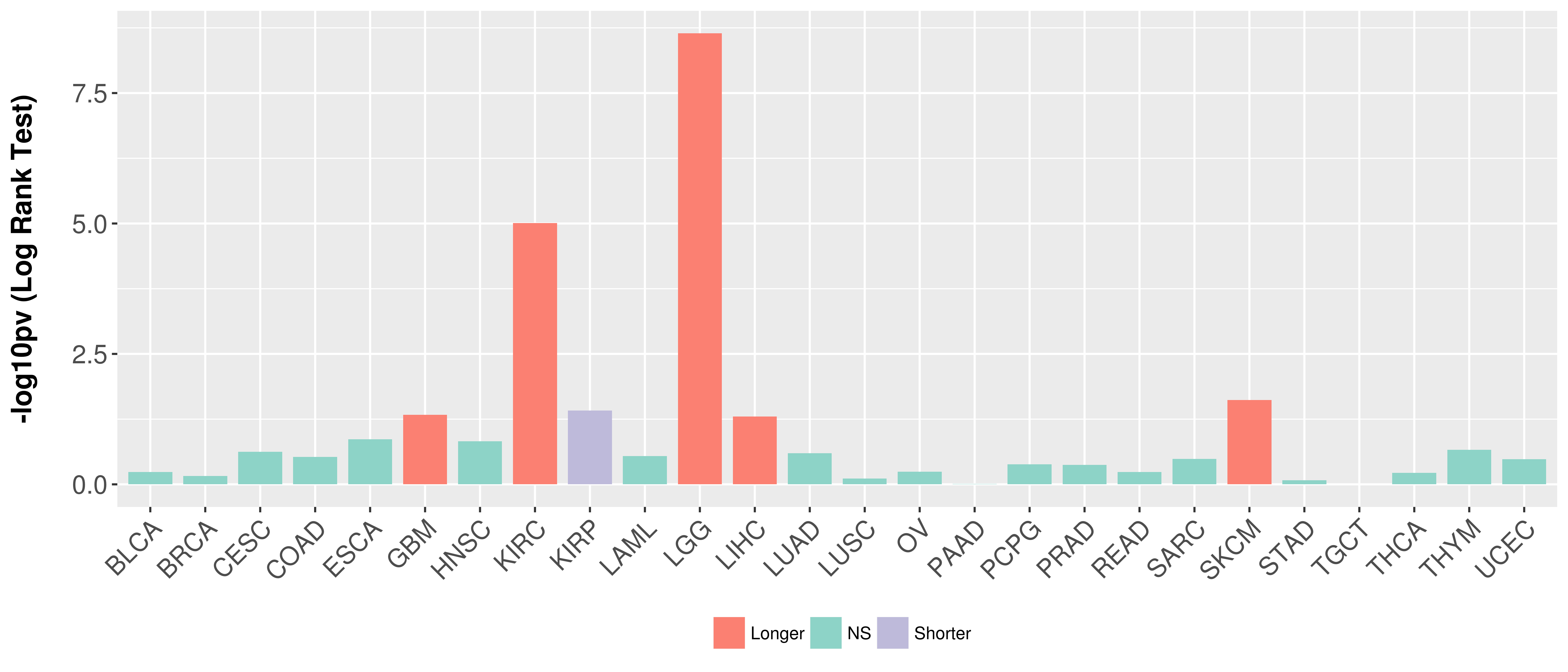
Overall survival analysis based on expression.
|
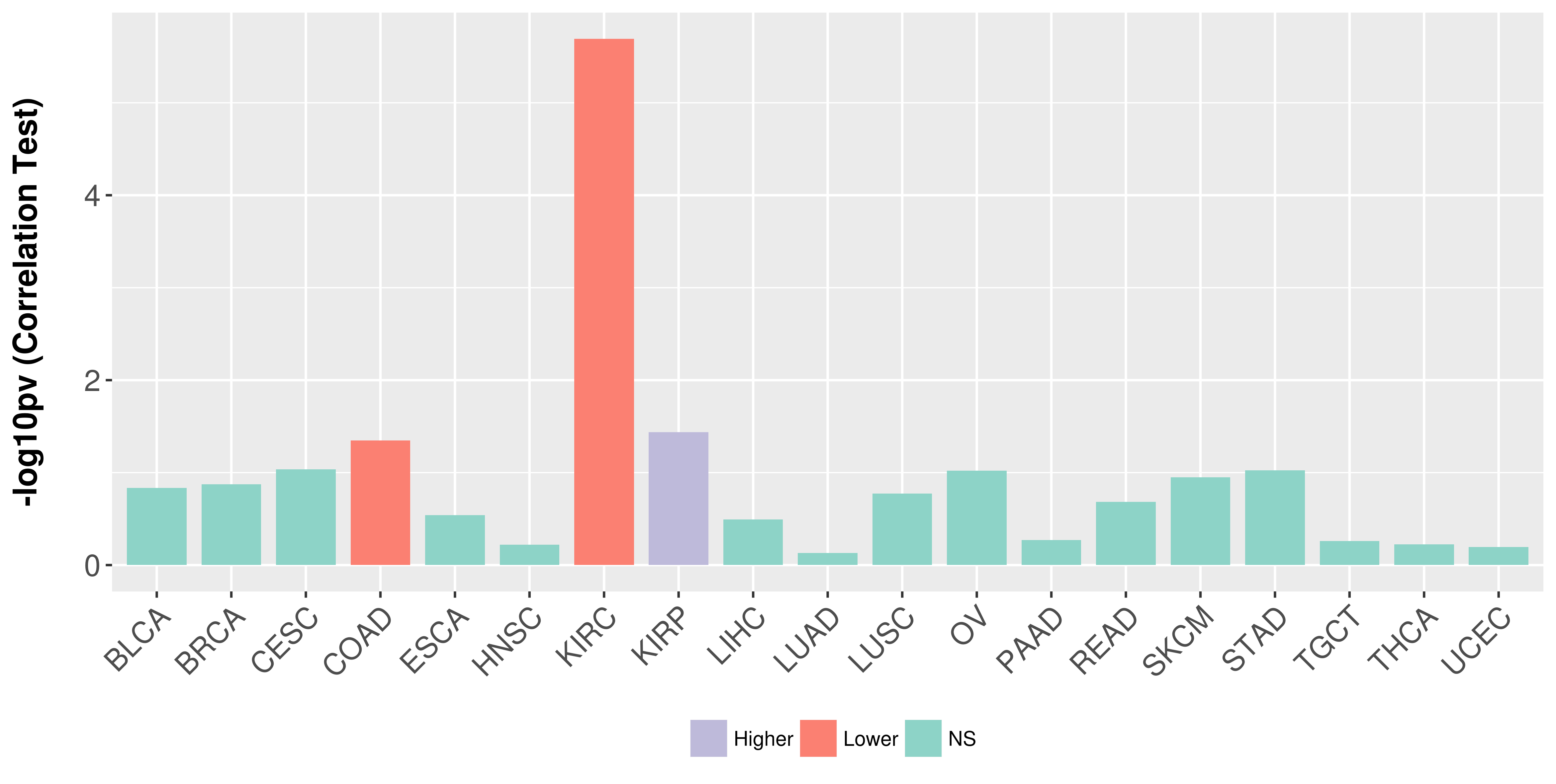
Association between expresson and stage.
|
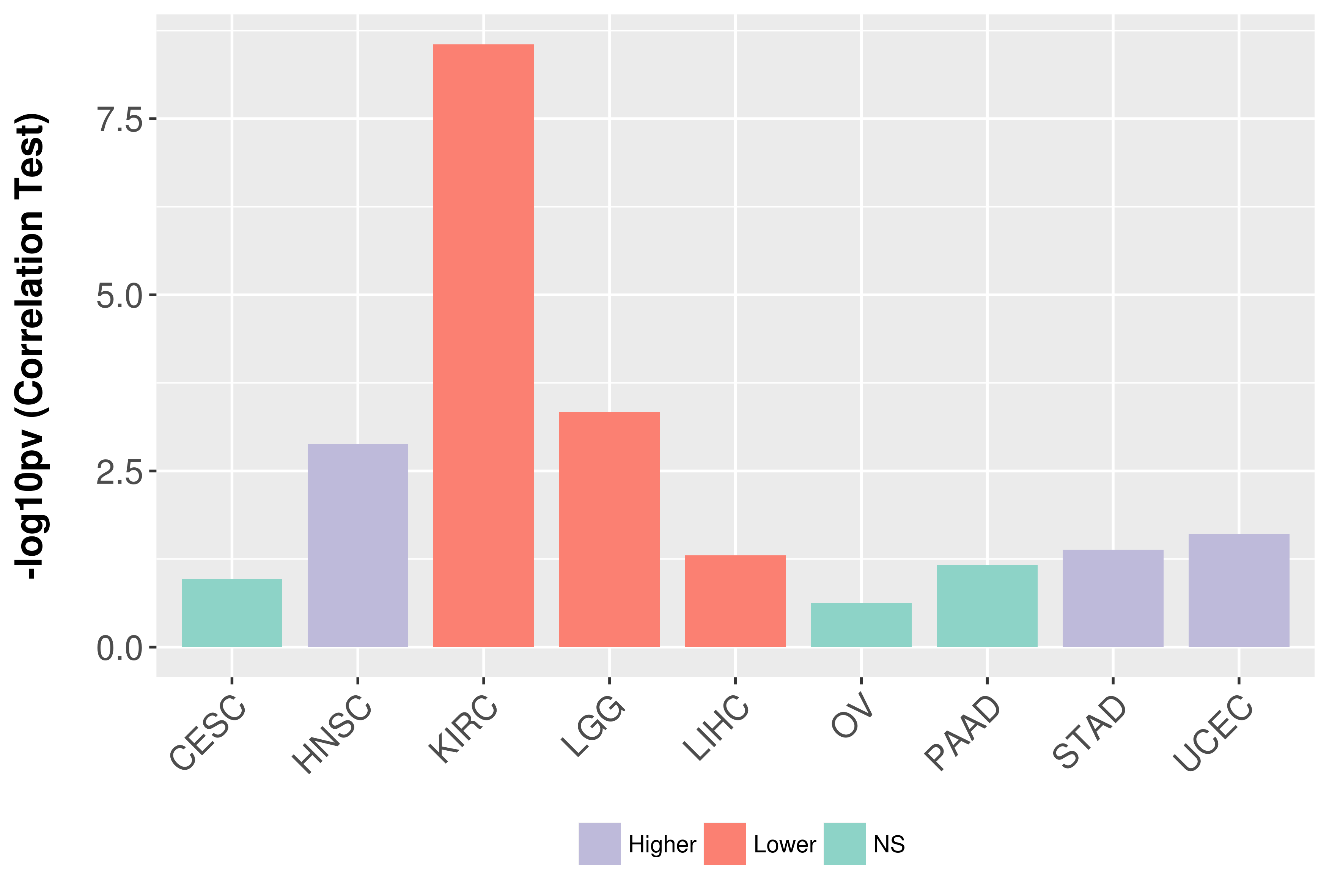
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
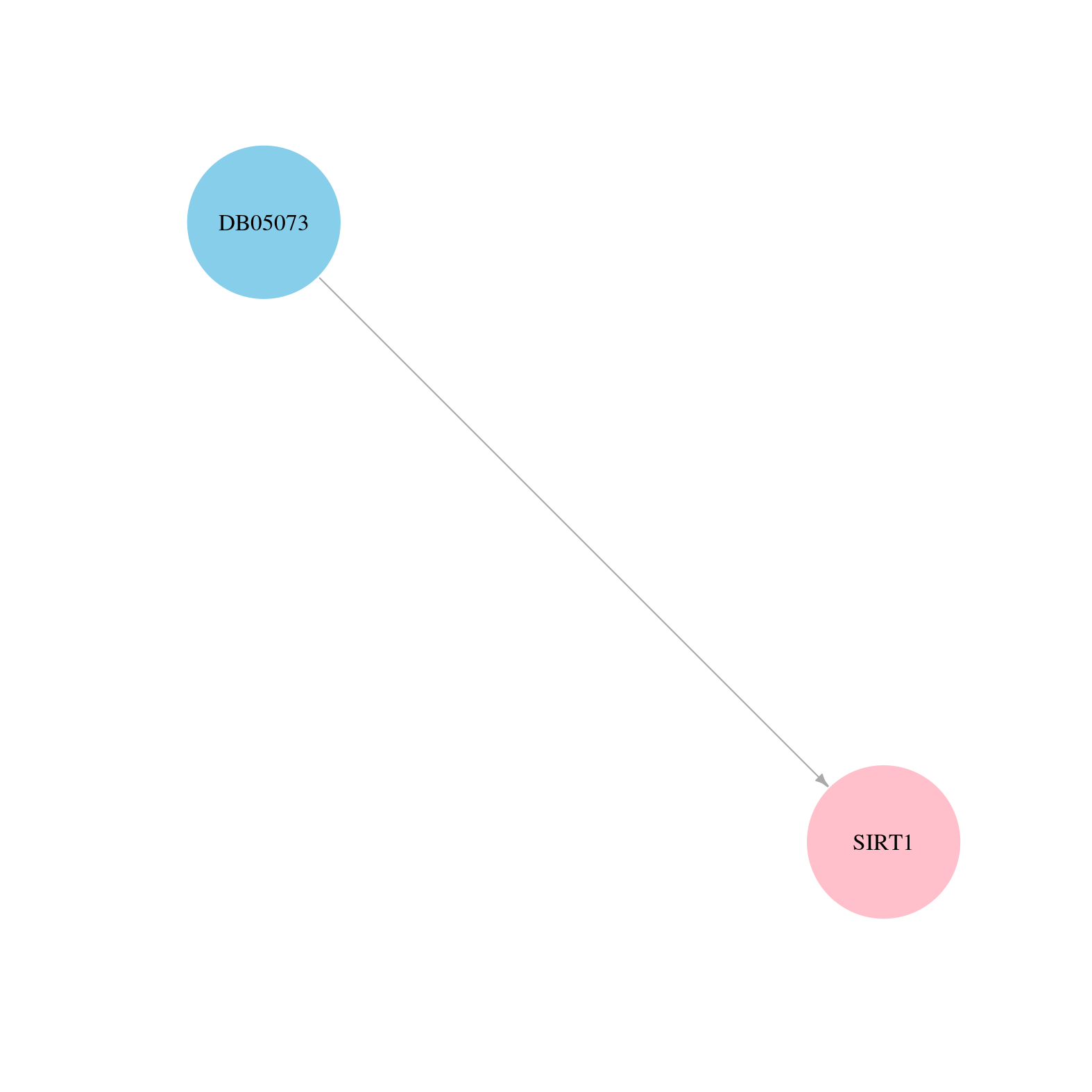
> Drugs from DrugBank database |
|
| Summary | |
|---|---|
| Symbol | SIRT1 |
| Name | sirtuin 1 |
| Aliases | SIR2L1; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 1; sirtuin (silent mat ...... |
| Location | 10q21.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|