| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
82 |
1784 |
4.027e-29 |
1.874e-25 |
| 2 |
POSITIVE REGULATION OF GENE EXPRESSION |
78 |
1733 |
6.068e-27 |
1.412e-23 |
| 3 |
POSITIVE REGULATION OF BIOSYNTHETIC PROCESS |
75 |
1805 |
1.185e-23 |
1.837e-20 |
| 4 |
POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
53 |
1004 |
5.82e-21 |
6.77e-18 |
| 5 |
CELLULAR RESPONSE TO DNA DAMAGE STIMULUS |
45 |
720 |
1.227e-20 |
1.142e-17 |
| 6 |
CHROMOSOME ORGANIZATION |
51 |
1009 |
2.403e-19 |
1.863e-16 |
| 7 |
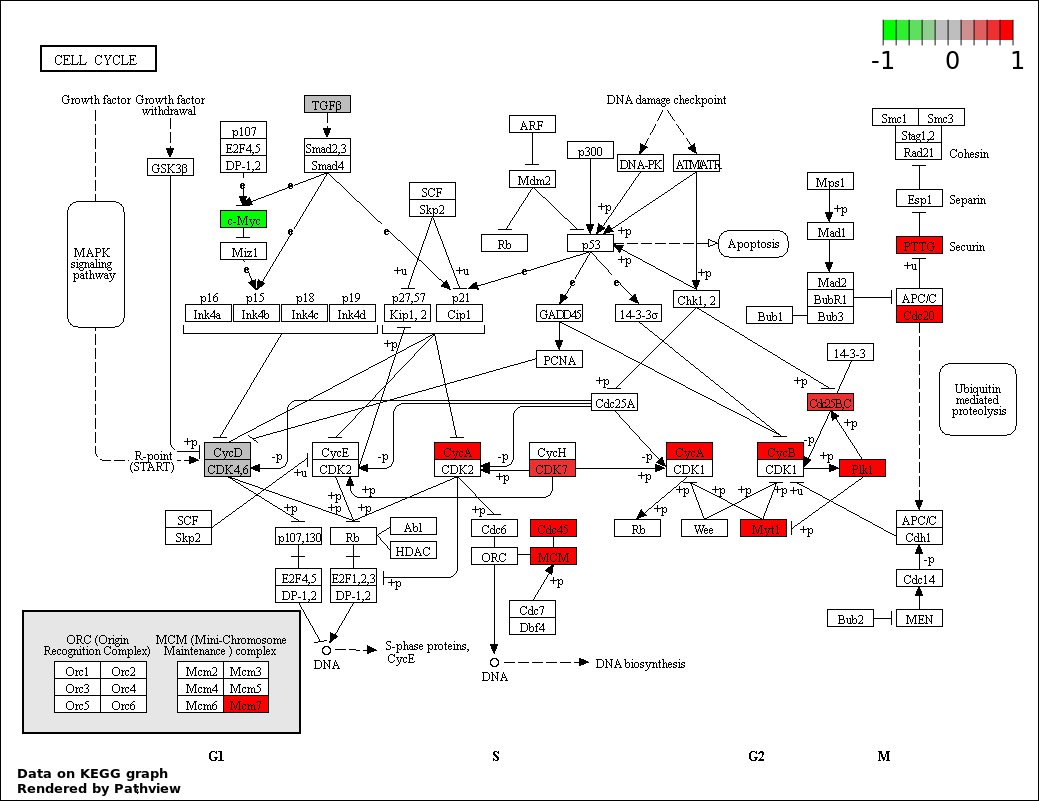
CELL CYCLE |
58 |
1316 |
3.385e-19 |
2.25e-16 |
| 8 |
CELLULAR RESPONSE TO STRESS |
63 |
1565 |
6.388e-19 |
3.716e-16 |
| 9 |
NEGATIVE REGULATION OF GENE EXPRESSION |
61 |
1493 |
1.317e-18 |
6.811e-16 |
| 10 |
TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
42 |
724 |
4.309e-18 |
1.9e-15 |
| 11 |
CELL CYCLE PROCESS |
51 |
1081 |
4.493e-18 |
1.9e-15 |
| 12 |
CHROMATIN ORGANIZATION |
40 |
663 |
7.695e-18 |
2.984e-15 |
| 13 |
REGULATION OF CELL CYCLE |
47 |
949 |
1.771e-17 |
6.338e-15 |
| 14 |
MITOTIC CELL CYCLE |
42 |
766 |
3.286e-17 |
1.092e-14 |
| 15 |
REGULATION OF CELL DEATH |
58 |
1472 |
6.149e-17 |
1.907e-14 |
| 16 |
NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS |
57 |
1517 |
1.009e-15 |
2.933e-13 |
| 17 |
CHROMATIN MODIFICATION |
33 |
539 |
5.157e-15 |
1.412e-12 |
| 18 |
INTERSPECIES INTERACTION BETWEEN ORGANISMS |
36 |
662 |
1.013e-14 |
2.481e-12 |
| 19 |
SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM |
36 |
662 |
1.013e-14 |
2.481e-12 |
| 20 |
NEGATIVE REGULATION OF CELL DEATH |
41 |
872 |
1.586e-14 |
3.691e-12 |
| 21 |
REGULATION OF BINDING |
24 |
283 |
3.146e-14 |
6.971e-12 |
| 22 |
NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
37 |
740 |
5.574e-14 |
1.179e-11 |
| 23 |
CELLULAR RESPONSE TO ORGANIC SUBSTANCE |
60 |
1848 |
1.046e-13 |
2.117e-11 |
| 24 |
PEPTIDYL AMINO ACID MODIFICATION |
39 |
841 |
1.18e-13 |
2.288e-11 |
| 25 |
REGULATION OF CELL PROLIFERATION |
53 |
1496 |
1.404e-13 |
2.614e-11 |
| 26 |
REGULATION OF CELLULAR RESPONSE TO STRESS |
35 |
691 |
1.993e-13 |
3.434e-11 |
| 27 |
REGULATION OF MITOTIC CELL CYCLE |
29 |
468 |
1.964e-13 |
3.434e-11 |
| 28 |
DNA TEMPLATED TRANSCRIPTION ELONGATION |
15 |
97 |
3.922e-13 |
6.518e-11 |
| 29 |
REGULATION OF CELL CYCLE PROCESS |
31 |
558 |
4.939e-13 |
7.924e-11 |
| 30 |
TRANSCRIPTION ELONGATION FROM RNA POLYMERASE II PROMOTER |
14 |
82 |
5.897e-13 |
9.146e-11 |
| 31 |
REGULATION OF PROTEIN MODIFICATION PROCESS |
56 |
1710 |
6.211e-13 |
9.323e-11 |
| 32 |
NEGATIVE REGULATION OF CELL CYCLE |
27 |
433 |
1.222e-12 |
1.777e-10 |
| 33 |
POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
44 |
1152 |
2.077e-12 |
2.929e-10 |
| 34 |
COVALENT CHROMATIN MODIFICATION |
24 |
345 |
2.344e-12 |
3.207e-10 |
| 35 |
DNA TEMPLATED TRANSCRIPTION INITIATION |
19 |
202 |
2.668e-12 |
3.547e-10 |
| 36 |
DNA METABOLIC PROCESS |
35 |
758 |
2.831e-12 |
3.659e-10 |
| 37 |
REGULATION OF CELL DIFFERENTIATION |
50 |
1492 |
6.329e-12 |
7.749e-10 |
| 38 |
POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS |
50 |
1492 |
6.329e-12 |
7.749e-10 |
| 39 |
CELL CYCLE CHECKPOINT |
18 |
194 |
1.288e-11 |
1.536e-09 |
| 40 |
PROTEIN PHOSPHORYLATION |
38 |
944 |
1.813e-11 |
2.109e-09 |
| 41 |
TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER |
16 |
153 |
3e-11 |
3.404e-09 |
| 42 |
RESPONSE TO ABIOTIC STIMULUS |
39 |
1024 |
4.958e-11 |
5.493e-09 |
| 43 |
DNA REPAIR |
26 |
480 |
7.508e-11 |
8.124e-09 |
| 44 |
NEGATIVE REGULATION OF CELL PROLIFERATION |
30 |
643 |
9.28e-11 |
9.814e-09 |
| 45 |
RESPONSE TO ENDOGENOUS STIMULUS |
47 |
1450 |
9.696e-11 |
1.003e-08 |
| 46 |
REGULATION OF RESPONSE TO STRESS |
47 |
1468 |
1.467e-10 |
1.484e-08 |
| 47 |
NEGATIVE REGULATION OF MOLECULAR FUNCTION |
39 |
1079 |
2.301e-10 |
2.229e-08 |
| 48 |
PHOSPHORYLATION |
42 |
1228 |
2.297e-10 |
2.229e-08 |
| 49 |
REGULATION OF PROTEOLYSIS |
31 |
711 |
2.347e-10 |
2.229e-08 |
| 50 |
REGULATION OF TRANSFERASE ACTIVITY |
36 |
946 |
3.116e-10 |
2.9e-08 |
| 51 |
POSITIVE REGULATION OF CELL CYCLE |
21 |
332 |
3.421e-10 |
3.122e-08 |
| 52 |
REGULATION OF CELL CYCLE ARREST |
13 |
108 |
3.827e-10 |
3.425e-08 |
| 53 |
RESPONSE TO ORGANIC CYCLIC COMPOUND |
35 |
917 |
5.249e-10 |
4.608e-08 |
| 54 |
POSITIVE REGULATION OF CELL CYCLE PROCESS |
18 |
247 |
6.92e-10 |
5.963e-08 |
| 55 |
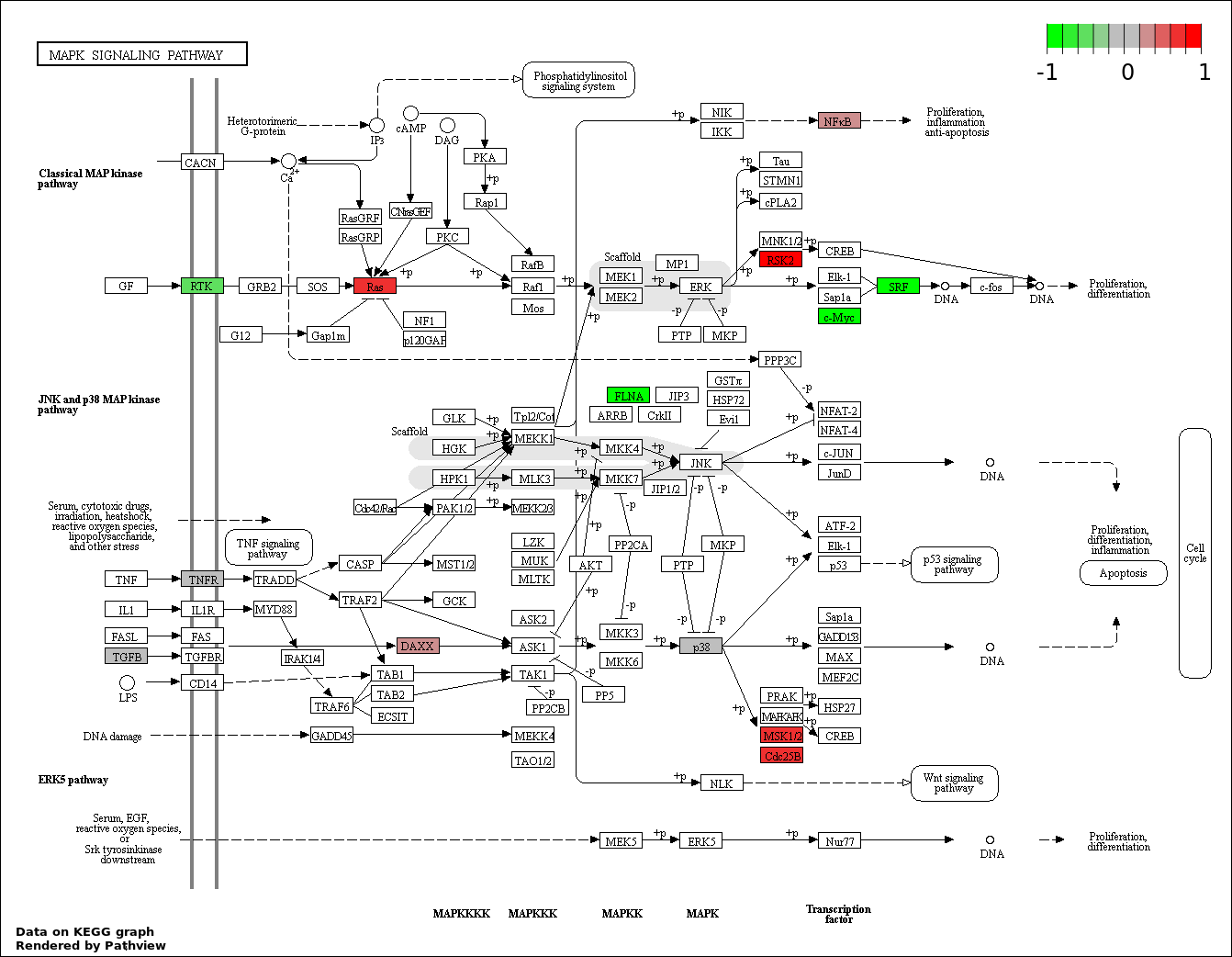
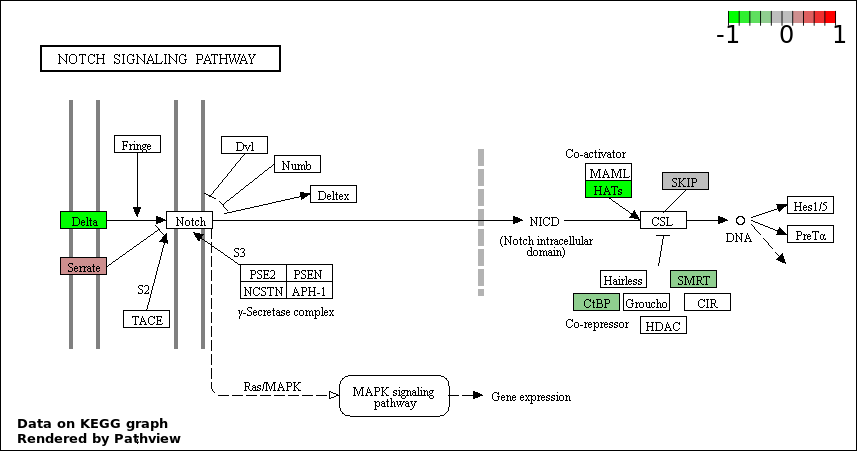
NOTCH SIGNALING PATHWAY |
13 |
114 |
7.575e-10 |
6.409e-08 |
| 56 |
RESPONSE TO STEROID HORMONE |
25 |
497 |
8.301e-10 |
6.897e-08 |
| 57 |
CELL DIVISION |
24 |
460 |
8.944e-10 |
7.176e-08 |
| 58 |
RESPONSE TO LIPID |
34 |
888 |
8.818e-10 |
7.176e-08 |
| 59 |
MACROMOLECULAR COMPLEX ASSEMBLY |
44 |
1398 |
1.088e-09 |
8.582e-08 |
| 60 |
CELL CYCLE PHASE TRANSITION |
18 |
255 |
1.155e-09 |
8.94e-08 |
| 61 |
POSITIVE REGULATION OF DEVELOPMENTAL PROCESS |
39 |
1142 |
1.172e-09 |
8.94e-08 |
| 62 |
REGULATION OF CELL CYCLE PHASE TRANSITION |
20 |
321 |
1.193e-09 |
8.953e-08 |
| 63 |
NEGATIVE REGULATION OF MITOTIC CELL CYCLE |
16 |
199 |
1.524e-09 |
1.125e-07 |
| 64 |
PROTEIN DNA COMPLEX SUBUNIT ORGANIZATION |
17 |
229 |
1.578e-09 |
1.147e-07 |
| 65 |
DNA INTEGRITY CHECKPOINT |
14 |
146 |
1.68e-09 |
1.202e-07 |
| 66 |
CHROMATIN REMODELING |
14 |
150 |
2.396e-09 |
1.689e-07 |
| 67 |
REGULATION OF ORGANELLE ORGANIZATION |
39 |
1178 |
2.807e-09 |
1.95e-07 |
| 68 |
TISSUE MORPHOGENESIS |
25 |
533 |
3.45e-09 |
2.361e-07 |
| 69 |
NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS |
37 |
1087 |
3.638e-09 |
2.453e-07 |
| 70 |
SKELETAL SYSTEM DEVELOPMENT |
23 |
455 |
3.736e-09 |
2.484e-07 |
| 71 |
TISSUE DEVELOPMENT |
45 |
1518 |
4.39e-09 |
2.877e-07 |
| 72 |
REGULATION OF CHROMOSOME ORGANIZATION |
18 |
278 |
4.55e-09 |
2.94e-07 |
| 73 |
PROTEIN COMPLEX SUBUNIT ORGANIZATION |
45 |
1527 |
5.263e-09 |
3.355e-07 |
| 74 |
CELLULAR RESPONSE TO STEROID HORMONE STIMULUS |
16 |
218 |
5.703e-09 |
3.586e-07 |
| 75 |
CELLULAR RESPONSE TO ENDOGENOUS STIMULUS |
35 |
1008 |
6.155e-09 |
3.818e-07 |
| 76 |
REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
14 |
162 |
6.528e-09 |
3.997e-07 |
| 77 |
POSITIVE REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION |
7 |
24 |
7.554e-09 |
4.565e-07 |
| 78 |
MITOTIC NUCLEAR DIVISION |
20 |
361 |
8.885e-09 |
5.233e-07 |
| 79 |
MITOTIC CELL CYCLE CHECKPOINT |
13 |
139 |
8.815e-09 |
5.233e-07 |
| 80 |
MORPHOGENESIS OF AN EPITHELIUM |
21 |
400 |
9.561e-09 |
5.561e-07 |
| 81 |
POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
42 |
1395 |
1.02e-08 |
5.861e-07 |
| 82 |
REGULATION OF PROTEIN BINDING |
14 |
168 |
1.044e-08 |
5.921e-07 |
| 83 |
POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
37 |
1135 |
1.145e-08 |
6.417e-07 |
| 84 |
ORGANELLE FISSION |
23 |
496 |
1.876e-08 |
1.039e-06 |
| 85 |
CELLULAR RESPONSE TO LIPID |
22 |
457 |
2e-08 |
1.095e-06 |
| 86 |
CELL PROLIFERATION |
27 |
672 |
2.089e-08 |
1.13e-06 |
| 87 |
DNA CONFORMATION CHANGE |
17 |
273 |
2.205e-08 |
1.179e-06 |
| 88 |
POSITIVE REGULATION OF CELL PROLIFERATION |
30 |
814 |
2.262e-08 |
1.196e-06 |
| 89 |
POSITIVE REGULATION OF TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER |
6 |
17 |
2.54e-08 |
1.328e-06 |
| 90 |
REGULATION OF CHROMATIN ORGANIZATION |
13 |
152 |
2.597e-08 |
1.343e-06 |
| 91 |
PEPTIDYL LYSINE MODIFICATION |
18 |
312 |
2.729e-08 |
1.395e-06 |
| 92 |
POSITIVE REGULATION OF BINDING |
12 |
127 |
2.967e-08 |
1.5e-06 |
| 93 |
NEGATIVE REGULATION OF CELL CYCLE PROCESS |
15 |
214 |
3.202e-08 |
1.585e-06 |
| 94 |
REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
17 |
280 |
3.198e-08 |
1.585e-06 |
| 95 |
REGULATION OF FIBROBLAST PROLIFERATION |
10 |
81 |
3.323e-08 |
1.627e-06 |
| 96 |
PROTEIN LOCALIZATION TO ORGANELLE |
24 |
556 |
3.54e-08 |
1.716e-06 |
| 97 |
ORGAN REGENERATION |
10 |
83 |
4.217e-08 |
2.023e-06 |
| 98 |
REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION |
7 |
31 |
5.356e-08 |
2.517e-06 |
| 99 |
POSITIVE REGULATION OF CHROMATIN MODIFICATION |
10 |
85 |
5.316e-08 |
2.517e-06 |
| 100 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
25 |
616 |
5.973e-08 |
2.779e-06 |
| 101 |
CELL CYCLE G1 S PHASE TRANSITION |
11 |
111 |
6.903e-08 |
3.149e-06 |
| 102 |
G1 S TRANSITION OF MITOTIC CELL CYCLE |
11 |
111 |
6.903e-08 |
3.149e-06 |
| 103 |
NCRNA METABOLIC PROCESS |
23 |
533 |
6.979e-08 |
3.153e-06 |
| 104 |
REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT |
45 |
1672 |
7.911e-08 |
3.539e-06 |
| 105 |
RESPONSE TO RADIATION |
20 |
413 |
8.247e-08 |
3.654e-06 |
| 106 |
REGULATION OF PHOSPHORUS METABOLIC PROCESS |
44 |
1618 |
8.471e-08 |
3.718e-06 |
| 107 |
VIRAL LATENCY |
5 |
11 |
8.781e-08 |
3.819e-06 |
| 108 |
POSITIVE REGULATION OF CELL DIFFERENTIATION |
29 |
823 |
1.035e-07 |
4.46e-06 |
| 109 |
REGULATION OF CATABOLIC PROCESS |
27 |
731 |
1.162e-07 |
4.96e-06 |
| 110 |
PROTEASOMAL PROTEIN CATABOLIC PROCESS |
16 |
271 |
1.206e-07 |
5.103e-06 |
| 111 |
CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND |
21 |
465 |
1.253e-07 |
5.254e-06 |
| 112 |
REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS |
12 |
145 |
1.298e-07 |
5.391e-06 |
| 113 |
POSITIVE REGULATION OF FIBROBLAST PROLIFERATION |
8 |
53 |
1.643e-07 |
6.765e-06 |
| 114 |
EPITHELIUM DEVELOPMENT |
31 |
945 |
1.771e-07 |
7.165e-06 |
| 115 |
EMBRYO DEVELOPMENT |
30 |
894 |
1.76e-07 |
7.165e-06 |
| 116 |
RESPONSE TO GROWTH FACTOR |
21 |
475 |
1.786e-07 |
7.165e-06 |
| 117 |
POSITIVE REGULATION OF CHROMOSOME ORGANIZATION |
12 |
150 |
1.882e-07 |
7.485e-06 |
| 118 |
RNA CAPPING |
7 |
37 |
1.977e-07 |
7.601e-06 |
| 119 |
X7 METHYLGUANOSINE RNA CAPPING |
7 |
37 |
1.977e-07 |
7.601e-06 |
| 120 |
GLAND DEVELOPMENT |
19 |
395 |
1.953e-07 |
7.601e-06 |
| 121 |
REGULATION OF TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER |
6 |
23 |
1.955e-07 |
7.601e-06 |
| 122 |
POSITIVE REGULATION OF MOLECULAR FUNCTION |
46 |
1791 |
2.164e-07 |
8.228e-06 |
| 123 |
REGULATION OF MUSCLE CELL DIFFERENTIATION |
12 |
152 |
2.175e-07 |
8.228e-06 |
| 124 |
MITOTIC DNA INTEGRITY CHECKPOINT |
10 |
100 |
2.527e-07 |
9.482e-06 |
| 125 |
ANAPHASE PROMOTING COMPLEX DEPENDENT CATABOLIC PROCESS |
9 |
77 |
2.648e-07 |
9.795e-06 |
| 126 |
POSITIVE REGULATION OF PROTEOLYSIS |
18 |
363 |
2.652e-07 |
9.795e-06 |
| 127 |
POSITIVE REGULATION OF RESPONSE TO STIMULUS |
48 |
1929 |
2.848e-07 |
1.043e-05 |
| 128 |
POSITIVE REGULATION OF VIRAL TRANSCRIPTION |
7 |
39 |
2.895e-07 |
1.052e-05 |
| 129 |
POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION |
20 |
448 |
3.043e-07 |
1.098e-05 |
| 130 |
NEGATIVE REGULATION OF BINDING |
11 |
131 |
3.777e-07 |
1.342e-05 |
| 131 |
BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE |
11 |
131 |
3.777e-07 |
1.342e-05 |
| 132 |
NEGATIVE REGULATION OF RESPONSE TO STIMULUS |
38 |
1360 |
3.857e-07 |
1.36e-05 |
| 133 |
SNRNA METABOLIC PROCESS |
9 |
82 |
4.575e-07 |
1.601e-05 |
| 134 |
CELLULAR RESPONSE TO ABIOTIC STIMULUS |
15 |
263 |
4.701e-07 |
1.633e-05 |
| 135 |
AGING |
15 |
264 |
4.934e-07 |
1.701e-05 |
| 136 |
REGULATION OF VIRAL TRANSCRIPTION |
8 |
61 |
5.04e-07 |
1.724e-05 |
| 137 |
REGULATION OF DNA METABOLIC PROCESS |
17 |
340 |
5.129e-07 |
1.742e-05 |
| 138 |
POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY |
13 |
197 |
5.403e-07 |
1.822e-05 |
| 139 |
ORGAN MORPHOGENESIS |
28 |
841 |
5.499e-07 |
1.841e-05 |
| 140 |
EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING |
22 |
554 |
5.612e-07 |
1.865e-05 |
| 141 |
PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS |
48 |
1977 |
5.85e-07 |
1.931e-05 |
| 142 |
POSITIVE REGULATION OF LIGASE ACTIVITY |
10 |
110 |
6.188e-07 |
2.028e-05 |
| 143 |
POSITIVE REGULATION OF CELL CYCLE ARREST |
9 |
85 |
6.238e-07 |
2.03e-05 |
| 144 |
INTRACELLULAR RECEPTOR SIGNALING PATHWAY |
12 |
168 |
6.415e-07 |
2.073e-05 |
| 145 |
REGULATION OF CELL DIVISION |
15 |
272 |
7.201e-07 |
2.311e-05 |
| 146 |
RESPONSE TO DRUG |
19 |
431 |
7.355e-07 |
2.344e-05 |
| 147 |
IN UTERO EMBRYONIC DEVELOPMENT |
16 |
311 |
7.69e-07 |
2.434e-05 |
| 148 |
REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
15 |
274 |
7.898e-07 |
2.483e-05 |
| 149 |
REGULATION OF PROTEIN CATABOLIC PROCESS |
18 |
393 |
8.42e-07 |
2.63e-05 |
| 150 |
REGULATION OF ORGAN MORPHOGENESIS |
14 |
242 |
9.648e-07 |
2.993e-05 |
| 151 |
POSITIVE REGULATION OF ORGANELLE ORGANIZATION |
22 |
573 |
9.84e-07 |
3.013e-05 |
| 152 |
PROTEIN COMPLEX BIOGENESIS |
33 |
1132 |
9.907e-07 |
3.013e-05 |
| 153 |
PROTEIN COMPLEX ASSEMBLY |
33 |
1132 |
9.907e-07 |
3.013e-05 |
| 154 |
RESPONSE TO IONIZING RADIATION |
11 |
145 |
1.045e-06 |
3.157e-05 |
| 155 |
CHROMATIN ASSEMBLY OR DISASSEMBLY |
12 |
177 |
1.118e-06 |
3.337e-05 |
| 156 |
NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION |
11 |
146 |
1.119e-06 |
3.337e-05 |
| 157 |
MACROMOLECULE CATABOLIC PROCESS |
29 |
926 |
1.175e-06 |
3.475e-05 |
| 158 |
POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION |
8 |
68 |
1.18e-06 |
3.475e-05 |
| 159 |
CARTILAGE DEVELOPMENT |
11 |
147 |
1.197e-06 |
3.503e-05 |
| 160 |
CELL DEVELOPMENT |
38 |
1426 |
1.233e-06 |
3.586e-05 |
| 161 |
REGULATION OF KINASE ACTIVITY |
26 |
776 |
1.283e-06 |
3.707e-05 |
| 162 |
REGULATION OF EPITHELIAL CELL PROLIFERATION |
15 |
285 |
1.293e-06 |
3.715e-05 |
| 163 |
CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY |
25 |
727 |
1.307e-06 |
3.732e-05 |
| 164 |
REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS |
12 |
181 |
1.416e-06 |
4.017e-05 |
| 165 |
RESPONSE TO OXYGEN CONTAINING COMPOUND |
37 |
1381 |
1.533e-06 |
4.322e-05 |
| 166 |
REGULATION OF CELL DEVELOPMENT |
27 |
836 |
1.59e-06 |
4.457e-05 |
| 167 |
INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY |
8 |
71 |
1.648e-06 |
4.593e-05 |
| 168 |
RESPONSE TO GAMMA RADIATION |
7 |
50 |
1.687e-06 |
4.673e-05 |
| 169 |
RESPONSE TO HORMONE |
28 |
893 |
1.785e-06 |
4.913e-05 |
| 170 |
CELLULAR SENESCENCE |
6 |
33 |
1.946e-06 |
5.327e-05 |
| 171 |
STEROID HORMONE MEDIATED SIGNALING PATHWAY |
10 |
125 |
2.012e-06 |
5.473e-05 |
| 172 |
G1 DNA DAMAGE CHECKPOINT |
8 |
73 |
2.042e-06 |
5.523e-05 |
| 173 |
POSITIVE REGULATION OF CELL DEATH |
22 |
605 |
2.396e-06 |
6.444e-05 |
| 174 |
PROTEIN ACYLATION |
11 |
159 |
2.588e-06 |
6.922e-05 |
| 175 |
PROTEIN ACETYLATION |
10 |
129 |
2.679e-06 |
7.041e-05 |
| 176 |
CELL FATE COMMITMENT |
13 |
227 |
2.653e-06 |
7.041e-05 |
| 177 |
NEGATIVE REGULATION OF CELL DIFFERENTIATION |
22 |
609 |
2.665e-06 |
7.041e-05 |
| 178 |
REGULATION OF CYTOKINE PRODUCTION |
21 |
563 |
2.764e-06 |
7.226e-05 |
| 179 |
TRANSCRIPTION FROM RNA POLYMERASE I PROMOTER |
6 |
35 |
2.798e-06 |
7.272e-05 |
| 180 |
REGULATION OF LIGASE ACTIVITY |
10 |
130 |
2.873e-06 |
7.426e-05 |
| 181 |
REGENERATION |
11 |
161 |
2.923e-06 |
7.438e-05 |
| 182 |
CONNECTIVE TISSUE DEVELOPMENT |
12 |
194 |
2.925e-06 |
7.438e-05 |
| 183 |
DNA PACKAGING |
12 |
194 |
2.925e-06 |
7.438e-05 |
| 184 |
NEGATIVE REGULATION OF CELL COMMUNICATION |
33 |
1192 |
3.038e-06 |
7.682e-05 |
| 185 |
POSITIVE REGULATION OF TRANSFERASE ACTIVITY |
22 |
616 |
3.204e-06 |
8.059e-05 |
| 186 |
NEGATIVE REGULATION OF MUSCLE CELL DIFFERENTIATION |
7 |
55 |
3.263e-06 |
8.119e-05 |
| 187 |
POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
12 |
196 |
3.253e-06 |
8.119e-05 |
| 188 |
REGULATION OF CELLULAR COMPONENT BIOGENESIS |
25 |
767 |
3.388e-06 |
8.385e-05 |
| 189 |
NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
29 |
983 |
3.818e-06 |
9.399e-05 |
| 190 |
REGULATION OF CELLULAR AMIDE METABOLIC PROCESS |
16 |
354 |
4.127e-06 |
0.0001011 |
| 191 |
MORPHOGENESIS OF A BRANCHING STRUCTURE |
11 |
167 |
4.167e-06 |
0.0001015 |
| 192 |
CELLULAR RESPONSE TO RADIATION |
10 |
137 |
4.607e-06 |
0.0001117 |
| 193 |
REGULATION OF GROWTH |
22 |
633 |
4.951e-06 |
0.0001194 |
| 194 |
CELL DEATH |
29 |
1001 |
5.42e-06 |
0.00013 |
| 195 |
RESPONSE TO ALCOHOL |
16 |
362 |
5.479e-06 |
0.0001307 |
| 196 |
POSITIVE REGULATION OF CELL COMMUNICATION |
38 |
1532 |
6.691e-06 |
0.0001589 |
| 197 |
NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS |
25 |
801 |
7.199e-06 |
0.00017 |
| 198 |
REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY |
12 |
213 |
7.637e-06 |
0.0001795 |
| 199 |
POSITIVE REGULATION OF DNA BINDING |
6 |
42 |
8.449e-06 |
0.0001966 |
| 200 |
NCRNA TRANSCRIPTION |
8 |
88 |
8.41e-06 |
0.0001966 |
| 201 |
REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
10 |
147 |
8.626e-06 |
0.0001997 |
| 202 |
CELLULAR RESPONSE TO EPIDERMAL GROWTH FACTOR STIMULUS |
5 |
25 |
8.843e-06 |
0.0002037 |
| 203 |
MAMMARY GLAND DEVELOPMENT |
9 |
117 |
9.077e-06 |
0.0002081 |
| 204 |
PEPTIDYL SERINE MODIFICATION |
10 |
148 |
9.158e-06 |
0.0002089 |
| 205 |
REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
12 |
218 |
9.66e-06 |
0.0002193 |
| 206 |
CELLULAR RESPONSE TO UV |
7 |
66 |
1.124e-05 |
0.0002539 |
| 207 |
REGULATION OF RNA POLYMERASE II TRANSCRIPTIONAL PREINITIATION COMPLEX ASSEMBLY |
4 |
13 |
1.162e-05 |
0.0002599 |
| 208 |
REGULATION OF HISTONE PHOSPHORYLATION |
4 |
13 |
1.162e-05 |
0.0002599 |
| 209 |
POSITIVE REGULATION OF VIRAL PROCESS |
8 |
92 |
1.17e-05 |
0.0002606 |
| 210 |
CELL AGING |
7 |
67 |
1.243e-05 |
0.0002753 |
| 211 |
REGULATION OF DNA BINDING |
8 |
93 |
1.268e-05 |
0.0002796 |
| 212 |
MUSCLE STRUCTURE DEVELOPMENT |
17 |
432 |
1.279e-05 |
0.0002808 |
| 213 |
POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS |
13 |
263 |
1.308e-05 |
0.0002857 |
| 214 |
POSITIVE REGULATION OF MITOTIC CELL CYCLE |
9 |
123 |
1.362e-05 |
0.0002961 |
| 215 |
RNA PROCESSING |
25 |
835 |
1.46e-05 |
0.0003159 |
| 216 |
PROTEIN CATABOLIC PROCESS |
20 |
579 |
1.483e-05 |
0.0003166 |
| 217 |
CANONICAL WNT SIGNALING PATHWAY |
8 |
95 |
1.483e-05 |
0.0003166 |
| 218 |
REGULATION OF PEPTIDASE ACTIVITY |
16 |
392 |
1.479e-05 |
0.0003166 |
| 219 |
POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
11 |
192 |
1.569e-05 |
0.0003334 |
| 220 |
HORMONE MEDIATED SIGNALING PATHWAY |
10 |
158 |
1.624e-05 |
0.0003436 |
| 221 |
REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
39 |
1656 |
1.651e-05 |
0.0003466 |
| 222 |
RESPONSE TO UV |
9 |
126 |
1.654e-05 |
0.0003466 |
| 223 |
POSITIVE REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS |
8 |
98 |
1.864e-05 |
0.0003872 |
| 224 |
NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
8 |
98 |
1.864e-05 |
0.0003872 |
| 225 |
REGULATION OF NUCLEAR DIVISION |
10 |
163 |
2.129e-05 |
0.0004402 |
| 226 |
RESPONSE TO EPIDERMAL GROWTH FACTOR |
5 |
30 |
2.262e-05 |
0.0004656 |
| 227 |
POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS |
16 |
406 |
2.271e-05 |
0.0004656 |
| 228 |
HISTONE H4 ACETYLATION |
6 |
50 |
2.369e-05 |
0.0004834 |
| 229 |
REGULATION OF APOPTOTIC SIGNALING PATHWAY |
15 |
363 |
2.381e-05 |
0.0004838 |
| 230 |
TUBE DEVELOPMENT |
19 |
552 |
2.562e-05 |
0.0005183 |
| 231 |
TUBE MORPHOGENESIS |
14 |
323 |
2.649e-05 |
0.0005336 |
| 232 |
REGULATION OF PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
8 |
103 |
2.68e-05 |
0.0005376 |
| 233 |
RESPONSE TO EXTERNAL STIMULUS |
41 |
1821 |
2.776e-05 |
0.0005543 |
| 234 |
REGULATION OF SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM |
11 |
205 |
2.883e-05 |
0.0005707 |
| 235 |
APOPTOTIC PROCESS INVOLVED IN MORPHOGENESIS |
4 |
16 |
2.878e-05 |
0.0005707 |
| 236 |
NUCLEUS ORGANIZATION |
9 |
136 |
3.041e-05 |
0.0005997 |
| 237 |
REGULATION OF FAT CELL DIFFERENTIATION |
8 |
106 |
3.3e-05 |
0.0006452 |
| 238 |
DNA REPLICATION |
11 |
208 |
3.294e-05 |
0.0006452 |
| 239 |
MAMMARY GLAND EPITHELIUM DEVELOPMENT |
6 |
53 |
3.325e-05 |
0.0006472 |
| 240 |
REGULATION OF PROTEIN COMPLEX ASSEMBLY |
15 |
375 |
3.463e-05 |
0.0006687 |
| 241 |
REGULATION OF CELLULAR COMPONENT MOVEMENT |
23 |
771 |
3.459e-05 |
0.0006687 |
| 242 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
9 |
139 |
3.613e-05 |
0.0006933 |
| 243 |
NEGATIVE REGULATION OF PHOSPHORYLATION |
16 |
422 |
3.621e-05 |
0.0006933 |
| 244 |
NEGATIVE REGULATION OF PROTEIN BINDING |
7 |
79 |
3.682e-05 |
0.0007021 |
| 245 |
NEGATIVE REGULATION OF CATALYTIC ACTIVITY |
24 |
829 |
3.742e-05 |
0.0007108 |
| 246 |
POSITIVE REGULATION OF CELL DEVELOPMENT |
17 |
472 |
3.933e-05 |
0.000744 |
| 247 |
CELLULAR MACROMOLECULE LOCALIZATION |
31 |
1234 |
4.13e-05 |
0.000778 |
| 248 |
DNA GEOMETRIC CHANGE |
7 |
81 |
4.331e-05 |
0.0008126 |
| 249 |
CYTOSKELETON ORGANIZATION |
24 |
838 |
4.439e-05 |
0.0008294 |
| 250 |
REGULATION OF PROTEIN LOCALIZATION |
26 |
950 |
4.509e-05 |
0.0008393 |
| 251 |
EPITHELIAL TO MESENCHYMAL TRANSITION |
6 |
56 |
4.568e-05 |
0.0008467 |
| 252 |
POSITIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS |
8 |
111 |
4.598e-05 |
0.0008489 |
| 253 |
NEGATIVE REGULATION OF ORGANELLE ORGANIZATION |
15 |
387 |
4.958e-05 |
0.0009119 |
| 254 |
REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY |
9 |
145 |
5.034e-05 |
0.0009221 |
| 255 |
IMMUNE SYSTEM DEVELOPMENT |
19 |
582 |
5.234e-05 |
0.0009514 |
| 256 |
NUCLEOTIDE EXCISION REPAIR |
8 |
113 |
5.224e-05 |
0.0009514 |
| 257 |
REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT |
11 |
220 |
5.489e-05 |
0.0009938 |
| 258 |
ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY |
21 |
689 |
5.662e-05 |
0.001013 |
| 259 |
REGULATION OF ERYTHROCYTE DIFFERENTIATION |
5 |
36 |
5.656e-05 |
0.001013 |
| 260 |
POSITIVE REGULATION OF PROTEIN ACETYLATION |
5 |
36 |
5.656e-05 |
0.001013 |
| 261 |
REGULATION OF PROTEIN IMPORT |
10 |
183 |
5.714e-05 |
0.001018 |
| 262 |
REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS |
27 |
1021 |
5.733e-05 |
0.001018 |
| 263 |
REGULATION OF PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
9 |
148 |
5.905e-05 |
0.001045 |
| 264 |
MICROTUBULE CYTOSKELETON ORGANIZATION |
14 |
348 |
5.952e-05 |
0.001047 |
| 265 |
LIVER REGENERATION |
4 |
19 |
5.964e-05 |
0.001047 |
| 266 |
REGULATION OF CELL CYCLE G2 M PHASE TRANSITION |
6 |
59 |
6.158e-05 |
0.001077 |
| 267 |
EMBRYONIC MORPHOGENESIS |
18 |
539 |
6.198e-05 |
0.00108 |
| 268 |
POSITIVE REGULATION OF CATABOLIC PROCESS |
15 |
395 |
6.246e-05 |
0.001084 |
| 269 |
INTRACELLULAR SIGNAL TRANSDUCTION |
36 |
1572 |
6.532e-05 |
0.00113 |
| 270 |
RNA SPLICING VIA TRANSESTERIFICATION REACTIONS |
12 |
267 |
7.089e-05 |
0.001222 |
| 271 |
REGULATION OF HOMEOSTATIC PROCESS |
16 |
447 |
7.157e-05 |
0.001229 |
| 272 |
REGULATION OF WNT SIGNALING PATHWAY |
13 |
310 |
7.199e-05 |
0.001232 |
| 273 |
NUCLEAR TRANSPORT |
14 |
355 |
7.363e-05 |
0.00125 |
| 274 |
REGULATION OF STEM CELL PROLIFERATION |
7 |
88 |
7.383e-05 |
0.00125 |
| 275 |
MESENCHYME MORPHOGENESIS |
5 |
38 |
7.389e-05 |
0.00125 |
| 276 |
CELL CYCLE ARREST |
9 |
154 |
8.031e-05 |
0.001349 |
| 277 |
POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION |
9 |
154 |
8.031e-05 |
0.001349 |
| 278 |
NEUROGENESIS |
33 |
1402 |
8.127e-05 |
0.00136 |
| 279 |
DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION |
19 |
602 |
8.184e-05 |
0.001365 |
| 280 |
PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
24 |
873 |
8.39e-05 |
0.001384 |
| 281 |
CELLULAR RESPONSE TO HORMONE STIMULUS |
18 |
552 |
8.388e-05 |
0.001384 |
| 282 |
REGULATION OF CELLULAR PROTEIN LOCALIZATION |
18 |
552 |
8.388e-05 |
0.001384 |
| 283 |
EMBRYONIC ORGAN DEVELOPMENT |
15 |
406 |
8.491e-05 |
0.001396 |
| 284 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE |
14 |
361 |
8.796e-05 |
0.001441 |
| 285 |
PROTEIN LOCALIZATION TO NUCLEUS |
9 |
156 |
8.869e-05 |
0.001448 |
| 286 |
APOPTOTIC PROCESS INVOLVED IN DEVELOPMENT |
4 |
21 |
9.042e-05 |
0.001471 |
| 287 |
CELLULAR RESPONSE TO LIGHT STIMULUS |
7 |
91 |
9.14e-05 |
0.001482 |
| 288 |
RESPONSE TO ENDOPLASMIC RETICULUM STRESS |
11 |
233 |
9.183e-05 |
0.001484 |
| 289 |
ANTERIOR POSTERIOR PATTERN SPECIFICATION |
10 |
194 |
9.305e-05 |
0.001498 |
| 290 |
MUSCLE TISSUE DEVELOPMENT |
12 |
275 |
9.385e-05 |
0.001501 |
| 291 |
RESPONSE TO CYTOKINE |
21 |
714 |
9.364e-05 |
0.001501 |
| 292 |
ENDOCRINE SYSTEM DEVELOPMENT |
8 |
123 |
9.522e-05 |
0.001517 |
| 293 |
EAR DEVELOPMENT |
10 |
195 |
9.71e-05 |
0.001542 |
| 294 |
REGULATION OF PROTEIN ACETYLATION |
6 |
64 |
9.764e-05 |
0.001545 |
| 295 |
MRNA METABOLIC PROCESS |
19 |
611 |
9.936e-05 |
0.001567 |
| 296 |
REGULATION OF CANONICAL WNT SIGNALING PATHWAY |
11 |
236 |
0.0001029 |
0.001617 |
| 297 |
MUSCLE CELL DIFFERENTIATION |
11 |
237 |
0.0001068 |
0.001673 |
| 298 |
ENDOCARDIAL CUSHION MORPHOGENESIS |
4 |
22 |
0.0001095 |
0.00171 |
| 299 |
RESPONSE TO LIGHT STIMULUS |
12 |
280 |
0.0001112 |
0.001731 |
| 300 |
CELLULAR COMPONENT DISASSEMBLY |
17 |
515 |
0.0001143 |
0.001773 |
| 301 |
PATTERN SPECIFICATION PROCESS |
15 |
418 |
0.0001172 |
0.001811 |
| 302 |
SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
8 |
127 |
0.0001191 |
0.001835 |
| 303 |
HEPATICOBILIARY SYSTEM DEVELOPMENT |
8 |
128 |
0.0001258 |
0.001932 |
| 304 |
SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE |
7 |
96 |
0.0001282 |
0.001962 |
| 305 |
SINGLE ORGANISM CELLULAR LOCALIZATION |
24 |
898 |
0.000129 |
0.001968 |
| 306 |
NEGATIVE REGULATION OF PROTEOLYSIS |
13 |
329 |
0.0001305 |
0.001985 |
| 307 |
REGULATION OF NUCLEASE ACTIVITY |
4 |
23 |
0.0001314 |
0.001991 |
| 308 |
REGULATION OF MICROTUBULE BASED PROCESS |
11 |
243 |
0.0001331 |
0.002011 |
| 309 |
MICROTUBULE BASED PROCESS |
17 |
522 |
0.0001343 |
0.002011 |
| 310 |
PROTEIN LOCALIZATION TO MEMBRANE |
14 |
376 |
0.0001349 |
0.002011 |
| 311 |
CARDIOVASCULAR SYSTEM DEVELOPMENT |
22 |
788 |
0.0001347 |
0.002011 |
| 312 |
CIRCULATORY SYSTEM DEVELOPMENT |
22 |
788 |
0.0001347 |
0.002011 |
| 313 |
GLAND MORPHOGENESIS |
7 |
97 |
0.0001368 |
0.002027 |
| 314 |
REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY |
7 |
97 |
0.0001368 |
0.002027 |
| 315 |
MITOTIC SPINDLE ORGANIZATION |
6 |
69 |
0.0001488 |
0.002198 |
| 316 |
REGULATION OF DNA TEMPLATED TRANSCRIPTION ELONGATION |
5 |
44 |
0.0001511 |
0.002225 |
| 317 |
RESPONSE TO VIRUS |
11 |
247 |
0.0001536 |
0.002254 |
| 318 |
DNA DEPENDENT DNA REPLICATION |
7 |
99 |
0.0001555 |
0.002275 |
| 319 |
REGULATION OF TRANSCRIPTION ELONGATION FROM RNA POLYMERASE II PROMOTER |
4 |
24 |
0.0001562 |
0.002279 |
| 320 |
RESPONSE TO INORGANIC SUBSTANCE |
16 |
479 |
0.0001589 |
0.00231 |
| 321 |
PROTEOLYSIS |
29 |
1208 |
0.0001636 |
0.002371 |
| 322 |
DNA TEMPLATED TRANSCRIPTION TERMINATION |
7 |
101 |
0.0001761 |
0.002545 |
| 323 |
HOMEOSTATIC PROCESS |
31 |
1337 |
0.0001783 |
0.002568 |
| 324 |
HISTONE PHOSPHORYLATION |
4 |
25 |
0.0001843 |
0.002646 |
| 325 |
IMMUNE SYSTEM PROCESS |
41 |
1984 |
0.0001861 |
0.002664 |
| 326 |
NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
15 |
437 |
0.0001901 |
0.002714 |
| 327 |
HEART MORPHOGENESIS |
10 |
212 |
0.0001925 |
0.002739 |
| 328 |
REGULATION OF MUSCLE TISSUE DEVELOPMENT |
7 |
103 |
0.000199 |
0.002814 |
| 329 |
REGULATION OF MUSCLE ORGAN DEVELOPMENT |
7 |
103 |
0.000199 |
0.002814 |
| 330 |
TRANSCRIPTION COUPLED NUCLEOTIDE EXCISION REPAIR |
6 |
73 |
0.0002033 |
0.002864 |
| 331 |
NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
17 |
541 |
0.000205 |
0.002864 |
| 332 |
RIBONUCLEOPROTEIN COMPLEX BIOGENESIS |
15 |
440 |
0.0002047 |
0.002864 |
| 333 |
NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
17 |
541 |
0.000205 |
0.002864 |
| 334 |
CELL CYCLE G2 M PHASE TRANSITION |
8 |
138 |
0.0002115 |
0.002946 |
| 335 |
GAMETE GENERATION |
18 |
595 |
0.0002127 |
0.002954 |
| 336 |
DEVELOPMENTAL PROGRAMMED CELL DEATH |
4 |
26 |
0.0002158 |
0.002962 |
| 337 |
REGULATION OF CELLULAR SENESCENCE |
4 |
26 |
0.0002158 |
0.002962 |
| 338 |
NEGATIVE REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION |
4 |
26 |
0.0002158 |
0.002962 |
| 339 |
DNA RECOMBINATION |
10 |
215 |
0.0002156 |
0.002962 |
| 340 |
CENTRAL NERVOUS SYSTEM DEVELOPMENT |
23 |
872 |
0.0002166 |
0.002964 |
| 341 |
ATP DEPENDENT CHROMATIN REMODELING |
6 |
74 |
0.0002191 |
0.00299 |
| 342 |
SENSORY ORGAN DEVELOPMENT |
16 |
493 |
0.0002199 |
0.002992 |
| 343 |
RESPONSE TO CORTICOSTEROID |
9 |
176 |
0.0002211 |
0.002999 |
| 344 |
REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT |
5 |
48 |
0.0002294 |
0.003103 |
| 345 |
REGULATION OF PROTEIN COMPLEX DISASSEMBLY |
10 |
217 |
0.0002323 |
0.003133 |
| 346 |
REGULATION OF DNA REPAIR |
6 |
75 |
0.0002359 |
0.003172 |
| 347 |
EPITHELIAL TO MESENCHYMAL TRANSITION INVOLVED IN ENDOCARDIAL CUSHION FORMATION |
3 |
11 |
0.0002373 |
0.003183 |
| 348 |
REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION |
9 |
178 |
0.0002405 |
0.003215 |
| 349 |
WNT SIGNALING PATHWAY |
13 |
351 |
0.0002459 |
0.003269 |
| 350 |
NEGATIVE REGULATION OF TRANSFERASE ACTIVITY |
13 |
351 |
0.0002459 |
0.003269 |
| 351 |
NEGATIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE |
4 |
27 |
0.0002511 |
0.003309 |
| 352 |
REGULATION OF CATENIN IMPORT INTO NUCLEUS |
4 |
27 |
0.0002511 |
0.003309 |
| 353 |
NEGATIVE REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
4 |
27 |
0.0002511 |
0.003309 |
| 354 |
REGULATION OF PROTEIN TARGETING |
12 |
307 |
0.0002609 |
0.003429 |
| 355 |
CELLULAR RESPONSE TO CYTOKINE STIMULUS |
18 |
606 |
0.0002655 |
0.00348 |
| 356 |
REGULATION OF SODIUM ION TRANSPORT |
6 |
77 |
0.0002724 |
0.003559 |
| 357 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO MEMBRANE |
11 |
264 |
0.0002731 |
0.003559 |
| 358 |
POSITIVE REGULATION OF STEM CELL DIFFERENTIATION |
5 |
50 |
0.0002786 |
0.003621 |
| 359 |
REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
4 |
28 |
0.0002902 |
0.003762 |
| 360 |
POSITIVE REGULATION OF GENE EXPRESSION EPIGENETIC |
6 |
78 |
0.0002923 |
0.003762 |
| 361 |
RESPONSE TO OXYGEN LEVELS |
12 |
311 |
0.0002935 |
0.003762 |
| 362 |
SOMITE DEVELOPMENT |
6 |
78 |
0.0002923 |
0.003762 |
| 363 |
REGIONALIZATION |
12 |
311 |
0.0002935 |
0.003762 |
| 364 |
REGULATION OF MYELOID CELL DIFFERENTIATION |
9 |
183 |
0.0002953 |
0.003774 |
| 365 |
CARTILAGE MORPHOGENESIS |
3 |
12 |
0.0003137 |
0.003978 |
| 366 |
REGULATION OF VITAMIN METABOLIC PROCESS |
3 |
12 |
0.0003137 |
0.003978 |
| 367 |
BONE MORPHOGENESIS |
6 |
79 |
0.0003133 |
0.003978 |
| 368 |
REGULATION OF TYPE I INTERFERON PRODUCTION |
7 |
111 |
0.0003154 |
0.003988 |
| 369 |
POSITIVE REGULATION OF DNA METABOLIC PROCESS |
9 |
185 |
0.0003199 |
0.004034 |
| 370 |
ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS |
24 |
957 |
0.0003307 |
0.004159 |
| 371 |
REGULATION OF HEART MORPHOGENESIS |
4 |
29 |
0.0003336 |
0.004184 |
| 372 |
NEGATIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS |
5 |
52 |
0.0003354 |
0.004195 |
| 373 |
REGULATION OF STEM CELL DIFFERENTIATION |
7 |
113 |
0.0003517 |
0.004388 |
| 374 |
REGULATION OF GENE EXPRESSION EPIGENETIC |
10 |
229 |
0.0003568 |
0.004439 |
| 375 |
REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY |
13 |
365 |
0.0003578 |
0.004439 |
| 376 |
REGULATION OF EMBRYONIC DEVELOPMENT |
7 |
114 |
0.0003711 |
0.004593 |
| 377 |
HEART DEVELOPMENT |
15 |
466 |
0.0003765 |
0.004647 |
| 378 |
MESENCHYME DEVELOPMENT |
9 |
190 |
0.000389 |
0.004788 |
| 379 |
ESTABLISHMENT OR MAINTENANCE OF TRANSMEMBRANE ELECTROCHEMICAL GRADIENT |
3 |
13 |
0.0004044 |
0.004913 |
| 380 |
WHITE FAT CELL DIFFERENTIATION |
3 |
13 |
0.0004044 |
0.004913 |
| 381 |
REGULATION OF IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE |
3 |
13 |
0.0004044 |
0.004913 |
| 382 |
RESPONSE TO NUTRIENT |
9 |
191 |
0.0004041 |
0.004913 |
| 383 |
NEGATIVE REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
3 |
13 |
0.0004044 |
0.004913 |
| 384 |
INTRINSIC APOPTOTIC SIGNALING PATHWAY |
8 |
152 |
0.0004068 |
0.004929 |
| 385 |
MUSCLE ORGAN DEVELOPMENT |
11 |
277 |
0.0004106 |
0.004959 |
| 386 |
RESPONSE TO FIBROBLAST GROWTH FACTOR |
7 |
116 |
0.0004124 |
0.004959 |
| 387 |
REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY |
15 |
470 |
0.0004117 |
0.004959 |
| 388 |
NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
19 |
684 |
0.0004138 |
0.004963 |
| 389 |
PALLIUM DEVELOPMENT |
8 |
153 |
0.000425 |
0.005083 |
| 390 |
POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
25 |
1036 |
0.0004389 |
0.005209 |
| 391 |
HISTONE METHYLATION |
6 |
84 |
0.0004367 |
0.005209 |
| 392 |
POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
25 |
1036 |
0.0004389 |
0.005209 |
| 393 |
REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION |
7 |
118 |
0.0004573 |
0.005414 |
| 394 |
REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION |
6 |
85 |
0.0004654 |
0.005482 |
| 395 |
PALATE DEVELOPMENT |
6 |
85 |
0.0004654 |
0.005482 |
| 396 |
POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY |
7 |
119 |
0.0004812 |
0.005654 |
| 397 |
ESTABLISHMENT OF LOCALIZATION IN CELL |
35 |
1676 |
0.0004841 |
0.005674 |
| 398 |
ENDOCARDIAL CUSHION DEVELOPMENT |
4 |
32 |
0.0004916 |
0.005718 |
| 399 |
PATTERNING OF BLOOD VESSELS |
4 |
32 |
0.0004916 |
0.005718 |
| 400 |
INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS |
4 |
32 |
0.0004916 |
0.005718 |
| 401 |
REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE |
9 |
197 |
0.0005059 |
0.005848 |
| 402 |
HISTONE H3 K9 MODIFICATION |
3 |
14 |
0.0005103 |
0.005848 |
| 403 |
POSITIVE REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING |
3 |
14 |
0.0005103 |
0.005848 |
| 404 |
POSITIVE REGULATION OF MULTI ORGANISM PROCESS |
8 |
157 |
0.0005047 |
0.005848 |
| 405 |
PEPTIDYL ARGININE METHYLATION |
3 |
14 |
0.0005103 |
0.005848 |
| 406 |
POST EMBRYONIC ORGAN DEVELOPMENT |
3 |
14 |
0.0005103 |
0.005848 |
| 407 |
REGULATION OF CYTOPLASMIC TRANSPORT |
15 |
481 |
0.0005234 |
0.005984 |
| 408 |
REPRODUCTION |
29 |
1297 |
0.0005287 |
0.006029 |
| 409 |
MRNA PROCESSING |
14 |
432 |
0.0005501 |
0.006259 |
| 410 |
REGULATION OF CELL AGING |
4 |
33 |
0.0005543 |
0.006291 |
| 411 |
REGULATION OF EPITHELIAL CELL DIFFERENTIATION |
7 |
122 |
0.0005588 |
0.006312 |
| 412 |
CELLULAR RESPONSE TO CORTICOSTEROID STIMULUS |
5 |
58 |
0.0005589 |
0.006312 |
| 413 |
NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY |
6 |
88 |
0.0005603 |
0.006313 |
| 414 |
NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY |
9 |
200 |
0.0005642 |
0.006341 |
| 415 |
MALE GAMETE GENERATION |
15 |
486 |
0.0005822 |
0.006528 |
| 416 |
SKELETAL SYSTEM MORPHOGENESIS |
9 |
201 |
0.0005848 |
0.006541 |
| 417 |
EPITHELIAL CELL PROLIFERATION |
6 |
89 |
0.0005951 |
0.00664 |
| 418 |
POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION |
8 |
162 |
0.0006208 |
0.006911 |
| 419 |
REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE |
4 |
34 |
0.0006226 |
0.006914 |
| 420 |
ENDOCARDIAL CUSHION FORMATION |
3 |
15 |
0.0006324 |
0.006924 |
| 421 |
NEGATIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY |
6 |
90 |
0.0006315 |
0.006924 |
| 422 |
MIDBRAIN DEVELOPMENT |
6 |
90 |
0.0006315 |
0.006924 |
| 423 |
HISTONE H2A ACETYLATION |
3 |
15 |
0.0006324 |
0.006924 |
| 424 |
POSITIVE REGULATION OF NUCLEASE ACTIVITY |
3 |
15 |
0.0006324 |
0.006924 |
| 425 |
ACTIVATION OF ANAPHASE PROMOTING COMPLEX ACTIVITY |
3 |
15 |
0.0006324 |
0.006924 |
| 426 |
HEAD DEVELOPMENT |
19 |
709 |
0.0006393 |
0.006983 |
| 427 |
CHONDROCYTE DIFFERENTIATION |
5 |
60 |
0.0006537 |
0.007124 |
| 428 |
MULTICELLULAR ORGANISM REPRODUCTION |
20 |
768 |
0.0006595 |
0.007169 |
| 429 |
RESPONSE TO BIOTIC STIMULUS |
22 |
886 |
0.0006713 |
0.007264 |
| 430 |
CATABOLIC PROCESS |
36 |
1773 |
0.0006704 |
0.007264 |
| 431 |
RESPONSE TO EXTRACELLULAR STIMULUS |
14 |
441 |
0.000673 |
0.007266 |
| 432 |
RESPONSE TO MINERALOCORTICOID |
4 |
35 |
0.0006965 |
0.007502 |
| 433 |
NEGATIVE REGULATION OF CATION TRANSMEMBRANE TRANSPORT |
5 |
61 |
0.0007055 |
0.007563 |
| 434 |
POSITIVE REGULATION OF STEM CELL PROLIFERATION |
5 |
61 |
0.0007055 |
0.007563 |
| 435 |
MITOCHONDRIAL MEMBRANE ORGANIZATION |
6 |
92 |
0.0007095 |
0.007589 |
| 436 |
CELLULAR CATABOLIC PROCESS |
29 |
1322 |
0.000716 |
0.007641 |
| 437 |
MULTI ORGANISM REPRODUCTIVE PROCESS |
22 |
891 |
0.0007229 |
0.007697 |
| 438 |
POSITIVE REGULATION OF CATALYTIC ACTIVITY |
32 |
1518 |
0.0007376 |
0.007835 |
| 439 |
TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY |
15 |
498 |
0.0007468 |
0.007916 |
| 440 |
CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM |
10 |
252 |
0.0007517 |
0.00793 |
| 441 |
RHYTHMIC PROCESS |
11 |
298 |
0.0007533 |
0.00793 |
| 442 |
REGULATION OF NEURON DEATH |
10 |
252 |
0.0007517 |
0.00793 |
| 443 |
SOMITOGENESIS |
5 |
62 |
0.0007602 |
0.007985 |
| 444 |
COPPER ION HOMEOSTASIS |
3 |
16 |
0.0007717 |
0.008063 |
| 445 |
CELL SURFACE RECEPTOR SIGNALING PATHWAY INVOLVED IN HEART DEVELOPMENT |
3 |
16 |
0.0007717 |
0.008063 |
| 446 |
LYMPHOCYTE DIFFERENTIATION |
9 |
209 |
0.0007728 |
0.008063 |
| 447 |
PROTEIN K63 LINKED UBIQUITINATION |
4 |
36 |
0.0007765 |
0.008065 |
| 448 |
REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING |
4 |
36 |
0.0007765 |
0.008065 |
| 449 |
POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
7 |
129 |
0.0007789 |
0.008071 |
| 450 |
REGULATION OF CYTOSKELETON ORGANIZATION |
15 |
502 |
0.0008098 |
0.008373 |
| 451 |
REGULATION OF CARTILAGE DEVELOPMENT |
5 |
63 |
0.000818 |
0.00844 |
| 452 |
NEGATIVE REGULATION OF CYTOKINE PRODUCTION |
9 |
211 |
0.0008268 |
0.008511 |
| 453 |
POSITIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS |
5 |
64 |
0.0008791 |
0.009029 |
| 454 |
POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY |
8 |
171 |
0.0008839 |
0.009059 |
| 455 |
POSITIVE REGULATION OF CELL DIVISION |
7 |
132 |
0.0008919 |
0.009121 |
| 456 |
SEXUAL REPRODUCTION |
19 |
730 |
0.0009041 |
0.009226 |
| 457 |
NUCLEOSOME DISASSEMBLY |
3 |
17 |
0.0009291 |
0.009357 |
| 458 |
PROTEIN DNA COMPLEX DISASSEMBLY |
3 |
17 |
0.0009291 |
0.009357 |
| 459 |
REGULATION OF CHROMATIN BINDING |
3 |
17 |
0.0009291 |
0.009357 |
| 460 |
INNER MITOCHONDRIAL MEMBRANE ORGANIZATION |
3 |
17 |
0.0009291 |
0.009357 |
| 461 |
CHROMATIN DISASSEMBLY |
3 |
17 |
0.0009291 |
0.009357 |
| 462 |
PROTEIN LOCALIZATION |
36 |
1805 |
0.0009261 |
0.009357 |
| 463 |
CELLULAR HOMEOSTASIS |
18 |
676 |
0.0009528 |
0.009575 |
| 464 |
RIBOSOME BIOGENESIS |
11 |
308 |
0.0009855 |
0.009882 |
| 465 |
SINGLE ORGANISM CELL ADHESION |
14 |
459 |
0.0009895 |
0.009901 |