| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
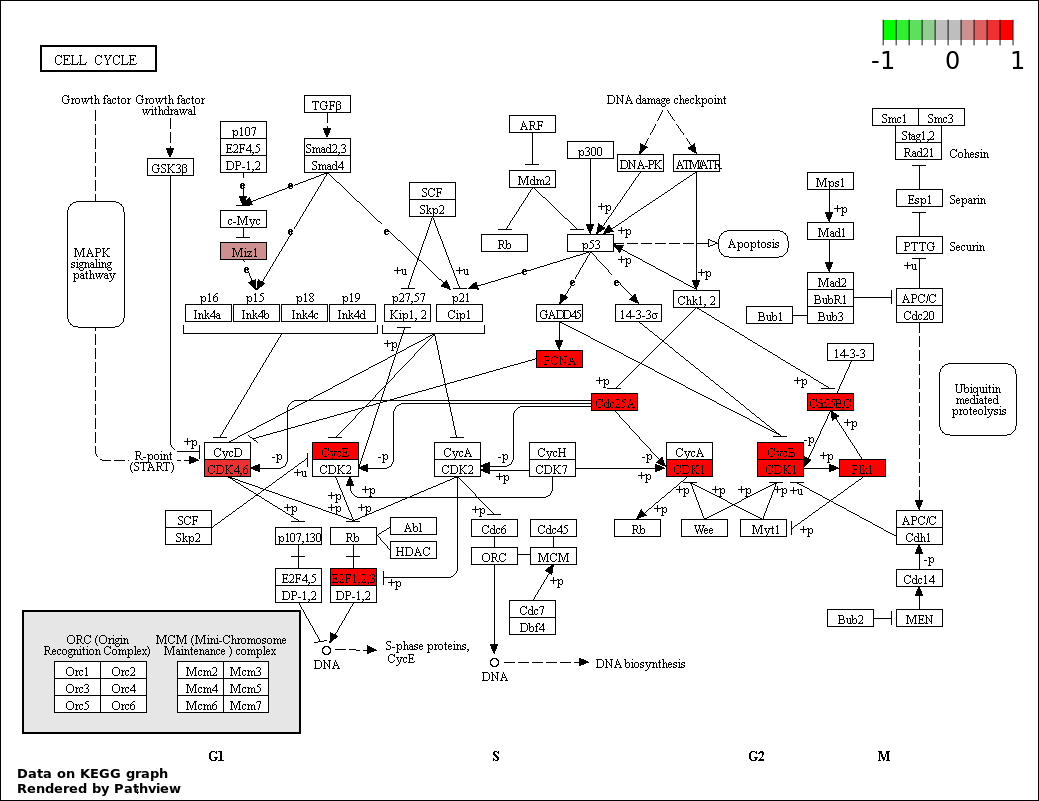
REGULATION OF CELL CYCLE |
27 |
949 |
1.39e-15 |
6.466e-12 |
| 2 |
CELL CYCLE |
30 |
1316 |
9.049e-15 |
2.105e-11 |
| 3 |
CELL CYCLE PHASE TRANSITION |
15 |
255 |
2.464e-13 |
3.821e-10 |
| 4 |
REGULATION OF CELL DEATH |
29 |
1472 |
1.168e-12 |
1.359e-09 |
| 5 |
REGULATION OF TRANSFERASE ACTIVITY |
23 |
946 |
7.034e-12 |
6.546e-09 |
| 6 |
CELL CYCLE PROCESS |
24 |
1081 |
1.491e-11 |
1.156e-08 |
| 7 |
CELL CYCLE G2 M PHASE TRANSITION |
11 |
138 |
1.775e-11 |
1.18e-08 |
| 8 |
MITOTIC CELL CYCLE |
20 |
766 |
6.101e-11 |
3.548e-08 |
| 9 |
CELL DEATH |
22 |
1001 |
1.545e-10 |
7.989e-08 |
| 10 |
TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY |
9 |
95 |
2.846e-10 |
1.324e-07 |
| 11 |
POSITIVE REGULATION OF DNA METABOLIC PROCESS |
11 |
185 |
4.215e-10 |
1.621e-07 |
| 12 |
NEGATIVE REGULATION OF CELL CYCLE |
15 |
433 |
4.528e-10 |
1.621e-07 |
| 13 |
REGULATION OF PHOSPHORUS METABOLIC PROCESS |
27 |
1618 |
3.884e-10 |
1.621e-07 |
| 14 |
REGULATION OF KINASE ACTIVITY |
19 |
776 |
5.856e-10 |
1.946e-07 |
| 15 |
POSITIVE REGULATION OF TRANSFERASE ACTIVITY |
17 |
616 |
8.961e-10 |
2.633e-07 |
| 16 |
REGULATION OF CELL CYCLE ARREST |
9 |
108 |
9.054e-10 |
2.633e-07 |
| 17 |
CELLULAR RESPONSE TO STRESS |
26 |
1565 |
1.003e-09 |
2.745e-07 |
| 18 |
REGULATION OF PROTEIN MODIFICATION PROCESS |
27 |
1710 |
1.319e-09 |
3.409e-07 |
| 19 |
REGULATION OF CELL CYCLE PROCESS |
16 |
558 |
1.732e-09 |
4.242e-07 |
| 20 |
POSITIVE REGULATION OF RESPONSE TO STIMULUS |
28 |
1929 |
3.893e-09 |
8.627e-07 |
| 21 |
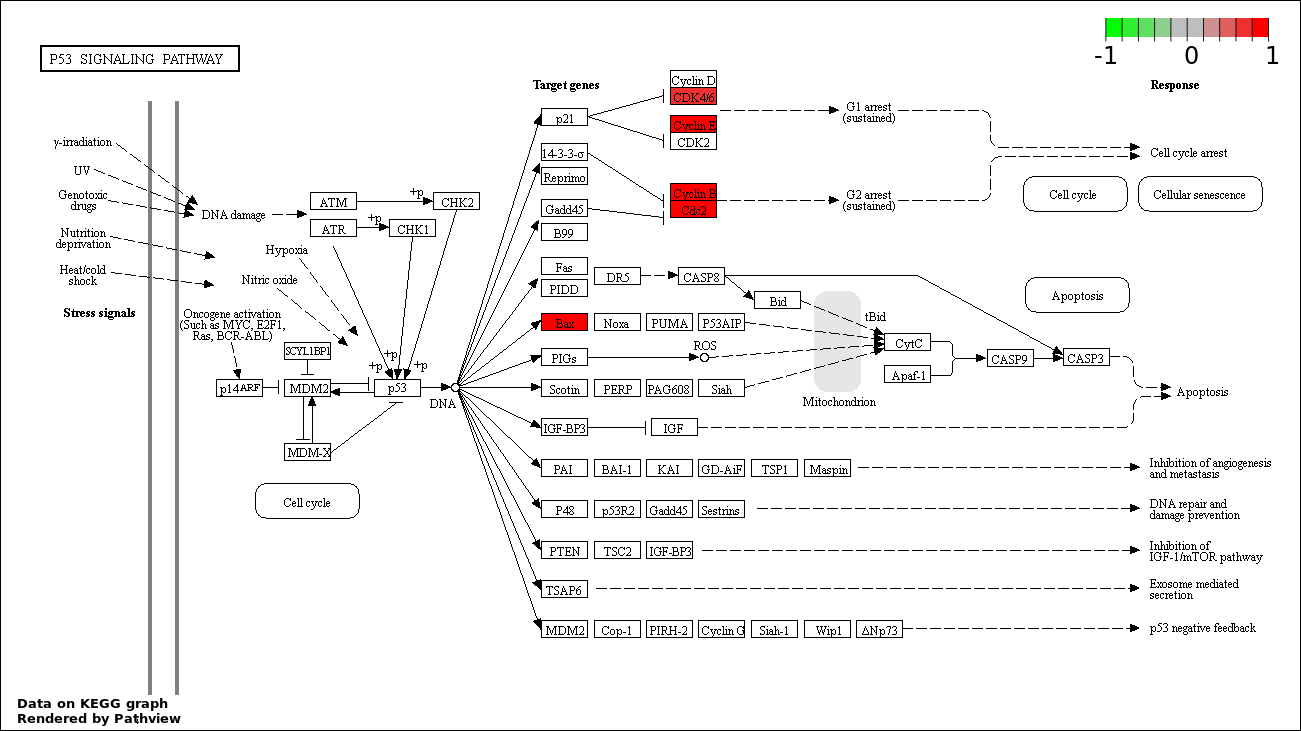
SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
9 |
127 |
3.84e-09 |
8.627e-07 |
| 22 |
REGULATION OF CELLULAR RESPONSE TO STRESS |
17 |
691 |
5.039e-09 |
1.066e-06 |
| 23 |
PHOSPHORYLATION |
22 |
1228 |
6.981e-09 |
1.412e-06 |
| 24 |
POSITIVE REGULATION OF CELL CYCLE PROCESS |
11 |
247 |
8.83e-09 |
1.643e-06 |
| 25 |
MITOTIC CELL CYCLE CHECKPOINT |
9 |
139 |
8.516e-09 |
1.643e-06 |
| 26 |
CELL DIVISION |
14 |
460 |
9.537e-09 |
1.644e-06 |
| 27 |
RESPONSE TO ABIOTIC STIMULUS |
20 |
1024 |
9.305e-09 |
1.644e-06 |
| 28 |
CELL CYCLE CHECKPOINT |
10 |
194 |
1.092e-08 |
1.815e-06 |
| 29 |
RESPONSE TO TRANSFORMING GROWTH FACTOR BETA |
9 |
144 |
1.161e-08 |
1.863e-06 |
| 30 |
ERROR FREE TRANSLESION SYNTHESIS |
5 |
19 |
1.449e-08 |
2.107e-06 |
| 31 |
NEGATIVE REGULATION OF MITOTIC CELL CYCLE |
10 |
199 |
1.393e-08 |
2.107e-06 |
| 32 |
ERROR PRONE TRANSLESION SYNTHESIS |
5 |
19 |
1.449e-08 |
2.107e-06 |
| 33 |
POSITIVE REGULATION OF CELL CYCLE |
12 |
332 |
1.829e-08 |
2.579e-06 |
| 34 |
REGULATION OF DNA METABOLIC PROCESS |
12 |
340 |
2.379e-08 |
3.255e-06 |
| 35 |
NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS |
20 |
1087 |
2.525e-08 |
3.356e-06 |
| 36 |
PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS |
27 |
1977 |
2.981e-08 |
3.853e-06 |
| 37 |
CELLULAR RESPONSE TO ORGANIC SUBSTANCE |
26 |
1848 |
3.223e-08 |
4.053e-06 |
| 38 |
POSITIVE REGULATION OF CELL DEATH |
15 |
605 |
4.094e-08 |
4.884e-06 |
| 39 |
MEMBRANE ORGANIZATION |
18 |
899 |
4.029e-08 |
4.884e-06 |
| 40 |
REGULATION OF PROTEIN BINDING |
9 |
168 |
4.439e-08 |
5.164e-06 |
| 41 |
MEMBRANE DISASSEMBLY |
6 |
47 |
4.937e-08 |
5.292e-06 |
| 42 |
NUCLEOTIDE EXCISION REPAIR DNA GAP FILLING |
5 |
24 |
5.208e-08 |
5.292e-06 |
| 43 |
DNA BIOSYNTHETIC PROCESS |
8 |
121 |
5.085e-08 |
5.292e-06 |
| 44 |
NUCLEAR ENVELOPE DISASSEMBLY |
6 |
47 |
4.937e-08 |
5.292e-06 |
| 45 |
REGULATION OF PROTEOLYSIS |
16 |
711 |
5.231e-08 |
5.292e-06 |
| 46 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
15 |
616 |
5.183e-08 |
5.292e-06 |
| 47 |
NUCLEAR ENVELOPE ORGANIZATION |
7 |
81 |
5.7e-08 |
5.643e-06 |
| 48 |
CELLULAR RESPONSE TO DNA DAMAGE STIMULUS |
16 |
720 |
6.219e-08 |
6.029e-06 |
| 49 |
POSITIVE REGULATION OF CATALYTIC ACTIVITY |
23 |
1518 |
6.662e-08 |
6.326e-06 |
| 50 |
REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
24 |
1656 |
7.379e-08 |
6.867e-06 |
| 51 |
REGULATION OF CATABOLIC PROCESS |
16 |
731 |
7.657e-08 |
6.985e-06 |
| 52 |
PROTEIN LOCALIZATION |
25 |
1805 |
8.781e-08 |
7.857e-06 |
| 53 |
REGULATION OF MITOTIC CELL CYCLE |
13 |
468 |
9.831e-08 |
8.63e-06 |
| 54 |
PROTEIN LOCALIZATION TO ORGANELLE |
14 |
556 |
1.01e-07 |
8.705e-06 |
| 55 |
NUCLEUS ORGANIZATION |
8 |
136 |
1.262e-07 |
1.042e-05 |
| 56 |
TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY |
9 |
190 |
1.277e-07 |
1.042e-05 |
| 57 |
DNA METABOLIC PROCESS |
16 |
758 |
1.256e-07 |
1.042e-05 |
| 58 |
REGULATION OF RESPONSE TO STRESS |
22 |
1468 |
1.679e-07 |
1.347e-05 |
| 59 |
SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE |
7 |
96 |
1.855e-07 |
1.463e-05 |
| 60 |
NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
7 |
98 |
2.138e-07 |
1.658e-05 |
| 61 |
NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION |
8 |
146 |
2.183e-07 |
1.665e-05 |
| 62 |
POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS |
22 |
1492 |
2.223e-07 |
1.669e-05 |
| 63 |
POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
19 |
1135 |
2.631e-07 |
1.943e-05 |
| 64 |
RESPONSE TO ORGANIC CYCLIC COMPOUND |
17 |
917 |
3.041e-07 |
2.211e-05 |
| 65 |
POSITIVE REGULATION OF MOLECULAR FUNCTION |
24 |
1791 |
3.177e-07 |
2.274e-05 |
| 66 |
POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
18 |
1036 |
3.349e-07 |
2.326e-05 |
| 67 |
POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
18 |
1036 |
3.349e-07 |
2.326e-05 |
| 68 |
NEGATIVE REGULATION OF CELL CYCLE PROCESS |
9 |
214 |
3.502e-07 |
2.397e-05 |
| 69 |
POSITIVE REGULATION OF BIOSYNTHETIC PROCESS |
24 |
1805 |
3.665e-07 |
2.472e-05 |
| 70 |
DNA DAMAGE RESPONSE DETECTION OF DNA DAMAGE |
5 |
36 |
4.436e-07 |
2.949e-05 |
| 71 |
PROTEIN PHOSPHORYLATION |
17 |
944 |
4.567e-07 |
2.982e-05 |
| 72 |
REGULATION OF DNA REPLICATION |
8 |
161 |
4.615e-07 |
2.982e-05 |
| 73 |
RESPONSE TO ENDOGENOUS STIMULUS |
21 |
1450 |
6.028e-07 |
3.842e-05 |
| 74 |
NUCLEOTIDE EXCISION REPAIR DNA INCISION |
5 |
39 |
6.707e-07 |
4.217e-05 |
| 75 |
G1 DNA DAMAGE CHECKPOINT |
6 |
73 |
7.16e-07 |
4.442e-05 |
| 76 |
DNA SYNTHESIS INVOLVED IN DNA REPAIR |
6 |
74 |
7.765e-07 |
4.717e-05 |
| 77 |
REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY |
12 |
470 |
7.806e-07 |
4.717e-05 |
| 78 |
TRANSLESION SYNTHESIS |
5 |
41 |
8.671e-07 |
5.172e-05 |
| 79 |
CELLULAR MACROMOLECULE LOCALIZATION |
19 |
1234 |
9.375e-07 |
5.522e-05 |
| 80 |
NEGATIVE REGULATION OF GENE EXPRESSION |
21 |
1493 |
9.703e-07 |
5.644e-05 |
| 81 |
POSITIVE REGULATION OF KINASE ACTIVITY |
12 |
482 |
1.017e-06 |
5.843e-05 |
| 82 |
REGULATION OF CELL CYCLE PHASE TRANSITION |
10 |
321 |
1.187e-06 |
6.738e-05 |
| 83 |
REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA |
5 |
44 |
1.244e-06 |
6.973e-05 |
| 84 |
ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY |
14 |
689 |
1.331e-06 |
7.375e-05 |
| 85 |
POSITIVE REGULATION OF CELL COMMUNICATION |
21 |
1532 |
1.471e-06 |
8.054e-05 |
| 86 |
REGULATION OF CELLULAR LOCALIZATION |
19 |
1277 |
1.564e-06 |
8.461e-05 |
| 87 |
POSITIVE REGULATION OF CELL CYCLE ARREST |
6 |
85 |
1.765e-06 |
9.441e-05 |
| 88 |
ESTABLISHMENT OF PROTEIN LOCALIZATION |
20 |
1423 |
1.892e-06 |
0.0001001 |
| 89 |
POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
8 |
196 |
2.036e-06 |
0.0001064 |
| 90 |
RESPONSE TO DRUG |
11 |
431 |
2.348e-06 |
0.0001214 |
| 91 |
NEGATIVE REGULATION OF MOLECULAR FUNCTION |
17 |
1079 |
2.857e-06 |
0.0001461 |
| 92 |
DNA INTEGRITY CHECKPOINT |
7 |
146 |
3.174e-06 |
0.0001605 |
| 93 |
REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
9 |
280 |
3.258e-06 |
0.000163 |
| 94 |
REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
7 |
147 |
3.321e-06 |
0.0001644 |
| 95 |
POSTREPLICATION REPAIR |
5 |
54 |
3.502e-06 |
0.000166 |
| 96 |
MITOTIC NUCLEAR DIVISION |
10 |
361 |
3.397e-06 |
0.000166 |
| 97 |
POSITIVE REGULATION OF PROTEOLYSIS |
10 |
363 |
3.568e-06 |
0.000166 |
| 98 |
REGULATION OF BINDING |
9 |
283 |
3.554e-06 |
0.000166 |
| 99 |
RESPONSE TO ALCOHOL |
10 |
362 |
3.481e-06 |
0.000166 |
| 100 |
REGULATION OF APOPTOTIC SIGNALING PATHWAY |
10 |
363 |
3.568e-06 |
0.000166 |
| 101 |
NEGATIVE REGULATION OF CELL DEATH |
15 |
872 |
4.179e-06 |
0.0001925 |
| 102 |
MITOTIC DNA INTEGRITY CHECKPOINT |
6 |
100 |
4.57e-06 |
0.0002085 |
| 103 |
REGULATION OF MAPK CASCADE |
13 |
660 |
4.649e-06 |
0.00021 |
| 104 |
CELLULAR GLUCAN METABOLIC PROCESS |
5 |
58 |
5.006e-06 |
0.0002218 |
| 105 |
GLUCAN METABOLIC PROCESS |
5 |
58 |
5.006e-06 |
0.0002218 |
| 106 |
REGULATION OF PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
6 |
103 |
5.426e-06 |
0.0002382 |
| 107 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM |
6 |
104 |
5.739e-06 |
0.0002496 |
| 108 |
REGENERATION |
7 |
161 |
6.059e-06 |
0.0002587 |
| 109 |
POSITIVE REGULATION OF ORGANELLE ORGANIZATION |
12 |
573 |
6.044e-06 |
0.0002587 |
| 110 |
DNA REPAIR |
11 |
480 |
6.56e-06 |
0.0002775 |
| 111 |
POSITIVE REGULATION OF CATABOLIC PROCESS |
10 |
395 |
7.505e-06 |
0.0003146 |
| 112 |
POSITIVE REGULATION OF LIGASE ACTIVITY |
6 |
110 |
7.936e-06 |
0.0003297 |
| 113 |
CELL CYCLE G1 S PHASE TRANSITION |
6 |
111 |
8.36e-06 |
0.0003412 |
| 114 |
G1 S TRANSITION OF MITOTIC CELL CYCLE |
6 |
111 |
8.36e-06 |
0.0003412 |
| 115 |
INTRACELLULAR SIGNAL TRANSDUCTION |
20 |
1572 |
8.539e-06 |
0.0003455 |
| 116 |
MACROMOLECULE CATABOLIC PROCESS |
15 |
926 |
8.613e-06 |
0.0003455 |
| 117 |
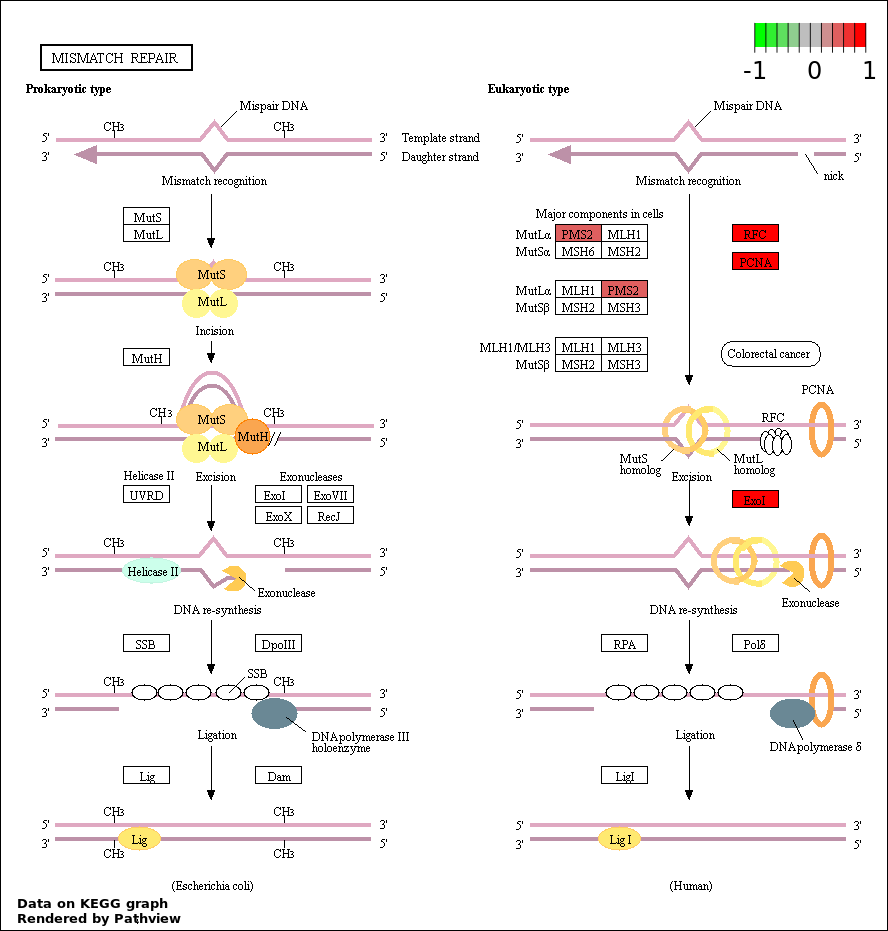
MISMATCH REPAIR |
4 |
31 |
9.175e-06 |
0.0003619 |
| 118 |
REGULATION OF ORGANELLE ORGANIZATION |
17 |
1178 |
9.177e-06 |
0.0003619 |
| 119 |
POSITIVE REGULATION OF GENE EXPRESSION |
21 |
1733 |
1.022e-05 |
0.0003994 |
| 120 |
NEGATIVE REGULATION OF CELL COMMUNICATION |
17 |
1192 |
1.071e-05 |
0.0004153 |
| 121 |
NUCLEAR TRANSCRIBED MRNA CATABOLIC PROCESS NONSENSE MEDIATED DECAY |
6 |
118 |
1.188e-05 |
0.0004568 |
| 122 |
MICROTUBULE BASED PROCESS |
11 |
522 |
1.44e-05 |
0.0005446 |
| 123 |
CELLULAR RESPONSE TO TOPOLOGICALLY INCORRECT PROTEIN |
6 |
122 |
1.437e-05 |
0.0005446 |
| 124 |
ENERGY RESERVE METABOLIC PROCESS |
5 |
72 |
1.458e-05 |
0.0005472 |
| 125 |
POSITIVE REGULATION OF MITOTIC CELL CYCLE |
6 |
123 |
1.506e-05 |
0.0005561 |
| 126 |
PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM |
6 |
123 |
1.506e-05 |
0.0005561 |
| 127 |
TRANSCRIPTION COUPLED NUCLEOTIDE EXCISION REPAIR |
5 |
73 |
1.56e-05 |
0.0005716 |
| 128 |
POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS |
8 |
263 |
1.76e-05 |
0.0006397 |
| 129 |
AGING |
8 |
264 |
1.808e-05 |
0.0006472 |
| 130 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO MEMBRANE |
8 |
264 |
1.808e-05 |
0.0006472 |
| 131 |
MICROTUBULE CYTOSKELETON ORGANIZATION |
9 |
348 |
1.867e-05 |
0.0006631 |
| 132 |
POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
7 |
192 |
1.908e-05 |
0.0006707 |
| 133 |
RESPONSE TO OXYGEN CONTAINING COMPOUND |
18 |
1381 |
1.917e-05 |
0.0006707 |
| 134 |
WNT SIGNALING PATHWAY |
9 |
351 |
1.998e-05 |
0.0006858 |
| 135 |
NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
11 |
541 |
2.004e-05 |
0.0006858 |
| 136 |
NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
11 |
541 |
2.004e-05 |
0.0006858 |
| 137 |
ANAPHASE PROMOTING COMPLEX DEPENDENT CATABOLIC PROCESS |
5 |
77 |
2.025e-05 |
0.0006877 |
| 138 |
REGULATION OF LIGASE ACTIVITY |
6 |
130 |
2.063e-05 |
0.0006957 |
| 139 |
VENTRICULAR CARDIAC MUSCLE CELL DEVELOPMENT |
3 |
13 |
2.128e-05 |
0.0007124 |
| 140 |
ESTABLISHMENT OF LOCALIZATION IN CELL |
20 |
1676 |
2.186e-05 |
0.0007213 |
| 141 |
PROTEASOMAL PROTEIN CATABOLIC PROCESS |
8 |
271 |
2.182e-05 |
0.0007213 |
| 142 |
REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
8 |
274 |
2.361e-05 |
0.0007736 |
| 143 |
POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION |
9 |
360 |
2.438e-05 |
0.0007826 |
| 144 |
POLYSACCHARIDE METABOLIC PROCESS |
5 |
80 |
2.439e-05 |
0.0007826 |
| 145 |
REGULATION OF CELLULAR PROTEIN LOCALIZATION |
11 |
552 |
2.412e-05 |
0.0007826 |
| 146 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE |
9 |
361 |
2.492e-05 |
0.0007941 |
| 147 |
POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND |
3 |
14 |
2.7e-05 |
0.0008547 |
| 148 |
RESPONSE TO LIGHT STIMULUS |
8 |
280 |
2.756e-05 |
0.0008665 |
| 149 |
INTRACELLULAR PROTEIN TRANSPORT |
13 |
781 |
2.78e-05 |
0.000868 |
| 150 |
POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
16 |
1152 |
2.807e-05 |
0.0008706 |
| 151 |
MULTI ORGANISM METABOLIC PROCESS |
6 |
138 |
2.892e-05 |
0.0008913 |
| 152 |
ORGAN REGENERATION |
5 |
83 |
2.916e-05 |
0.0008926 |
| 153 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
6 |
139 |
3.013e-05 |
0.0009162 |
| 154 |
REGULATION OF HEART GROWTH |
4 |
42 |
3.148e-05 |
0.0009512 |
| 155 |
REPRODUCTION |
17 |
1297 |
3.175e-05 |
0.0009512 |
| 156 |
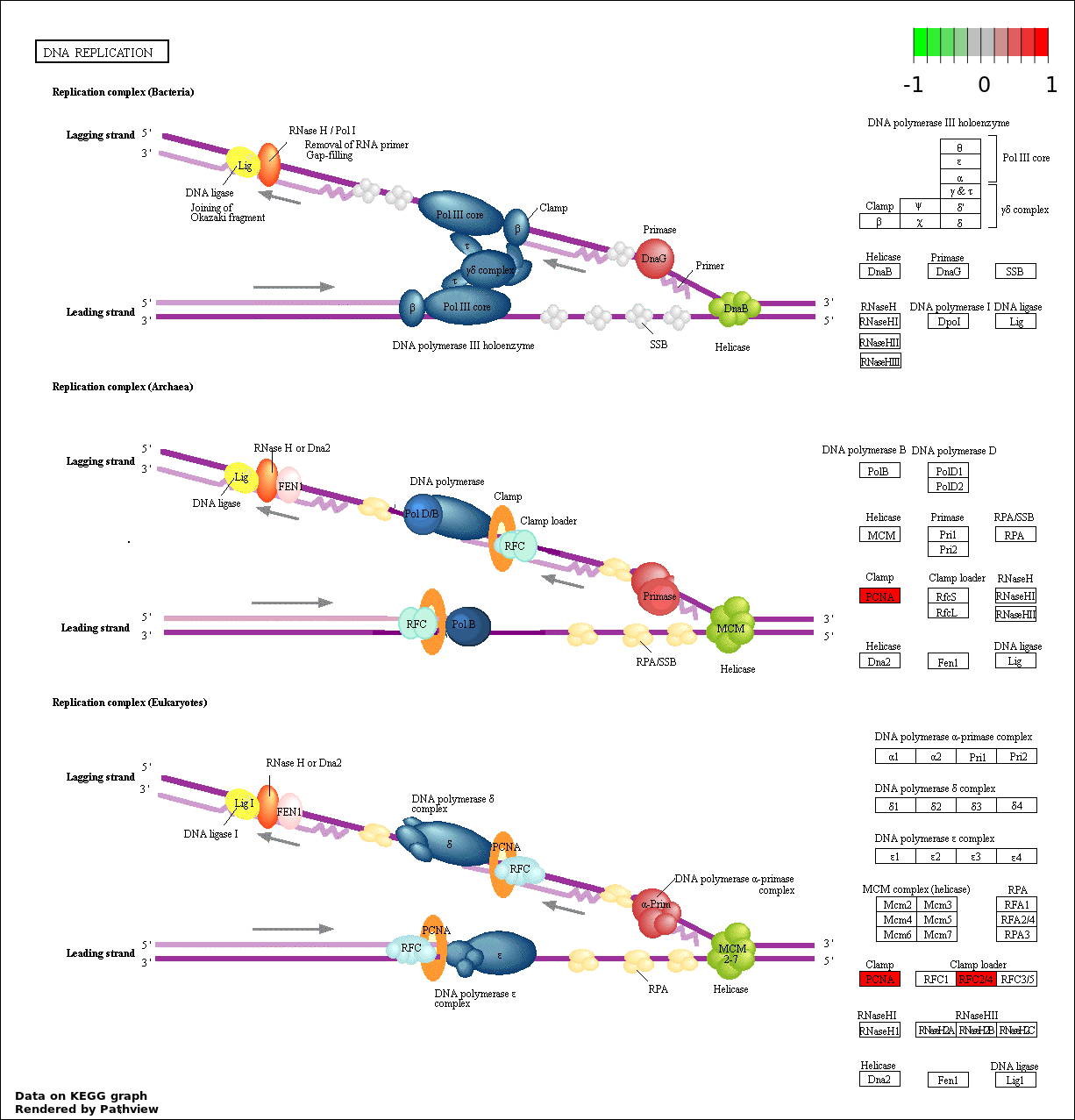
DNA REPLICATION |
7 |
208 |
3.189e-05 |
0.0009512 |
| 157 |
POSITIVE REGULATION OF NUCLEASE ACTIVITY |
3 |
15 |
3.365e-05 |
0.0009909 |
| 158 |
T CELL APOPTOTIC PROCESS |
3 |
15 |
3.365e-05 |
0.0009909 |
| 159 |
POSITIVE REGULATION OF DNA REPLICATION |
5 |
86 |
3.462e-05 |
0.001001 |
| 160 |
PROTEIN LOCALIZATION TO MEMBRANE |
9 |
376 |
3.425e-05 |
0.001001 |
| 161 |
APOPTOTIC SIGNALING PATHWAY |
8 |
289 |
3.451e-05 |
0.001001 |
| 162 |
CELLULAR RESPONSE TO OXYGEN LEVELS |
6 |
143 |
3.534e-05 |
0.001015 |
| 163 |
GENERATION OF PRECURSOR METABOLITES AND ENERGY |
8 |
292 |
3.714e-05 |
0.00106 |
| 164 |
RESPONSE TO INORGANIC SUBSTANCE |
10 |
479 |
3.931e-05 |
0.001115 |
| 165 |
TRANSLATIONAL INITIATION |
6 |
146 |
3.971e-05 |
0.00112 |
| 166 |
REGULATION OF CYTOPLASMIC TRANSPORT |
10 |
481 |
4.072e-05 |
0.001141 |
| 167 |
PROTEIN LOCALIZATION TO CHROMOSOME |
4 |
45 |
4.15e-05 |
0.001156 |
| 168 |
ENERGY DERIVATION BY OXIDATION OF ORGANIC COMPOUNDS |
7 |
217 |
4.177e-05 |
0.001157 |
| 169 |
REGULATION OF MITOCHONDRION ORGANIZATION |
7 |
218 |
4.3e-05 |
0.00117 |
| 170 |
RHYTHMIC PROCESS |
8 |
298 |
4.288e-05 |
0.00117 |
| 171 |
REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
7 |
218 |
4.3e-05 |
0.00117 |
| 172 |
REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND |
4 |
46 |
4.53e-05 |
0.001226 |
| 173 |
REGULATION OF PROTEIN STABILITY |
7 |
221 |
4.69e-05 |
0.001261 |
| 174 |
REGULATION OF PROTEIN CATABOLIC PROCESS |
9 |
393 |
4.827e-05 |
0.001291 |
| 175 |
REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY |
6 |
153 |
5.159e-05 |
0.001364 |
| 176 |
NEGATIVE REGULATION OF CATALYTIC ACTIVITY |
13 |
829 |
5.141e-05 |
0.001364 |
| 177 |
ORGANELLE FISSION |
10 |
496 |
5.269e-05 |
0.001385 |
| 178 |
REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT |
4 |
48 |
5.367e-05 |
0.001403 |
| 179 |
CYTOSKELETON ORGANIZATION |
13 |
838 |
5.739e-05 |
0.001489 |
| 180 |
RESPONSE TO OXYGEN LEVELS |
8 |
311 |
5.791e-05 |
0.001489 |
| 181 |
NEGATIVE REGULATION OF RESPONSE TO STIMULUS |
17 |
1360 |
5.761e-05 |
0.001489 |
| 182 |
PROTEIN TARGETING TO MEMBRANE |
6 |
157 |
5.957e-05 |
0.00152 |
| 183 |
LYMPHOCYTE APOPTOTIC PROCESS |
3 |
18 |
5.978e-05 |
0.00152 |
| 184 |
LYMPHOCYTE HOMEOSTASIS |
4 |
50 |
6.311e-05 |
0.001596 |
| 185 |
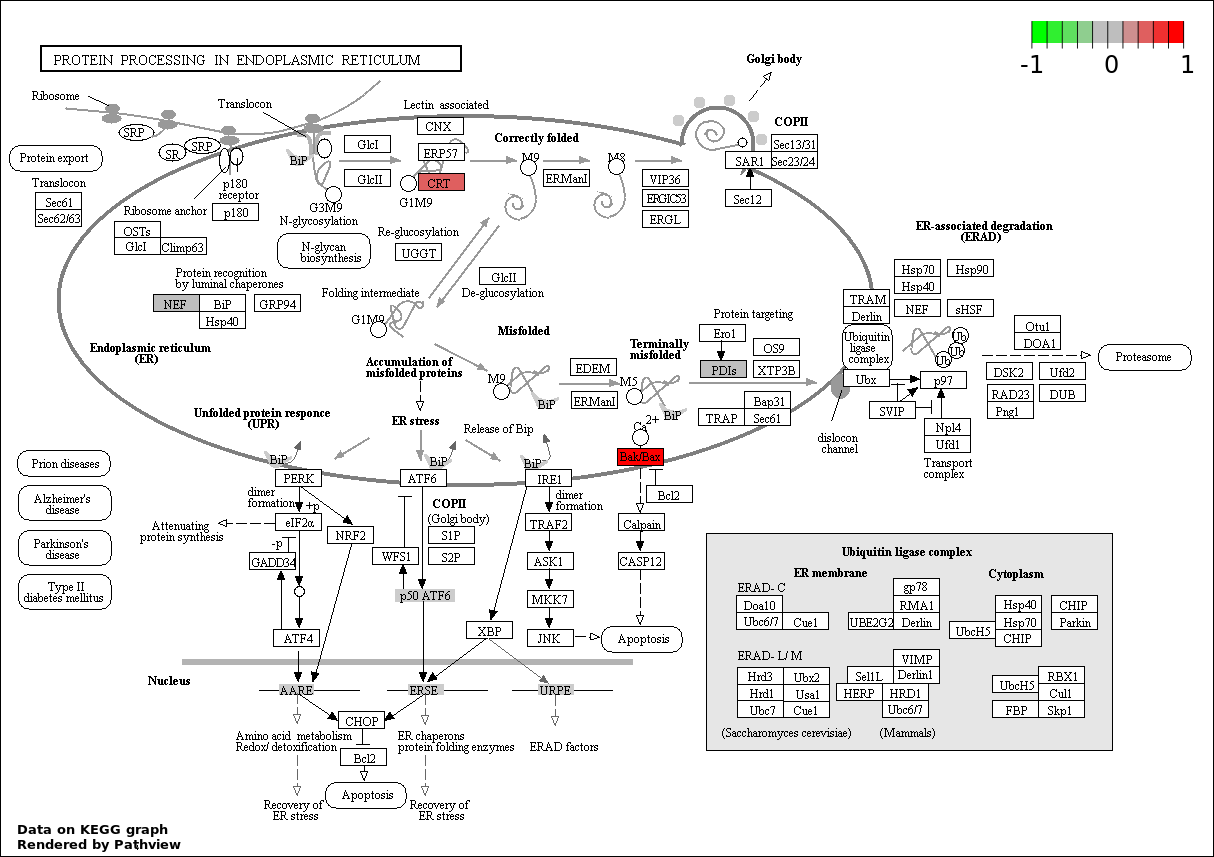
RESPONSE TO ENDOPLASMIC RETICULUM STRESS |
7 |
233 |
6.547e-05 |
0.001647 |
| 186 |
NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS |
18 |
1517 |
6.613e-05 |
0.001654 |
| 187 |
REGULATION OF MAP KINASE ACTIVITY |
8 |
319 |
6.918e-05 |
0.001721 |
| 188 |
VENTRICULAR CARDIAC MUSCLE CELL DIFFERENTIATION |
3 |
19 |
7.077e-05 |
0.001736 |
| 189 |
RESPONSE TO RADIATION |
9 |
413 |
7.07e-05 |
0.001736 |
| 190 |
REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
6 |
162 |
7.089e-05 |
0.001736 |
| 191 |
RESPONSE TO TOPOLOGICALLY INCORRECT PROTEIN |
6 |
163 |
7.335e-05 |
0.001787 |
| 192 |
POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY |
4 |
53 |
7.946e-05 |
0.001926 |
| 193 |
RESPONSE TO TOXIC SUBSTANCE |
7 |
241 |
8.091e-05 |
0.001951 |
| 194 |
NEGATIVE REGULATION OF PHOSPHORYLATION |
9 |
422 |
8.335e-05 |
0.001999 |
| 195 |
POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
13 |
876 |
8.99e-05 |
0.002145 |
| 196 |
RESPONSE TO METAL ION |
8 |
333 |
9.326e-05 |
0.002214 |
| 197 |
POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY |
6 |
171 |
9.557e-05 |
0.002257 |
| 198 |
CHROMOSOME ORGANIZATION |
14 |
1009 |
9.611e-05 |
0.002259 |
| 199 |
REGULATION OF PROTEIN HOMODIMERIZATION ACTIVITY |
3 |
22 |
0.0001114 |
0.002592 |
| 200 |
RELEASE OF CYTOCHROME C FROM MITOCHONDRIA |
3 |
22 |
0.0001114 |
0.002592 |
| 201 |
NUCLEIC ACID PHOSPHODIESTER BOND HYDROLYSIS |
7 |
254 |
0.0001123 |
0.002599 |
| 202 |
SINGLE ORGANISM CELLULAR LOCALIZATION |
13 |
898 |
0.0001152 |
0.002654 |
| 203 |
POSITIVE REGULATION OF DNA BIOSYNTHETIC PROCESS |
4 |
59 |
0.0001211 |
0.002776 |
| 204 |
REGULATION OF NUCLEASE ACTIVITY |
3 |
23 |
0.0001277 |
0.002885 |
| 205 |
NUCLEOTIDE EXCISION REPAIR |
5 |
113 |
0.0001274 |
0.002885 |
| 206 |
LEUKOCYTE APOPTOTIC PROCESS |
3 |
23 |
0.0001277 |
0.002885 |
| 207 |
LEUKOCYTE HOMEOSTASIS |
4 |
60 |
0.0001293 |
0.002907 |
| 208 |
NOTCH SIGNALING PATHWAY |
5 |
114 |
0.0001328 |
0.002972 |
| 209 |
NEGATIVE REGULATION OF TRANSFERASE ACTIVITY |
8 |
351 |
0.0001341 |
0.002985 |
| 210 |
CELLULAR CATABOLIC PROCESS |
16 |
1322 |
0.0001435 |
0.003179 |
| 211 |
PROTEIN MATURATION |
7 |
265 |
0.0001459 |
0.003218 |
| 212 |
REGULATION OF HYDROLASE ACTIVITY |
16 |
1327 |
0.0001499 |
0.003274 |
| 213 |
RESPONSE TO WOUNDING |
10 |
563 |
0.0001498 |
0.003274 |
| 214 |
GLYCOGEN BIOSYNTHETIC PROCESS |
3 |
25 |
0.0001649 |
0.003552 |
| 215 |
EPITHELIAL CELL APOPTOTIC PROCESS |
3 |
25 |
0.0001649 |
0.003552 |
| 216 |
GLUCAN BIOSYNTHETIC PROCESS |
3 |
25 |
0.0001649 |
0.003552 |
| 217 |
ENDOMEMBRANE SYSTEM ORGANIZATION |
9 |
465 |
0.0001732 |
0.003715 |
| 218 |
HEART DEVELOPMENT |
9 |
466 |
0.000176 |
0.003757 |
| 219 |
FEMALE MEIOTIC DIVISION |
3 |
26 |
0.0001858 |
0.003948 |
| 220 |
CELLULAR RESPONSE TO UV |
4 |
66 |
0.0001874 |
0.00395 |
| 221 |
POSITIVE REGULATION OF MAPK CASCADE |
9 |
470 |
0.0001876 |
0.00395 |
| 222 |
CELL AGING |
4 |
67 |
0.0001987 |
0.004164 |
| 223 |
REGULATION OF PROTEIN LOCALIZATION |
13 |
950 |
0.0002009 |
0.004192 |
| 224 |
RESPONSE TO GROWTH FACTOR |
9 |
475 |
0.000203 |
0.004216 |
| 225 |
REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE |
6 |
197 |
0.0002069 |
0.004278 |
| 226 |
REGULATION OF TRANSCRIPTION INVOLVED IN G1 S TRANSITION OF MITOTIC CELL CYCLE |
3 |
27 |
0.0002084 |
0.00429 |
| 227 |
MULTI ORGANISM TRANSPORT |
4 |
68 |
0.0002104 |
0.004294 |
| 228 |
MULTI ORGANISM LOCALIZATION |
4 |
68 |
0.0002104 |
0.004294 |
| 229 |
REGULATION OF G PROTEIN COUPLED RECEPTOR PROTEIN SIGNALING PATHWAY |
5 |
127 |
0.0002202 |
0.004455 |
| 230 |
TISSUE DEVELOPMENT |
17 |
1518 |
0.0002194 |
0.004455 |
| 231 |
RESPONSE TO COPPER ION |
3 |
28 |
0.0002327 |
0.004666 |
| 232 |
POSITIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA |
3 |
28 |
0.0002327 |
0.004666 |
| 233 |
POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
5 |
129 |
0.0002368 |
0.004729 |
| 234 |
PEPTIDYL AMINO ACID MODIFICATION |
12 |
841 |
0.000247 |
0.004911 |
| 235 |
POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY |
7 |
289 |
0.0002481 |
0.004913 |
| 236 |
VIRAL LIFE CYCLE |
7 |
290 |
0.0002534 |
0.004996 |
| 237 |
REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION |
3 |
29 |
0.0002587 |
0.005079 |
| 238 |
REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY |
6 |
207 |
0.0002701 |
0.005258 |
| 239 |
POSITIVE REGULATION OF MAP KINASE ACTIVITY |
6 |
207 |
0.0002701 |
0.005258 |
| 240 |
MACROMOLECULAR COMPLEX ASSEMBLY |
16 |
1398 |
0.0002714 |
0.005262 |
| 241 |
POSITIVE REGULATION OF PROTEIN BINDING |
4 |
73 |
0.0002767 |
0.005319 |
| 242 |
REGULATION OF ORGAN GROWTH |
4 |
73 |
0.0002767 |
0.005319 |
| 243 |
PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
5 |
134 |
0.0002825 |
0.005409 |
| 244 |
DNA STRAND ELONGATION |
3 |
30 |
0.0002866 |
0.005465 |
| 245 |
CARDIAC MUSCLE CELL DIFFERENTIATION |
4 |
74 |
0.0002915 |
0.005519 |
| 246 |
RESPONSE TO MECHANICAL STIMULUS |
6 |
210 |
0.0002918 |
0.005519 |
| 247 |
CELLULAR RESPONSE TO RADIATION |
5 |
137 |
0.0003129 |
0.005894 |
| 248 |
REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY |
6 |
213 |
0.0003148 |
0.005906 |
| 249 |
REGULATION OF INTRACELLULAR TRANSPORT |
10 |
621 |
0.0003286 |
0.006141 |
| 250 |
PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
12 |
873 |
0.0003468 |
0.006399 |
| 251 |
PATTERNING OF BLOOD VESSELS |
3 |
32 |
0.0003479 |
0.006399 |
| 252 |
TELOMERE MAINTENANCE VIA RECOMBINATION |
3 |
32 |
0.0003479 |
0.006399 |
| 253 |
CARDIAC MUSCLE TISSUE DEVELOPMENT |
5 |
140 |
0.0003458 |
0.006399 |
| 254 |
CELLULAR RESPONSE TO ENDOGENOUS STIMULUS |
13 |
1008 |
0.0003565 |
0.006531 |
| 255 |
PROTEIN TARGETING |
8 |
406 |
0.000359 |
0.00655 |
| 256 |
REGULATION OF WNT SIGNALING PATHWAY |
7 |
310 |
0.000379 |
0.006854 |
| 257 |
SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND |
3 |
33 |
0.0003815 |
0.006854 |
| 258 |
EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND |
3 |
33 |
0.0003815 |
0.006854 |
| 259 |
REGULATION OF CELL AGING |
3 |
33 |
0.0003815 |
0.006854 |
| 260 |
CELLULAR CARBOHYDRATE METABOLIC PROCESS |
5 |
144 |
0.0003935 |
0.007043 |
| 261 |
RESPONSE TO LIPID |
12 |
888 |
0.0004044 |
0.007209 |
| 262 |
T CELL HOMEOSTASIS |
3 |
34 |
0.0004172 |
0.007408 |
| 263 |
RNA CATABOLIC PROCESS |
6 |
227 |
0.0004416 |
0.007813 |
| 264 |
PEPTIDYL SERINE MODIFICATION |
5 |
148 |
0.0004462 |
0.007834 |
| 265 |
RESPONSE TO TRANSITION METAL NANOPARTICLE |
5 |
148 |
0.0004462 |
0.007834 |
| 266 |
NIK NF KAPPAB SIGNALING |
4 |
83 |
0.0004521 |
0.007897 |
| 267 |
REGULATION OF GLYCOGEN METABOLIC PROCESS |
3 |
35 |
0.0004548 |
0.007897 |
| 268 |
NEURON APOPTOTIC PROCESS |
3 |
35 |
0.0004548 |
0.007897 |
| 269 |
EPHRIN RECEPTOR SIGNALING PATHWAY |
4 |
85 |
0.0004949 |
0.008529 |
| 270 |
POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY |
3 |
36 |
0.0004947 |
0.008529 |
| 271 |
INTRINSIC APOPTOTIC SIGNALING PATHWAY |
5 |
152 |
0.0005039 |
0.008652 |
| 272 |
REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING |
6 |
233 |
0.0005069 |
0.008671 |
| 273 |
INTERSPECIES INTERACTION BETWEEN ORGANISMS |
10 |
662 |
0.0005424 |
0.009177 |
| 274 |
ORGANONITROGEN COMPOUND METABOLIC PROCESS |
18 |
1796 |
0.0005398 |
0.009177 |
| 275 |
SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM |
10 |
662 |
0.0005424 |
0.009177 |
| 276 |
CARDIOVASCULAR SYSTEM DEVELOPMENT |
11 |
788 |
0.0005482 |
0.009209 |
| 277 |
CIRCULATORY SYSTEM DEVELOPMENT |
11 |
788 |
0.0005482 |
0.009209 |
| 278 |
POSITIVE REGULATION OF DNA REPAIR |
3 |
38 |
0.0005808 |
0.009722 |