This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7b-5p | AARSD1 | -0.96 | 0 | 0.39 | 0.0002 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 2 | hsa-let-7b-5p | AATF | -0.96 | 0 | 0.62 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 3 | hsa-miR-148a-5p | ABCB5 | -0.77 | 0 | 1.74 | 1.0E-5 | mirMAP | -0.57 | 0 | NA | |
| 4 | hsa-let-7b-5p | ABCC5 | -0.96 | 0 | 1.23 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 5 | hsa-miR-148a-5p | ABCC5 | -0.77 | 0 | 1.23 | 0 | mirMAP | -0.21 | 0 | NA | |
| 6 | hsa-miR-3607-3p | ABCC5 | -2.16 | 0 | 1.23 | 0 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 7 | hsa-miR-199a-5p | ABCC6P2 | -1.99 | 0 | 0.58 | 0.00027 | miRanda | -0.13 | 0 | NA | |
| 8 | hsa-let-7b-5p | ABCF1 | -0.96 | 0 | 0.32 | 1.0E-5 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 9 | hsa-miR-148a-5p | ABI2 | -0.77 | 0 | 0.14 | 0.17339 | miRNATAP | -0.24 | 0 | NA | |
| 10 | hsa-miR-193b-3p | ABI2 | -0.17 | 0.27202 | 0.14 | 0.17339 | miRNAWalker2 validate; MirTarget | -0.24 | 0 | NA | |
| 11 | hsa-miR-374b-5p | ABI2 | -0.31 | 0.00301 | 0.14 | 0.17339 | mirMAP | -0.15 | 0.00242 | NA | |
| 12 | hsa-miR-30e-3p | ABL2 | -1.21 | 0 | 0.6 | 0 | mirMAP | -0.19 | 0 | NA | |
| 13 | hsa-miR-374b-5p | ABR | -0.31 | 0.00301 | -0.28 | 0.11187 | miRNATAP | -0.37 | 1.0E-5 | NA | |
| 14 | hsa-let-7b-5p | ACACA | -0.96 | 0 | 0.78 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 15 | hsa-miR-30a-3p | ACACA | -1.53 | 0 | 0.78 | 0 | mirMAP | -0.16 | 0 | NA | |
| 16 | hsa-miR-30e-3p | ACACA | -1.21 | 0 | 0.78 | 0 | mirMAP | -0.17 | 0.00073 | NA | |
| 17 | hsa-miR-193b-3p | ACBD6 | -0.17 | 0.27202 | 1.02 | 0 | miRNAWalker2 validate | -0.12 | 0.00029 | NA | |
| 18 | hsa-let-7b-3p | ACSL3 | -1.22 | 0 | 0.05 | 0.65044 | mirMAP | -0.13 | 0.00013 | NA | |
| 19 | hsa-miR-195-3p | ACSL3 | -1.09 | 0 | 0.05 | 0.65044 | mirMAP | -0.13 | 0 | NA | |
| 20 | hsa-miR-30a-3p | ACSL3 | -1.53 | 0 | 0.05 | 0.65044 | mirMAP | -0.12 | 3.0E-5 | NA | |
| 21 | hsa-miR-30e-3p | ACSL3 | -1.21 | 0 | 0.05 | 0.65044 | mirMAP | -0.14 | 0.00094 | NA | |
| 22 | hsa-miR-195-3p | ACSL4 | -1.09 | 0 | 2.07 | 0 | mirMAP | -0.39 | 4.0E-5 | NA | |
| 23 | hsa-miR-374b-5p | ACSL4 | -0.31 | 0.00301 | 2.07 | 0 | mirMAP | -0.48 | 0.00583 | NA | |
| 24 | hsa-miR-148a-5p | ACTG1 | -0.77 | 0 | -0.12 | 0.20938 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 25 | hsa-miR-193b-3p | ACTG1 | -0.17 | 0.27202 | -0.12 | 0.20938 | miRNAWalker2 validate | -0.18 | 0 | NA | |
| 26 | hsa-let-7b-3p | ACTL6A | -1.22 | 0 | 0.55 | 0 | MirTarget | -0.13 | 3.0E-5 | NA | |
| 27 | hsa-let-7f-1-3p | ACTL6A | -0.7 | 0 | 0.55 | 0 | MirTarget | -0.12 | 0.00039 | NA | |
| 28 | hsa-miR-199a-5p | ACTN2 | -1.99 | 0 | 1.96 | 2.0E-5 | miRanda | -0.23 | 0.00264 | NA | |
| 29 | hsa-miR-148a-5p | ACTR3 | -0.77 | 0 | 0.05 | 0.43435 | mirMAP | -0.12 | 0 | NA | |
| 30 | hsa-let-7b-3p | ACVR2B | -1.22 | 0 | -0.24 | 0.05503 | MirTarget | -0.17 | 3.0E-5 | NA | |
| 31 | hsa-miR-30a-3p | ADA | -1.53 | 0 | 0.81 | 0 | MirTarget | -0.2 | 0 | NA | |
| 32 | hsa-miR-30e-3p | ADA | -1.21 | 0 | 0.81 | 0 | MirTarget | -0.4 | 0 | NA | |
| 33 | hsa-miR-3607-3p | ADAM11 | -2.16 | 0 | 1.25 | 0 | miRNATAP | -0.25 | 0 | NA | |
| 34 | hsa-miR-3607-3p | ADAM12 | -2.16 | 0 | 0.93 | 0.0022 | mirMAP; miRNATAP | -0.27 | 3.0E-5 | NA | |
| 35 | hsa-miR-374b-5p | ADAM12 | -0.31 | 0.00301 | 0.93 | 0.0022 | mirMAP | -0.57 | 8.0E-5 | NA | |
| 36 | hsa-miR-3607-3p | ADAM22 | -2.16 | 0 | 1.17 | 0.00038 | mirMAP | -0.3 | 2.0E-5 | NA | |
| 37 | hsa-let-7b-3p | ADAM23 | -1.22 | 0 | 1.53 | 0.00013 | mirMAP | -0.37 | 0.00396 | NA | |
| 38 | hsa-let-7f-1-3p | ADAM23 | -0.7 | 0 | 1.53 | 0.00013 | mirMAP | -0.39 | 0.00446 | NA | |
| 39 | hsa-miR-374b-5p | ADAM28 | -0.31 | 0.00301 | -0.45 | 0.16409 | mirMAP | -0.52 | 0.00056 | NA | |
| 40 | hsa-miR-195-3p | ADAM9 | -1.09 | 0 | 0.47 | 0.00581 | mirMAP | -0.17 | 0.0001 | NA | |
| 41 | hsa-miR-148a-5p | ADAMTS15 | -0.77 | 0 | -0.88 | 0.00206 | mirMAP | -0.4 | 1.0E-5 | NA | |
| 42 | hsa-miR-148a-5p | ADAMTS5 | -0.77 | 0 | -0.16 | 0.36499 | mirMAP; miRNATAP | -0.31 | 0 | NA | |
| 43 | hsa-miR-193b-3p | ADARB1 | -0.17 | 0.27202 | -0.13 | 0.22208 | miRNAWalker2 validate; MirTarget | -0.11 | 0.00093 | NA | |
| 44 | hsa-miR-199a-5p | ADCK5 | -1.99 | 0 | 0.84 | 0 | miRanda | -0.11 | 0 | NA | |
| 45 | hsa-let-7b-5p | ADRM1 | -0.96 | 0 | 0.19 | 0.01952 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 46 | hsa-miR-148a-5p | ADSS | -0.77 | 0 | 0.31 | 0.00033 | MirTarget | -0.11 | 4.0E-5 | NA | |
| 47 | hsa-let-7b-5p | AEN | -0.96 | 0 | 0.28 | 0.01239 | MirTarget | -0.16 | 2.0E-5 | NA | |
| 48 | hsa-miR-3607-3p | AFAP1 | -2.16 | 0 | 0.33 | 0.05949 | mirMAP | -0.15 | 7.0E-5 | NA | |
| 49 | hsa-let-7b-5p | AFF2 | -0.96 | 0 | 1.4 | 0.00046 | MirTarget | -0.39 | 0.00293 | NA | |
| 50 | hsa-let-7f-1-3p | AFF2 | -0.7 | 0 | 1.4 | 0.00046 | mirMAP | -0.52 | 0.00013 | NA | |
| 51 | hsa-miR-3607-3p | AFF2 | -2.16 | 0 | 1.4 | 0.00046 | mirMAP | -0.29 | 0.00066 | NA | |
| 52 | hsa-miR-3607-3p | AFF3 | -2.16 | 0 | -0.32 | 0.28974 | mirMAP; miRNATAP | -0.17 | 0.0084 | NA | |
| 53 | hsa-miR-199a-5p | AGBL5 | -1.99 | 0 | 0.72 | 0 | miRanda | -0.13 | 0 | NA | |
| 54 | hsa-let-7b-5p | AGFG2 | -0.96 | 0 | -0.02 | 0.87061 | miRNAWalker2 validate | -0.17 | 1.0E-5 | NA | |
| 55 | hsa-miR-148a-5p | AGPAT4 | -0.77 | 0 | 0.87 | 7.0E-5 | mirMAP | -0.25 | 0.00016 | NA | |
| 56 | hsa-miR-3607-3p | AGPAT4 | -2.16 | 0 | 0.87 | 7.0E-5 | mirMAP | -0.16 | 0.00056 | NA | |
| 57 | hsa-miR-374b-5p | AHDC1 | -0.31 | 0.00301 | -0.23 | 0.02626 | miRNATAP | -0.14 | 0.0054 | NA | |
| 58 | hsa-miR-374b-5p | AHI1 | -0.31 | 0.00301 | 0.07 | 0.63637 | MirTarget | -0.2 | 0.00238 | NA | |
| 59 | hsa-miR-450b-5p | AIDA | -1.34 | 0 | 0.39 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 60 | hsa-miR-29b-1-5p | AIF1L | -0.54 | 0.00103 | 0.76 | 0.0005 | MirTarget | -0.23 | 0.00049 | NA | |
| 61 | hsa-miR-374b-5p | AIF1L | -0.31 | 0.00301 | 0.76 | 0.0005 | mirMAP | -0.32 | 0.00237 | NA | |
| 62 | hsa-miR-30e-3p | AKAP8 | -1.21 | 0 | 0.3 | 0 | MirTarget | -0.11 | 0 | NA | |
| 63 | hsa-miR-199a-5p | AKR1C2 | -1.99 | 0 | 0.25 | 0.39842 | miRanda | -0.19 | 8.0E-5 | NA | |
| 64 | hsa-miR-199a-5p | ALDH1L1 | -1.99 | 0 | -1.8 | 0 | miRanda | -0.21 | 0.00084 | NA | |
| 65 | hsa-miR-374b-5p | ALDH1L2 | -0.31 | 0.00301 | 0.46 | 0.04465 | mirMAP | -0.37 | 0.00046 | NA | |
| 66 | hsa-let-7b-5p | ALG3 | -0.96 | 0 | 0.6 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 67 | hsa-miR-199a-5p | ALKBH2 | -1.99 | 0 | 0.58 | 2.0E-5 | miRanda | -0.14 | 0 | NA | |
| 68 | hsa-miR-148a-5p | ALPK3 | -0.77 | 0 | 1.32 | 0 | mirMAP | -0.3 | 0.00022 | NA | |
| 69 | hsa-miR-30e-3p | ALPK3 | -1.21 | 0 | 1.32 | 0 | mirMAP | -0.36 | 0.00065 | NA | |
| 70 | hsa-miR-374b-5p | AMOTL1 | -0.31 | 0.00301 | -0.53 | 0.02603 | mirMAP | -0.48 | 2.0E-5 | NA | |
| 71 | hsa-let-7b-5p | AMT | -0.96 | 0 | -0.85 | 0 | MirTarget | -0.16 | 0.00457 | NA | |
| 72 | hsa-miR-450b-5p | ANGEL2 | -1.34 | 0 | 0.4 | 0 | mirMAP | -0.11 | 0 | NA | |
| 73 | hsa-miR-148a-5p | ANK2 | -0.77 | 0 | -0.94 | 8.0E-5 | MirTarget; miRNATAP | -0.3 | 4.0E-5 | NA | |
| 74 | hsa-miR-199a-5p | ANKAR | -1.99 | 0 | 0.96 | 0 | miRanda | -0.12 | 0 | NA | |
| 75 | hsa-miR-30e-3p | ANKDD1A | -1.21 | 0 | 0.53 | 0.0002 | MirTarget | -0.25 | 1.0E-5 | NA | |
| 76 | hsa-miR-199a-5p | ANKRD26P1 | -1.99 | 0 | 0.43 | 0.21485 | miRanda | -0.16 | 0.00457 | NA | |
| 77 | hsa-miR-450b-5p | ANKRD27 | -1.34 | 0 | 0.9 | 0 | PITA | -0.12 | 0.00037 | NA | |
| 78 | hsa-miR-30e-3p | ANKRD29 | -1.21 | 0 | 1.13 | 4.0E-5 | MirTarget; mirMAP | -0.33 | 0.00303 | NA | |
| 79 | hsa-let-7b-3p | ANKRD52 | -1.22 | 0 | 1.46 | 0 | mirMAP | -0.31 | 0 | NA | |
| 80 | hsa-let-7b-5p | ANKRD52 | -0.96 | 0 | 1.46 | 0 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.21 | 0 | NA | |
| 81 | hsa-let-7f-1-3p | ANKRD52 | -0.7 | 0 | 1.46 | 0 | mirMAP | -0.14 | 0.00159 | NA | |
| 82 | hsa-miR-148a-5p | ANKRD52 | -0.77 | 0 | 1.46 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 83 | hsa-miR-29b-1-5p | ANKRD52 | -0.54 | 0.00103 | 1.46 | 0 | mirMAP | -0.15 | 6.0E-5 | NA | |
| 84 | hsa-miR-3607-3p | ANKRD52 | -2.16 | 0 | 1.46 | 0 | miRNATAP | -0.22 | 0 | NA | |
| 85 | hsa-let-7b-3p | ANKS1B | -1.22 | 0 | 1.43 | 0.00012 | miRNATAP | -0.39 | 0.00133 | NA | |
| 86 | hsa-miR-450b-5p | ANKS1B | -1.34 | 0 | 1.43 | 0.00012 | PITA; miRNATAP | -0.33 | 0.00256 | NA | |
| 87 | hsa-let-7b-5p | ANKZF1 | -0.96 | 0 | 0.85 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 88 | hsa-miR-30a-3p | ANLN | -1.53 | 0 | 3.89 | 0 | MirTarget | -0.56 | 0 | NA | |
| 89 | hsa-miR-30e-3p | ANLN | -1.21 | 0 | 3.89 | 0 | MirTarget | -0.77 | 0 | NA | |
| 90 | hsa-miR-3607-3p | ANO4 | -2.16 | 0 | 1.36 | 0 | MirTarget; miRNATAP | -0.28 | 1.0E-5 | NA | |
| 91 | hsa-miR-193b-3p | ANP32B | -0.17 | 0.27202 | -0.02 | 0.80628 | miRNAWalker2 validate | -0.12 | 4.0E-5 | NA | |
| 92 | hsa-miR-374b-5p | ANTXR1 | -0.31 | 0.00301 | -0.48 | 0.04009 | MirTarget; mirMAP | -0.44 | 6.0E-5 | NA | |
| 93 | hsa-miR-374b-5p | ANXA4 | -0.31 | 0.00301 | -0.2 | 0.167 | mirMAP | -0.33 | 0 | NA | |
| 94 | hsa-let-7b-5p | AP1S1 | -0.96 | 0 | 0.55 | 0 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 95 | hsa-let-7b-3p | AP3B1 | -1.22 | 0 | 0.63 | 0 | mirMAP | -0.14 | 0 | NA | |
| 96 | hsa-let-7f-1-3p | AP3B1 | -0.7 | 0 | 0.63 | 0 | mirMAP | -0.1 | 0.00013 | NA | |
| 97 | hsa-miR-193b-3p | AP3M2 | -0.17 | 0.27202 | 0.87 | 0 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 98 | hsa-miR-199a-5p | APC2 | -1.99 | 0 | 1.72 | 0 | mirMAP | -0.1 | 0.00178 | NA | |
| 99 | hsa-miR-193b-3p | APCDD1 | -0.17 | 0.27202 | -1.3 | 0.00043 | miRNAWalker2 validate | -0.53 | 1.0E-5 | NA | |
| 100 | hsa-let-7b-5p | APRT | -0.96 | 0 | 0.19 | 0.12663 | miRNAWalker2 validate | -0.27 | 0 | NA | |
| 101 | hsa-let-7b-5p | ARFIP2 | -0.96 | 0 | 0.37 | 0 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 102 | hsa-let-7b-5p | ARG2 | -0.96 | 0 | 0.15 | 0.43591 | MirTarget | -0.18 | 0.00562 | NA | |
| 103 | hsa-miR-193b-3p | ARHGAP19 | -0.17 | 0.27202 | 0.49 | 0 | miRNAWalker2 validate; mirMAP | -0.14 | 0 | NA | |
| 104 | hsa-miR-29b-1-5p | ARHGAP19 | -0.54 | 0.00103 | 0.49 | 0 | mirMAP | -0.13 | 0 | NA | |
| 105 | hsa-let-7f-1-3p | ARHGAP28 | -0.7 | 0 | 0.59 | 0.03308 | mirMAP | -0.27 | 0.00485 | NA | |
| 106 | hsa-miR-148a-5p | ARHGAP31 | -0.77 | 0 | -0.42 | 0.00347 | mirMAP | -0.14 | 0.00129 | NA | |
| 107 | hsa-miR-374b-5p | ARHGAP6 | -0.31 | 0.00301 | 0.22 | 0.19399 | mirMAP; miRNATAP | -0.26 | 0.00099 | NA | |
| 108 | hsa-miR-148a-5p | ARHGEF18 | -0.77 | 0 | 0.5 | 0 | MirTarget; miRNATAP | -0.12 | 0 | NA | |
| 109 | hsa-miR-193b-3p | ARHGEF18 | -0.17 | 0.27202 | 0.5 | 0 | mirMAP | -0.17 | 0 | NA | |
| 110 | hsa-miR-3607-3p | ARHGEF3 | -2.16 | 0 | 0.51 | 0.00381 | MirTarget; miRNATAP | -0.12 | 0.00127 | NA | |
| 111 | hsa-let-7b-5p | ARID3A | -0.96 | 0 | 1.53 | 0 | miRNAWalker2 validate | -0.38 | 1.0E-5 | NA | |
| 112 | hsa-let-7b-5p | ARID3B | -0.96 | 0 | 0.46 | 0.00021 | miRNAWalker2 validate; miRNATAP | -0.14 | 0.00075 | NA | |
| 113 | hsa-miR-193b-3p | ARID3B | -0.17 | 0.27202 | 0.46 | 0.00021 | MirTarget | -0.19 | 0 | NA | |
| 114 | hsa-miR-148a-5p | ARID5B | -0.77 | 0 | -0.52 | 0.00012 | mirMAP | -0.17 | 3.0E-5 | NA | |
| 115 | hsa-miR-193b-3p | ARL10 | -0.17 | 0.27202 | -0.42 | 0.01556 | mirMAP | -0.18 | 0.00149 | NA | |
| 116 | hsa-miR-450b-5p | ARMC1 | -1.34 | 0 | 0.39 | 0.0001 | MirTarget; mirMAP | -0.11 | 0.00015 | NA | |
| 117 | hsa-miR-450b-5p | ARPC5 | -1.34 | 0 | 0.69 | 0 | MirTarget; PITA; mirMAP | -0.14 | 0 | NA | |
| 118 | hsa-let-7b-3p | ARSB | -1.22 | 0 | 0 | 0.99867 | mirMAP | -0.16 | 8.0E-5 | NA | |
| 119 | hsa-let-7f-1-3p | ARSB | -0.7 | 0 | 0 | 0.99867 | mirMAP | -0.16 | 0.00033 | NA | |
| 120 | hsa-miR-148a-5p | ASAH1 | -0.77 | 0 | -0.44 | 0.00056 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 121 | hsa-miR-3607-3p | ASAP1 | -2.16 | 0 | 0.6 | 1.0E-5 | MirTarget; mirMAP; miRNATAP | -0.1 | 0.00049 | NA | |
| 122 | hsa-let-7f-1-3p | ASAP2 | -0.7 | 0 | 0.69 | 2.0E-5 | MirTarget | -0.15 | 0.00778 | NA | |
| 123 | hsa-miR-30e-3p | ASB1 | -1.21 | 0 | 0.21 | 0.0408 | mirMAP | -0.18 | 0 | NA | |
| 124 | hsa-miR-199a-5p | ASB4 | -1.99 | 0 | -0.62 | 0.06683 | miRanda | -0.17 | 0.0018 | NA | |
| 125 | hsa-miR-374b-5p | ASCC3 | -0.31 | 0.00301 | 0.03 | 0.78425 | mirMAP | -0.21 | 0.0001 | NA | |
| 126 | hsa-let-7b-5p | ASNA1 | -0.96 | 0 | 0.58 | 0 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 127 | hsa-miR-450b-5p | ASPH | -1.34 | 0 | 0.47 | 0.00525 | MirTarget | -0.15 | 0.00224 | NA | |
| 128 | hsa-let-7b-5p | ASPSCR1 | -0.96 | 0 | 0.72 | 1.0E-5 | miRNAWalker2 validate | -0.43 | 0 | NA | |
| 129 | hsa-let-7b-3p | ASXL1 | -1.22 | 0 | 0.64 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 130 | hsa-miR-193b-3p | ASXL1 | -0.17 | 0.27202 | 0.64 | 0 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 131 | hsa-miR-193b-3p | ATAD2 | -0.17 | 0.27202 | 1.43 | 0 | miRNAWalker2 validate | -0.21 | 0.00024 | NA | |
| 132 | hsa-let-7b-5p | ATAD3B | -0.96 | 0 | 0.76 | 0 | miRNAWalker2 validate | -0.29 | 0 | NA | |
| 133 | hsa-let-7b-3p | ATAD5 | -1.22 | 0 | 1.62 | 0 | MirTarget | -0.3 | 0 | NA | |
| 134 | hsa-let-7f-1-3p | ATAD5 | -0.7 | 0 | 1.62 | 0 | MirTarget | -0.22 | 0.00173 | NA | |
| 135 | hsa-let-7b-5p | ATG10 | -0.96 | 0 | 0.35 | 0.00052 | MirTarget | -0.2 | 0 | NA | |
| 136 | hsa-let-7b-5p | ATG4B | -0.96 | 0 | 0.37 | 0 | miRNAWalker2 validate; miRNATAP | -0.12 | 0 | NA | |
| 137 | hsa-let-7b-5p | ATOX1 | -0.96 | 0 | 0.43 | 0.00496 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 138 | hsa-miR-199a-5p | ATOX1 | -1.99 | 0 | 0.43 | 0.00496 | mirMAP | -0.11 | 0 | NA | |
| 139 | hsa-miR-3607-3p | ATP10B | -2.16 | 0 | -0.24 | 0.56639 | MirTarget | -0.24 | 0.00744 | NA | |
| 140 | hsa-miR-193b-3p | ATP13A2 | -0.17 | 0.27202 | 0.92 | 0 | miRNAWalker2 validate | -0.15 | 0.00012 | NA | |
| 141 | hsa-miR-193b-3p | ATP1A1 | -0.17 | 0.27202 | 0.44 | 0.00034 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 142 | hsa-miR-374b-5p | ATP1A1 | -0.31 | 0.00301 | 0.44 | 0.00034 | miRNATAP | -0.19 | 0.00112 | NA | |
| 143 | hsa-let-7b-3p | ATP2B1 | -1.22 | 0 | -0.16 | 0.1608 | MirTarget; miRNATAP | -0.11 | 0.00385 | NA | |
| 144 | hsa-miR-30a-3p | ATP2B2 | -1.53 | 0 | -0.4 | 0.15518 | mirMAP | -0.22 | 0.00279 | NA | |
| 145 | hsa-let-7b-3p | ATP5E | -1.22 | 0 | 0.38 | 0.00141 | mirMAP | -0.24 | 0 | NA | |
| 146 | hsa-miR-450b-5p | ATP5E | -1.34 | 0 | 0.38 | 0.00141 | mirMAP | -0.18 | 0 | NA | |
| 147 | hsa-let-7b-5p | ATP6V0A1 | -0.96 | 0 | 0.28 | 0.00395 | miRNAWalker2 validate | -0.12 | 0.00032 | NA | |
| 148 | hsa-let-7b-3p | ATP6V1C1 | -1.22 | 0 | 0.88 | 0 | mirMAP | -0.23 | 0 | NA | |
| 149 | hsa-miR-195-3p | ATP6V1C1 | -1.09 | 0 | 0.88 | 0 | mirMAP | -0.19 | 0 | NA | |
| 150 | hsa-miR-450b-5p | ATP6V1C1 | -1.34 | 0 | 0.88 | 0 | mirMAP | -0.2 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL CYCLE | 185 | 1316 | 2.884e-17 | 1.342e-13 |
| 2 | CELL CYCLE PROCESS | 158 | 1081 | 2.411e-16 | 5.61e-13 |
| 3 | MITOTIC CELL CYCLE | 121 | 766 | 3.678e-15 | 5.704e-12 |
| 4 | CELL DIVISION | 77 | 460 | 2.629e-11 | 3.058e-08 |
| 5 | NEGATIVE REGULATION OF GENE EXPRESSION | 178 | 1493 | 3.274e-10 | 3.047e-07 |
| 6 | MITOTIC NUCLEAR DIVISION | 62 | 361 | 8.113e-10 | 6.292e-07 |
| 7 | CYTOSKELETON ORGANIZATION | 112 | 838 | 1.736e-09 | 1.042e-06 |
| 8 | REGULATION OF CELL CYCLE | 123 | 949 | 1.792e-09 | 1.042e-06 |
| 9 | MICROTUBULE CYTOSKELETON ORGANIZATION | 59 | 348 | 3.433e-09 | 1.775e-06 |
| 10 | NEGATIVE REGULATION OF CELL CYCLE | 68 | 433 | 5.925e-09 | 2.757e-06 |
| 11 | DNA REPLICATION | 41 | 208 | 1.126e-08 | 4.763e-06 |
| 12 | DNA METABOLIC PROCESS | 101 | 758 | 1.32e-08 | 5.12e-06 |
| 13 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 173 | 1517 | 1.632e-08 | 5.841e-06 |
| 14 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 41 | 214 | 2.626e-08 | 8.727e-06 |
| 15 | CELL CYCLE CHECKPOINT | 38 | 194 | 4.567e-08 | 1.417e-05 |
| 16 | CELL CYCLE PHASE TRANSITION | 45 | 255 | 7.35e-08 | 2.026e-05 |
| 17 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 60 | 387 | 7.402e-08 | 2.026e-05 |
| 18 | MICROTUBULE BASED PROCESS | 74 | 522 | 1.034e-07 | 2.673e-05 |
| 19 | EMBRYO DEVELOPMENT | 111 | 894 | 1.15e-07 | 2.678e-05 |
| 20 | NCRNA METABOLIC PROCESS | 75 | 533 | 1.151e-07 | 2.678e-05 |
| 21 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 123 | 1021 | 1.258e-07 | 2.787e-05 |
| 22 | RIBONUCLEOPROTEIN COMPLEX BIOGENESIS | 65 | 440 | 1.358e-07 | 2.872e-05 |
| 23 | RIBOSOME BIOGENESIS | 50 | 308 | 2.149e-07 | 4.347e-05 |
| 24 | RNA PROCESSING | 104 | 835 | 2.51e-07 | 4.685e-05 |
| 25 | CELL PROLIFERATION | 88 | 672 | 2.517e-07 | 4.685e-05 |
| 26 | ORGANELLE FISSION | 70 | 496 | 2.714e-07 | 4.761e-05 |
| 27 | PROTEIN LOCALIZATION | 193 | 1805 | 2.763e-07 | 4.761e-05 |
| 28 | REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION | 34 | 178 | 4.269e-07 | 7.094e-05 |
| 29 | NEUROGENESIS | 155 | 1402 | 6.722e-07 | 0.0001079 |
| 30 | NCRNA PROCESSING | 57 | 386 | 8.034e-07 | 0.0001246 |
| 31 | REGULATION OF MICROTUBULE BASED PROCESS | 41 | 243 | 9.375e-07 | 0.0001343 |
| 32 | REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 38 | 217 | 9.07e-07 | 0.0001343 |
| 33 | CELLULAR MACROMOLECULE LOCALIZATION | 139 | 1234 | 9.528e-07 | 0.0001343 |
| 34 | MACROMOLECULAR COMPLEX ASSEMBLY | 153 | 1398 | 1.447e-06 | 0.0001923 |
| 35 | REGULATION OF CELL CYCLE PROCESS | 74 | 558 | 1.416e-06 | 0.0001923 |
| 36 | EPITHELIUM DEVELOPMENT | 111 | 945 | 1.815e-06 | 0.0002346 |
| 37 | TISSUE DEVELOPMENT | 163 | 1518 | 2.021e-06 | 0.0002541 |
| 38 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 35 | 199 | 2.175e-06 | 0.000262 |
| 39 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 141 | 1275 | 2.196e-06 | 0.000262 |
| 40 | REGULATION OF MITOTIC CELL CYCLE | 64 | 468 | 2.596e-06 | 0.000302 |
| 41 | EMBRYONIC MORPHOGENESIS | 71 | 539 | 2.942e-06 | 0.0003339 |
| 42 | REGULATION OF CELL MORPHOGENESIS | 72 | 552 | 3.58e-06 | 0.0003966 |
| 43 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 31 | 170 | 3.761e-06 | 0.0003988 |
| 44 | NEGATIVE REGULATION OF CYTOSKELETON ORGANIZATION | 37 | 221 | 3.771e-06 | 0.0003988 |
| 45 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 72 | 554 | 4.075e-06 | 0.0004214 |
| 46 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 88 | 720 | 4.598e-06 | 0.0004651 |
| 47 | MORPHOGENESIS OF AN EPITHELIUM | 56 | 400 | 5.33e-06 | 0.0005167 |
| 48 | REGULATION OF CELL PROJECTION ORGANIZATION | 72 | 558 | 5.263e-06 | 0.0005167 |
| 49 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 110 | 957 | 5.511e-06 | 0.0005233 |
| 50 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 84 | 684 | 6.295e-06 | 0.0005858 |
| 51 | TUBE DEVELOPMENT | 71 | 552 | 6.824e-06 | 0.0006226 |
| 52 | CELL CYCLE G1 S PHASE TRANSITION | 23 | 111 | 7.527e-06 | 0.0006486 |
| 53 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 23 | 111 | 7.527e-06 | 0.0006486 |
| 54 | REGULATION OF CELL PROLIFERATION | 158 | 1496 | 7.516e-06 | 0.0006486 |
| 55 | CHROMOSOME ORGANIZATION | 114 | 1009 | 8.227e-06 | 0.0006931 |
| 56 | SKELETAL SYSTEM DEVELOPMENT | 61 | 455 | 8.342e-06 | 0.0006931 |
| 57 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 173 | 1672 | 9.067e-06 | 0.0007073 |
| 58 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 39 | 247 | 8.851e-06 | 0.0007073 |
| 59 | PROTEIN LOCALIZATION TO ORGANELLE | 71 | 556 | 8.76e-06 | 0.0007073 |
| 60 | REGULATION OF CELL CYCLE PHASE TRANSITION | 47 | 321 | 9.12e-06 | 0.0007073 |
| 61 | REGULATION OF CELL DIFFERENTIATION | 157 | 1492 | 9.854e-06 | 0.0007278 |
| 62 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 56 | 408 | 9.678e-06 | 0.0007278 |
| 63 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 45 | 303 | 9.698e-06 | 0.0007278 |
| 64 | ORGAN MORPHOGENESIS | 98 | 841 | 1.028e-05 | 0.0007358 |
| 65 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 160 | 1527 | 1.02e-05 | 0.0007358 |
| 66 | TUBE MORPHOGENESIS | 47 | 323 | 1.077e-05 | 0.0007594 |
| 67 | DNA INTEGRITY CHECKPOINT | 27 | 146 | 1.191e-05 | 0.0008268 |
| 68 | SPINDLE ASSEMBLY | 17 | 70 | 1.291e-05 | 0.0008834 |
| 69 | CELL PROJECTION ORGANIZATION | 103 | 902 | 1.452e-05 | 0.000979 |
| 70 | REGULATION OF CARDIAC MUSCLE CELL CONTRACTION | 10 | 27 | 1.495e-05 | 0.0009936 |
| 71 | ESTABLISHMENT OF LOCALIZATION IN CELL | 172 | 1676 | 1.543e-05 | 0.001004 |
| 72 | REGULATION OF ORGANELLE ORGANIZATION | 128 | 1178 | 1.553e-05 | 0.001004 |
| 73 | ORGANELLE ASSEMBLY | 64 | 495 | 1.626e-05 | 0.001037 |
| 74 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 86 | 724 | 1.759e-05 | 0.001106 |
| 75 | RRNA METABOLIC PROCESS | 39 | 255 | 1.884e-05 | 0.001169 |
| 76 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 59 | 448 | 1.998e-05 | 0.001223 |
| 77 | SISTER CHROMATID SEGREGATION | 30 | 176 | 2.117e-05 | 0.001279 |
| 78 | REGULATION OF CELL DEVELOPMENT | 96 | 836 | 2.275e-05 | 0.001357 |
| 79 | REGULATION OF CYTOSKELETON ORGANIZATION | 64 | 502 | 2.523e-05 | 0.001486 |
| 80 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 49 | 354 | 2.733e-05 | 0.001589 |
| 81 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 148 | 1423 | 3.266e-05 | 0.001876 |
| 82 | TRANSLATIONAL INITIATION | 26 | 146 | 3.393e-05 | 0.001889 |
| 83 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 26 | 146 | 3.393e-05 | 0.001889 |
| 84 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 123 | 1142 | 3.41e-05 | 0.001889 |
| 85 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 179 | 1784 | 3.724e-05 | 0.002039 |
| 86 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 87 | 750 | 3.799e-05 | 0.002055 |
| 87 | MITOTIC CELL CYCLE CHECKPOINT | 25 | 139 | 4.017e-05 | 0.002148 |
| 88 | DNA DEPENDENT DNA REPLICATION | 20 | 99 | 4.259e-05 | 0.002226 |
| 89 | MITOTIC SPINDLE ORGANIZATION | 16 | 69 | 4.257e-05 | 0.002226 |
| 90 | MICROTUBULE ORGANIZING CENTER ORGANIZATION | 18 | 84 | 4.485e-05 | 0.002319 |
| 91 | POSITIVE REGULATION OF GENE EXPRESSION | 174 | 1733 | 4.656e-05 | 0.002355 |
| 92 | POSITIVE REGULATION OF CELL CYCLE | 46 | 332 | 4.637e-05 | 0.002355 |
| 93 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 90 | 788 | 4.989e-05 | 0.002469 |
| 94 | CIRCULATORY SYSTEM DEVELOPMENT | 90 | 788 | 4.989e-05 | 0.002469 |
| 95 | PROTEIN COMPLEX BIOGENESIS | 121 | 1132 | 5.514e-05 | 0.002673 |
| 96 | PROTEIN COMPLEX ASSEMBLY | 121 | 1132 | 5.514e-05 | 0.002673 |
| 97 | CELLULAR COMPONENT MORPHOGENESIS | 100 | 900 | 5.739e-05 | 0.002753 |
| 98 | MRNA METABOLIC PROCESS | 73 | 611 | 6.211e-05 | 0.002949 |
| 99 | CHROMATIN MODIFICATION | 66 | 539 | 6.637e-05 | 0.00312 |
| 100 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 179 | 1805 | 7.085e-05 | 0.003297 |
| 101 | ORGANIC ACID TRANSMEMBRANE TRANSPORT | 20 | 103 | 7.666e-05 | 0.003531 |
| 102 | REGULATION OF ACTIN FILAMENT BASED MOVEMENT | 10 | 32 | 8.064e-05 | 0.003678 |
| 103 | TISSUE MORPHOGENESIS | 65 | 533 | 8.481e-05 | 0.003822 |
| 104 | REGULATION OF NEURON DIFFERENTIATION | 67 | 554 | 8.543e-05 | 0.003822 |
| 105 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM | 20 | 104 | 8.827e-05 | 0.003872 |
| 106 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 143 | 1395 | 8.904e-05 | 0.003872 |
| 107 | NEGATIVE REGULATION OF CHROMOSOME ORGANIZATION | 19 | 96 | 8.781e-05 | 0.003872 |
| 108 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 92 | 823 | 9.087e-05 | 0.003915 |
| 109 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 14 | 59 | 9.687e-05 | 0.004135 |
| 110 | LOCOMOTION | 118 | 1114 | 9.987e-05 | 0.004225 |
| 111 | G2 DNA DAMAGE CHECKPOINT | 10 | 33 | 0.0001079 | 0.004524 |
| 112 | NEGATIVE REGULATION OF CELL DIVISION | 14 | 60 | 0.0001177 | 0.004845 |
| 113 | CELLULAR RESPONSE TO STRESS | 157 | 1565 | 0.0001168 | 0.004845 |
| 114 | PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM | 22 | 123 | 0.0001239 | 0.005057 |
| 115 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 45 | 337 | 0.0001342 | 0.005393 |
| 116 | MITOTIC SISTER CHROMATID SEGREGATION | 18 | 91 | 0.0001344 | 0.005393 |
| 117 | VASCULATURE DEVELOPMENT | 58 | 469 | 0.0001376 | 0.005473 |
| 118 | CELL MOTILITY | 92 | 835 | 0.0001521 | 0.005947 |
| 119 | LOCALIZATION OF CELL | 92 | 835 | 0.0001521 | 0.005947 |
| 120 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 164 | 1656 | 0.0001557 | 0.005988 |
| 121 | MITOTIC DNA INTEGRITY CHECKPOINT | 19 | 100 | 0.000155 | 0.005988 |
| 122 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 62 | 513 | 0.0001576 | 0.006011 |
| 123 | APPENDAGE DEVELOPMENT | 27 | 169 | 0.0001657 | 0.006167 |
| 124 | LIMB DEVELOPMENT | 27 | 169 | 0.0001657 | 0.006167 |
| 125 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 86 | 771 | 0.0001645 | 0.006167 |
| 126 | CHROMOSOME SEGREGATION | 38 | 272 | 0.0001721 | 0.006343 |
| 127 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 120 | 1152 | 0.0001745 | 0.006343 |
| 128 | REGULATION OF CELL GROWTH | 50 | 391 | 0.0001735 | 0.006343 |
| 129 | PALATE DEVELOPMENT | 17 | 85 | 0.0001776 | 0.006358 |
| 130 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 83 | 740 | 0.0001776 | 0.006358 |
| 131 | HEAD DEVELOPMENT | 80 | 709 | 0.0001911 | 0.006787 |
| 132 | DNA REPLICATION INITIATION | 9 | 29 | 0.0001947 | 0.006862 |
| 133 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 26 | 162 | 0.0002013 | 0.007043 |
| 134 | NEURAL TUBE FORMATION | 18 | 94 | 0.0002065 | 0.007171 |
| 135 | NUCLEAR CHROMOSOME SEGREGATION | 33 | 228 | 0.0002321 | 0.008 |
| 136 | RESPONSE TO NUTRIENT | 29 | 191 | 0.0002392 | 0.008182 |
| 137 | CELL DEVELOPMENT | 143 | 1426 | 0.000242 | 0.00822 |
| 138 | CARTILAGE DEVELOPMENT | 24 | 147 | 0.0002675 | 0.009019 |
| 139 | ACTIN FILAMENT BASED PROCESS | 55 | 450 | 0.0002729 | 0.009136 |
| 140 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 94 | 872 | 0.0002784 | 0.009252 |
| 141 | EMBRYONIC ORGAN MORPHOGENESIS | 38 | 279 | 0.0002895 | 0.009529 |
| 142 | POSITIVE REGULATION OF CELL DEVELOPMENT | 57 | 472 | 0.0002908 | 0.009529 |
| 143 | PROTEIN TARGETING TO MEMBRANE | 25 | 157 | 0.0002993 | 0.00974 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RNA BINDING | 202 | 1598 | 6.533e-14 | 6.069e-11 |
| 2 | POLY A RNA BINDING | 154 | 1170 | 4.008e-12 | 1.862e-09 |
| 3 | RIBONUCLEOTIDE BINDING | 210 | 1860 | 8.807e-10 | 2.727e-07 |
| 4 | PROTEIN DOMAIN SPECIFIC BINDING | 85 | 624 | 7.215e-08 | 1.676e-05 |
| 5 | ADENYL NUCLEOTIDE BINDING | 168 | 1514 | 1.711e-07 | 3.179e-05 |
| 6 | MACROMOLECULAR COMPLEX BINDING | 154 | 1399 | 9.476e-07 | 0.0001467 |
| 7 | MRNA BINDING | 29 | 155 | 4.525e-06 | 0.0006005 |
| 8 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 73 | 588 | 1.746e-05 | 0.001802 |
| 9 | ENZYME BINDING | 177 | 1737 | 1.73e-05 | 0.001802 |
| 10 | TRANSCRIPTION COACTIVATOR ACTIVITY | 43 | 296 | 2.616e-05 | 0.00243 |
| 11 | CHAPERONE BINDING | 17 | 81 | 9.525e-05 | 0.008044 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NUCLEOLUS | 122 | 848 | 2.416e-12 | 1.411e-09 |
| 2 | CHROMOSOME | 111 | 880 | 5.025e-08 | 1.467e-05 |
| 3 | SPINDLE | 48 | 289 | 1.858e-07 | 3.617e-05 |
| 4 | PCG PROTEIN COMPLEX | 15 | 43 | 2.76e-07 | 4.029e-05 |
| 5 | MICROTUBULE CYTOSKELETON | 125 | 1068 | 4.788e-07 | 5.593e-05 |
| 6 | CELL JUNCTION | 130 | 1151 | 1.901e-06 | 0.000185 |
| 7 | RIBONUCLEOPROTEIN COMPLEX | 88 | 721 | 4.86e-06 | 0.0004055 |
| 8 | ANCHORING JUNCTION | 65 | 489 | 5.67e-06 | 0.0004139 |
| 9 | CYTOSKELETON | 199 | 1967 | 7.719e-06 | 0.0005009 |
| 10 | EUCHROMATIN | 11 | 31 | 9.015e-06 | 0.0005265 |
| 11 | MICROTUBULE | 55 | 405 | 1.578e-05 | 0.000838 |
| 12 | CELL SUBSTRATE JUNCTION | 54 | 398 | 1.928e-05 | 0.0009385 |
| 13 | EXCITATORY SYNAPSE | 32 | 197 | 3.096e-05 | 0.001292 |
| 14 | HETEROCHROMATIN | 16 | 67 | 2.887e-05 | 0.001292 |
| 15 | CHROMOSOME CENTROMERIC REGION | 29 | 174 | 4.421e-05 | 0.001721 |
| 16 | NUCLEAR CHROMOSOME | 65 | 523 | 4.808e-05 | 0.001755 |
| 17 | SPINDLE MICROTUBULE | 14 | 58 | 7.934e-05 | 0.002726 |
| 18 | SPINDLE MIDZONE | 9 | 27 | 0.0001046 | 0.003392 |
| 19 | CYTOSKELETAL PART | 146 | 1436 | 0.0001104 | 0.003394 |
| 20 | CONDENSED CHROMOSOME | 30 | 195 | 0.0001486 | 0.004338 |
| 21 | CHROMATIN | 55 | 441 | 0.0001629 | 0.004531 |
| 22 | POSTSYNAPSE | 48 | 378 | 0.0002728 | 0.007242 |
| 23 | CENTROSOME | 58 | 487 | 0.0003697 | 0.009244 |
| 24 | SUPRAMOLECULAR FIBER | 75 | 670 | 0.0003799 | 0.009244 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
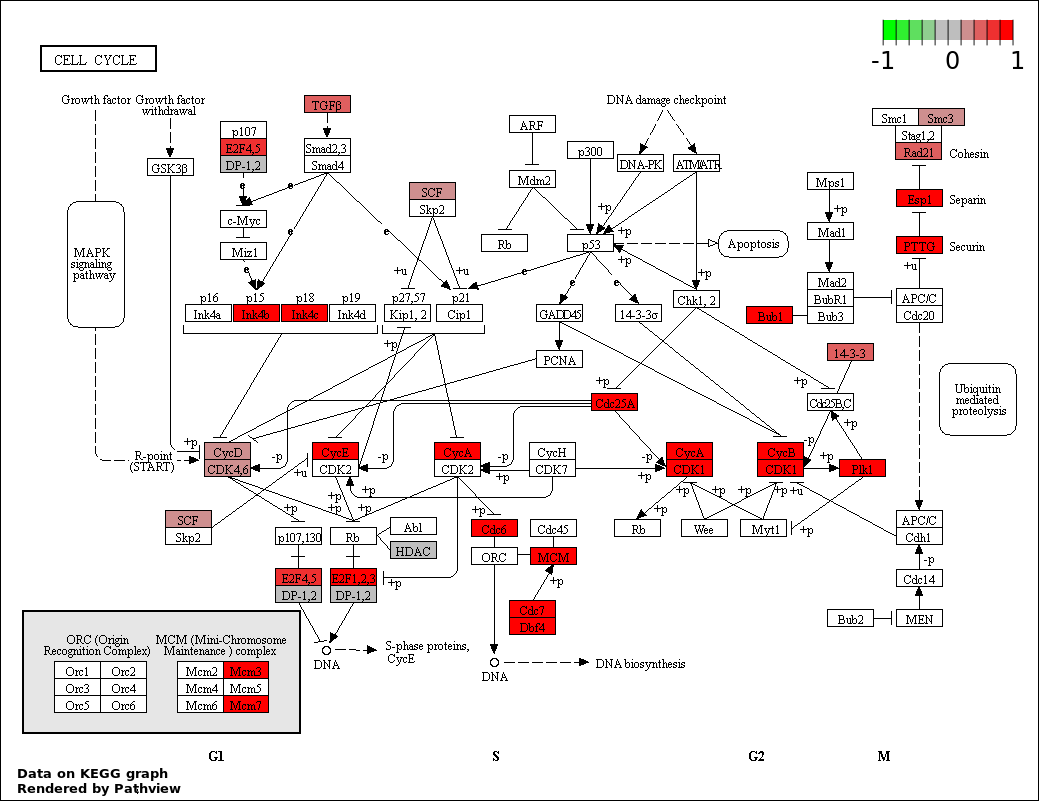
| 1 | hsa04110_Cell_cycle | 28 | 128 | 2.483e-07 | 4.47e-05 | |
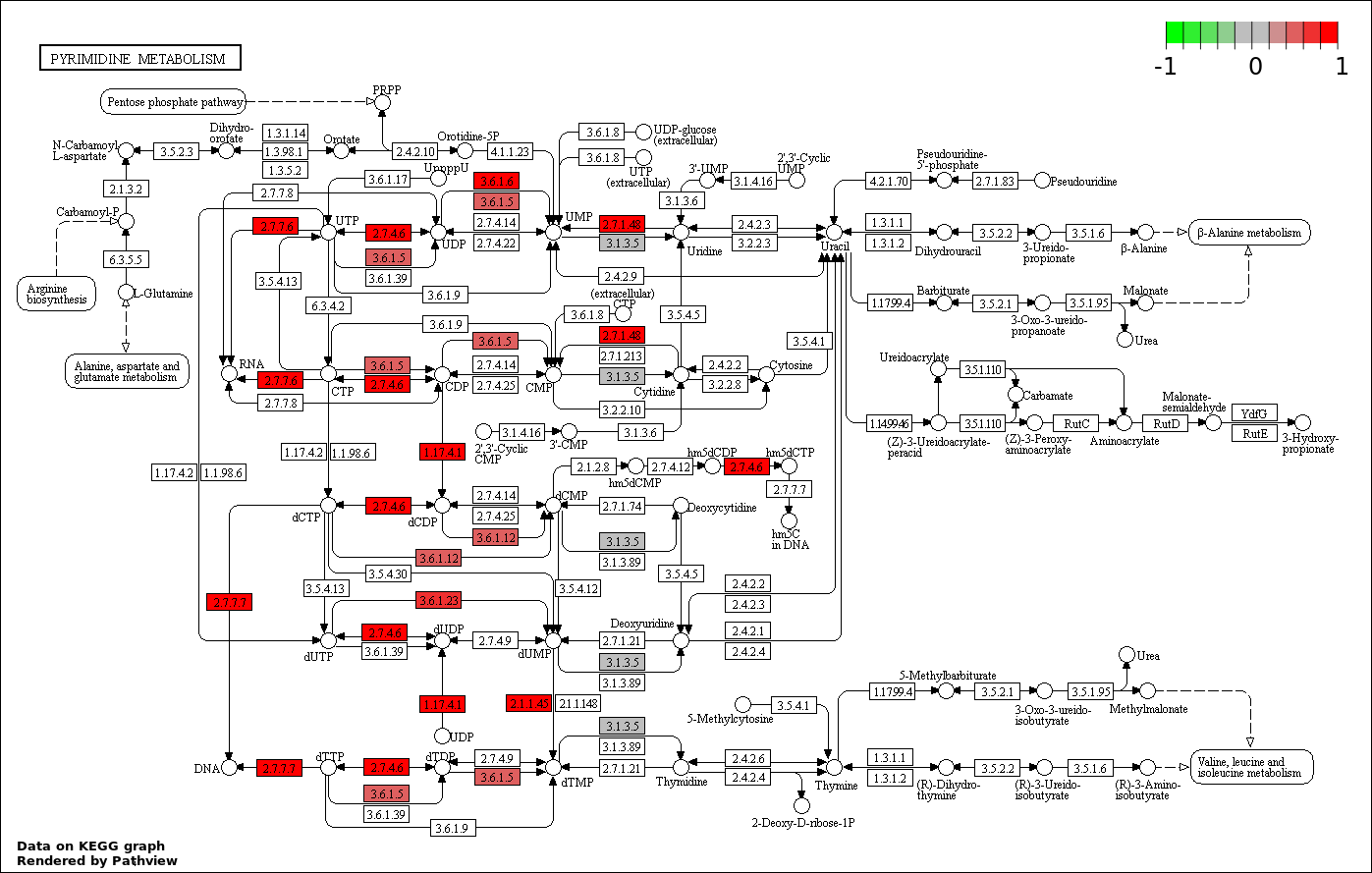
| 2 | hsa00240_Pyrimidine_metabolism | 19 | 99 | 0.000135 | 0.01215 | |
| 3 | hsa04390_Hippo_signaling_pathway | 25 | 154 | 0.0002206 | 0.01324 | |
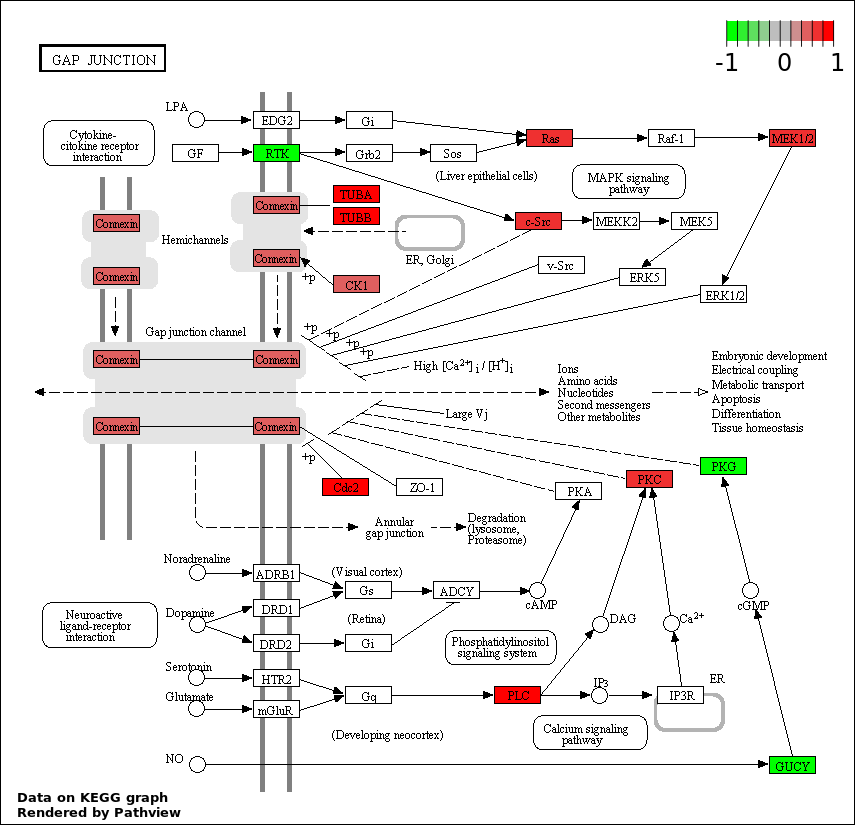
| 4 | hsa04540_Gap_junction | 17 | 90 | 0.000363 | 0.01459 | |
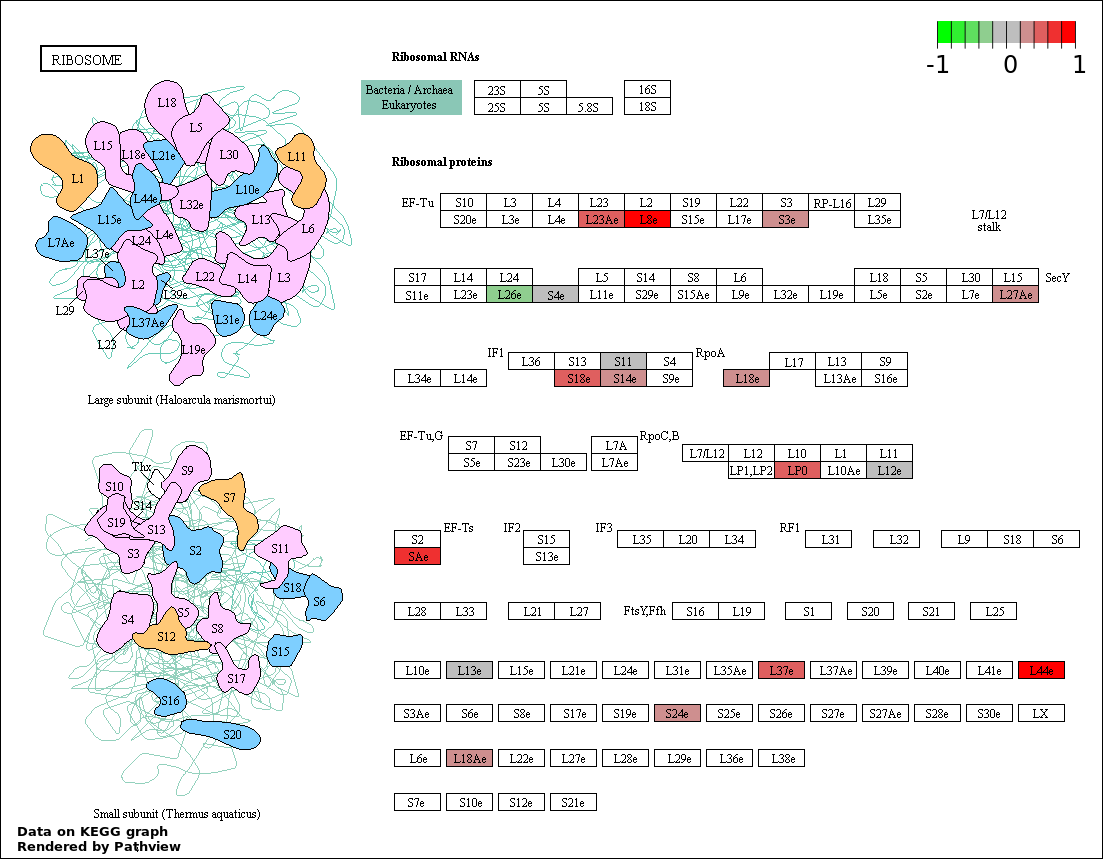
| 5 | hsa03010_Ribosome | 17 | 92 | 0.0004745 | 0.01459 | |
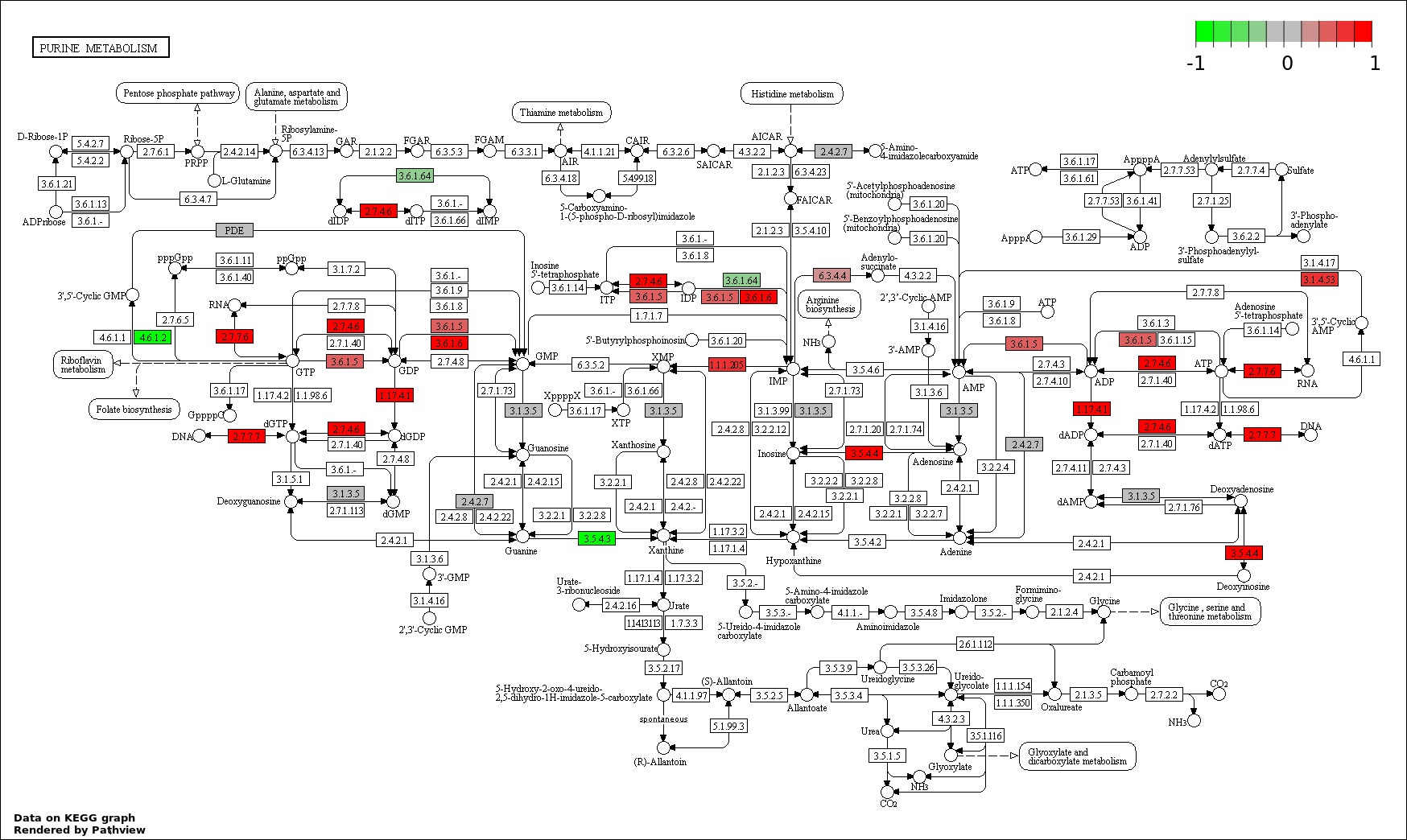
| 6 | hsa00230_Purine_metabolism | 25 | 162 | 0.0004863 | 0.01459 | |
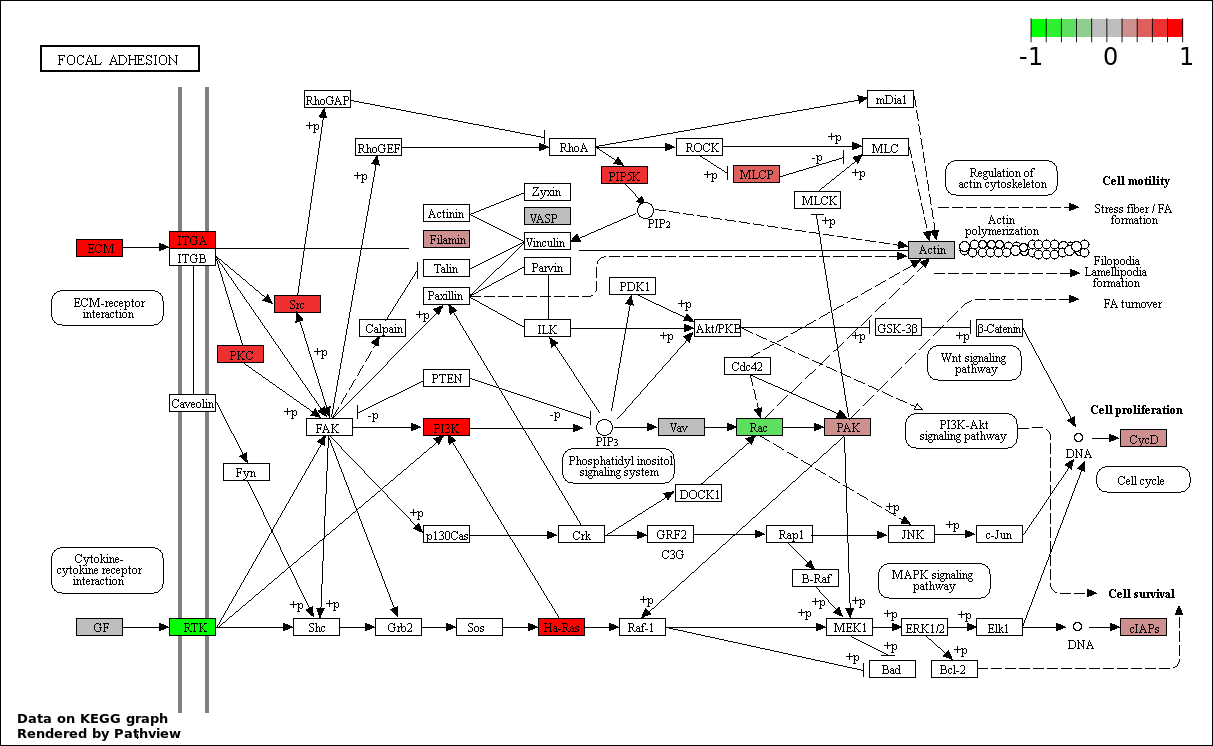
| 7 | hsa04510_Focal_adhesion | 28 | 200 | 0.001125 | 0.02893 | |
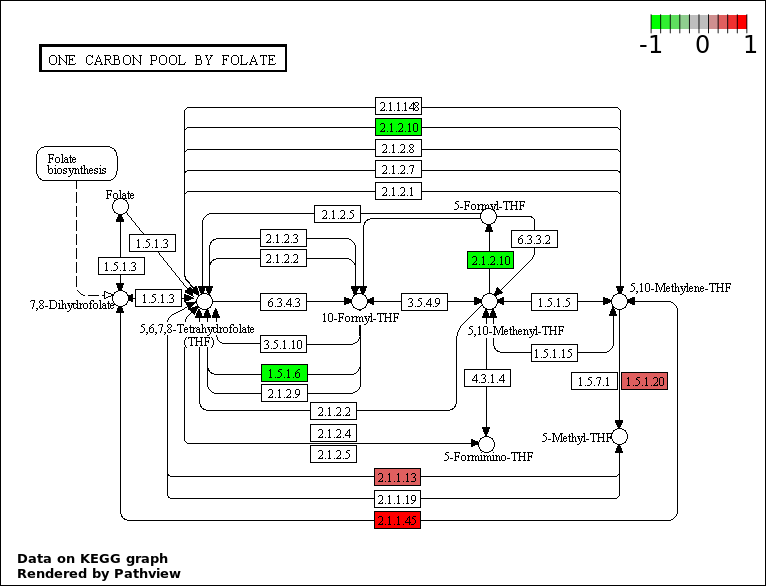
| 8 | hsa00670_One_carbon_pool_by_folate | 6 | 18 | 0.001541 | 0.03468 | |
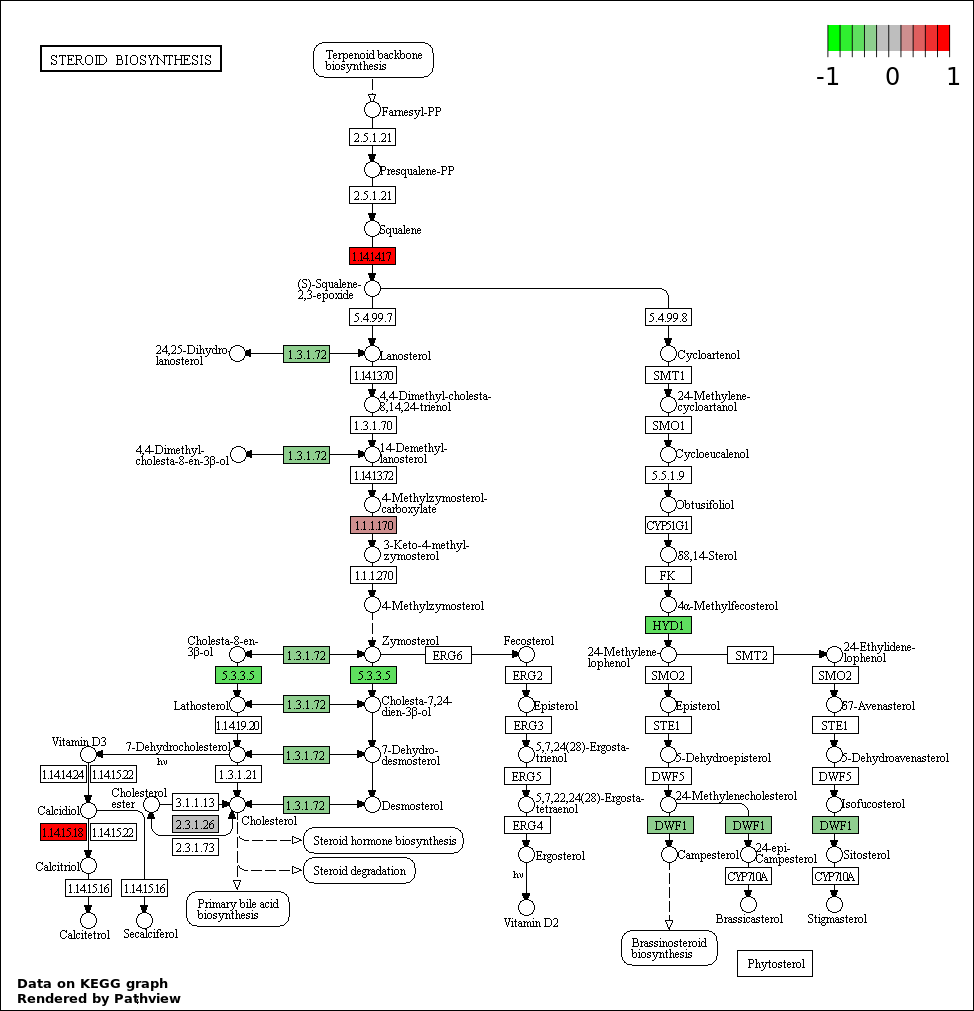
| 9 | hsa00100_Steroid_biosynthesis | 6 | 19 | 0.00211 | 0.0395 | |
| 10 | hsa04310_Wnt_signaling_pathway | 22 | 151 | 0.002194 | 0.0395 | |
| 11 | hsa03030_DNA_replication | 8 | 36 | 0.004652 | 0.06644 | |
| 12 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 15 | 95 | 0.004914 | 0.06644 | |
| 13 | hsa03020_RNA_polymerase | 7 | 29 | 0.004915 | 0.06644 | |
| 14 | hsa04115_p53_signaling_pathway | 12 | 69 | 0.005168 | 0.06644 | |
| 15 | hsa04730_Long.term_depression | 12 | 70 | 0.005818 | 0.06806 | |
| 16 | hsa00910_Nitrogen_metabolism | 6 | 23 | 0.00605 | 0.06806 | |
| 17 | hsa04360_Axon_guidance | 18 | 130 | 0.008937 | 0.09434 | |
| 18 | hsa04142_Lysosome | 17 | 121 | 0.009434 | 0.09434 | |
| 19 | hsa04810_Regulation_of_actin_cytoskeleton | 26 | 214 | 0.01099 | 0.09935 | |
| 20 | hsa04512_ECM.receptor_interaction | 13 | 85 | 0.01104 | 0.09935 | |
| 21 | hsa04114_Oocyte_meiosis | 16 | 114 | 0.01167 | 0.1 | |
| 22 | hsa04014_Ras_signaling_pathway | 27 | 236 | 0.02013 | 0.1606 | |
| 23 | hsa04520_Adherens_junction | 11 | 73 | 0.02052 | 0.1606 | |
| 24 | hsa04330_Notch_signaling_pathway | 8 | 47 | 0.02322 | 0.1741 | |
| 25 | hsa04151_PI3K_AKT_signaling_pathway | 37 | 351 | 0.02476 | 0.1782 | |
| 26 | hsa04710_Circadian_rhythm_._mammal | 5 | 23 | 0.0261 | 0.1807 | |
| 27 | hsa04012_ErbB_signaling_pathway | 12 | 87 | 0.03007 | 0.2005 | |
| 28 | hsa04070_Phosphatidylinositol_signaling_system | 11 | 78 | 0.03192 | 0.2052 | |
| 29 | hsa04664_Fc_epsilon_RI_signaling_pathway | 11 | 79 | 0.03465 | 0.209 | |
| 30 | hsa00450_Selenocompound_metabolism | 4 | 17 | 0.03483 | 0.209 | |
| 31 | hsa04916_Melanogenesis | 13 | 101 | 0.04042 | 0.2347 | |
| 32 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 5 | 26 | 0.04235 | 0.2382 | |
| 33 | hsa04145_Phagosome | 18 | 156 | 0.04719 | 0.2574 | |
| 34 | hsa04144_Endocytosis | 22 | 203 | 0.05516 | 0.292 | |
| 35 | hsa03040_Spliceosome | 15 | 128 | 0.05894 | 0.2955 | |
| 36 | hsa04370_VEGF_signaling_pathway | 10 | 76 | 0.05911 | 0.2955 | |
| 37 | hsa04010_MAPK_signaling_pathway | 27 | 268 | 0.07657 | 0.3502 | |
| 38 | hsa04530_Tight_junction | 15 | 133 | 0.07677 | 0.3502 | |
| 39 | hsa04912_GnRH_signaling_pathway | 12 | 101 | 0.07809 | 0.3502 | |
| 40 | hsa00564_Glycerophospholipid_metabolism | 10 | 80 | 0.07814 | 0.3502 | |
| 41 | hsa04720_Long.term_potentiation | 9 | 70 | 0.07977 | 0.3502 | |
| 42 | hsa03450_Non.homologous_end.joining | 3 | 14 | 0.08357 | 0.3582 | |
| 43 | hsa03060_Protein_export | 4 | 23 | 0.09081 | 0.3715 | |
| 44 | hsa03430_Mismatch_repair | 4 | 23 | 0.09081 | 0.3715 | |
| 45 | hsa00900_Terpenoid_backbone_biosynthesis | 3 | 15 | 0.09884 | 0.3954 | |
| 46 | hsa04120_Ubiquitin_mediated_proteolysis | 15 | 139 | 0.1024 | 0.4007 | |
| 47 | hsa04350_TGF.beta_signaling_pathway | 10 | 85 | 0.1067 | 0.4086 | |
| 48 | hsa03013_RNA_transport | 16 | 152 | 0.1107 | 0.415 | |
| 49 | hsa04914_Progesterone.mediated_oocyte_maturation | 10 | 87 | 0.1196 | 0.4393 | |
| 50 | hsa00562_Inositol_phosphate_metabolism | 7 | 57 | 0.1362 | 0.4904 | |
| 51 | hsa04972_Pancreatic_secretion | 11 | 101 | 0.1398 | 0.4933 | |
| 52 | hsa04974_Protein_digestion_and_absorption | 9 | 81 | 0.1564 | 0.5414 | |
| 53 | hsa04270_Vascular_smooth_muscle_contraction | 12 | 116 | 0.1653 | 0.5569 | |
| 54 | hsa00531_Glycosaminoglycan_degradation | 3 | 19 | 0.1691 | 0.5569 | |
| 55 | hsa00430_Taurine_and_hypotaurine_metabolism | 2 | 10 | 0.1714 | 0.5569 | |
| 56 | hsa03015_mRNA_surveillance_pathway | 9 | 83 | 0.1733 | 0.5569 | |
| 57 | hsa04660_T_cell_receptor_signaling_pathway | 11 | 108 | 0.1915 | 0.6016 | |
| 58 | hsa04971_Gastric_acid_secretion | 8 | 74 | 0.1938 | 0.6016 | |
| 59 | hsa04662_B_cell_receptor_signaling_pathway | 8 | 75 | 0.2037 | 0.6215 | |
| 60 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 5 | 42 | 0.2081 | 0.6243 | |
| 61 | hsa04970_Salivary_secretion | 9 | 89 | 0.2283 | 0.6737 | |
| 62 | hsa03410_Base_excision_repair | 4 | 34 | 0.2529 | 0.7341 | |
| 63 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 8 | 81 | 0.2668 | 0.7621 | |
| 64 | hsa00051_Fructose_and_mannose_metabolism | 4 | 36 | 0.2872 | 0.7832 | |
| 65 | hsa00270_Cysteine_and_methionine_metabolism | 4 | 36 | 0.2872 | 0.7832 | |
| 66 | hsa00563_Glycosylphosphatidylinositol.GPI..anchor_biosynthesis | 3 | 25 | 0.2915 | 0.7832 | |
| 67 | hsa04320_Dorso.ventral_axis_formation | 3 | 25 | 0.2915 | 0.7832 | |
| 68 | hsa00561_Glycerolipid_metabolism | 5 | 50 | 0.3254 | 0.8578 | |
| 69 | hsa00030_Pentose_phosphate_pathway | 3 | 27 | 0.334 | 0.8578 | |
| 70 | hsa04966_Collecting_duct_acid_secretion | 3 | 27 | 0.334 | 0.8578 | |
| 71 | hsa00970_Aminoacyl.tRNA_biosynthesis | 6 | 63 | 0.3395 | 0.8578 | |
| 72 | hsa00770_Pantothenate_and_CoA_biosynthesis | 2 | 16 | 0.3431 | 0.8578 | |
| 73 | hsa03440_Homologous_recombination | 3 | 28 | 0.3553 | 0.8605 | |
| 74 | hsa04020_Calcium_signaling_pathway | 15 | 177 | 0.3594 | 0.8605 | |
| 75 | hsa04260_Cardiac_muscle_contraction | 7 | 77 | 0.3624 | 0.8605 | |
| 76 | hsa04722_Neurotrophin_signaling_pathway | 11 | 127 | 0.3633 | 0.8605 | |
| 77 | hsa04670_Leukocyte_transendothelial_migration | 10 | 117 | 0.3896 | 0.8863 | |
| 78 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 14 | 168 | 0.3906 | 0.8863 | |
| 79 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 3 | 30 | 0.3974 | 0.8863 | |
| 80 | hsa00630_Glyoxylate_and_dicarboxylate_metabolism | 2 | 18 | 0.3988 | 0.8863 | |
| 81 | hsa04340_Hedgehog_signaling_pathway | 5 | 56 | 0.4173 | 0.9157 | |
| 82 | hsa02010_ABC_transporters | 4 | 44 | 0.4273 | 0.9157 | |
| 83 | hsa03320_PPAR_signaling_pathway | 6 | 70 | 0.4359 | 0.923 | |
| 84 | hsa01040_Biosynthesis_of_unsaturated_fatty_acids | 2 | 21 | 0.4779 | 1 | |
| 85 | hsa00532_Glycosaminoglycan_biosynthesis_._chondroitin_sulfate | 2 | 22 | 0.5028 | 1 | |
| 86 | hsa00565_Ether_lipid_metabolism | 3 | 36 | 0.5178 | 1 | |
| 87 | hsa04964_Proximal_tubule_bicarbonate_reclamation | 2 | 23 | 0.527 | 1 | |
| 88 | hsa04977_Vitamin_digestion_and_absorption | 2 | 24 | 0.5503 | 1 | |
| 89 | hsa04150_mTOR_signaling_pathway | 4 | 52 | 0.5594 | 1 | |
| 90 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 10 | 136 | 0.5817 | 1 | |
| 91 | hsa00330_Arginine_and_proline_metabolism | 4 | 54 | 0.5898 | 1 | |
| 92 | hsa00620_Pyruvate_metabolism | 3 | 40 | 0.5904 | 1 | |
| 93 | hsa04920_Adipocytokine_signaling_pathway | 5 | 68 | 0.5906 | 1 | |
| 94 | hsa04910_Insulin_signaling_pathway | 10 | 138 | 0.6005 | 1 | |
| 95 | hsa00052_Galactose_metabolism | 2 | 27 | 0.6153 | 1 | |
| 96 | hsa03018_RNA_degradation | 5 | 71 | 0.6294 | 1 | |
| 97 | hsa00340_Histidine_metabolism | 2 | 29 | 0.6545 | 1 | |
| 98 | hsa04973_Carbohydrate_digestion_and_absorption | 3 | 44 | 0.6556 | 1 | |
| 99 | hsa00590_Arachidonic_acid_metabolism | 4 | 59 | 0.6601 | 1 | |
| 100 | hsa04621_NOD.like_receptor_signaling_pathway | 4 | 59 | 0.6601 | 1 | |
| 101 | hsa03420_Nucleotide_excision_repair | 3 | 45 | 0.6707 | 1 | |
| 102 | hsa00020_Citrate_cycle_.TCA_cycle. | 2 | 30 | 0.6729 | 1 | |
| 103 | hsa04062_Chemokine_signaling_pathway | 13 | 189 | 0.677 | 1 | |
| 104 | hsa00250_Alanine._aspartate_and_glutamate_metabolism | 2 | 32 | 0.7072 | 1 | |
| 105 | hsa00260_Glycine._serine_and_threonine_metabolism | 2 | 32 | 0.7072 | 1 | |
| 106 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 3 | 48 | 0.713 | 1 | |
| 107 | hsa00510_N.Glycan_biosynthesis | 3 | 49 | 0.7261 | 1 | |
| 108 | hsa00010_Glycolysis_._Gluconeogenesis | 4 | 65 | 0.7331 | 1 | |
| 109 | hsa04140_Regulation_of_autophagy | 2 | 34 | 0.7385 | 1 | |
| 110 | hsa00480_Glutathione_metabolism | 3 | 50 | 0.7387 | 1 | |
| 111 | hsa04210_Apoptosis | 5 | 89 | 0.8113 | 1 | |
| 112 | hsa04514_Cell_adhesion_molecules_.CAMs. | 8 | 136 | 0.8144 | 1 | |
| 113 | hsa00600_Sphingolipid_metabolism | 2 | 40 | 0.8156 | 1 | |
| 114 | hsa00071_Fatty_acid_metabolism | 2 | 43 | 0.846 | 1 | |
| 115 | hsa00860_Porphyrin_and_chlorophyll_metabolism | 2 | 43 | 0.846 | 1 | |
| 116 | hsa04612_Antigen_processing_and_presentation | 4 | 78 | 0.8497 | 1 | |
| 117 | hsa00280_Valine._leucine_and_isoleucine_degradation | 2 | 44 | 0.8551 | 1 | |
| 118 | hsa00310_Lysine_degradation | 2 | 44 | 0.8551 | 1 | |
| 119 | hsa04146_Peroxisome | 4 | 79 | 0.8565 | 1 | |
| 120 | hsa00190_Oxidative_phosphorylation | 7 | 132 | 0.8783 | 1 | |
| 121 | hsa04610_Complement_and_coagulation_cascades | 3 | 69 | 0.9014 | 1 | |
| 122 | hsa04640_Hematopoietic_cell_lineage | 4 | 88 | 0.907 | 1 | |
| 123 | hsa04976_Bile_secretion | 3 | 71 | 0.9117 | 1 | |
| 124 | hsa00983_Drug_metabolism_._other_enzymes | 2 | 52 | 0.9118 | 1 | |
| 125 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.9316 | 1 | |
| 126 | hsa00140_Steroid_hormone_biosynthesis | 2 | 57 | 0.9358 | 1 | |
| 127 | hsa04630_Jak.STAT_signaling_pathway | 6 | 155 | 0.9796 | 1 | |
| 128 | hsa04380_Osteoclast_differentiation | 4 | 128 | 0.9893 | 1 | |
| 129 | hsa04620_Toll.like_receptor_signaling_pathway | 2 | 102 | 0.9969 | 1 | |
| 130 | hsa04740_Olfactory_transduction | 3 | 388 | 1 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SNHG1 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | HELLS | Sponge network | 2.013 | 0 | 2.623 | 0 | 0.703 |
| 2 | RP5-1074L1.4 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | HELLS | Sponge network | 2.302 | 0 | 2.623 | 0 | 0.638 |
| 3 | RP11-196G18.22 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 10 | HELLS | Sponge network | 2.705 | 0 | 2.623 | 0 | 0.597 |
| 4 | AP001469.9 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 11 | HELLS | Sponge network | 2.428 | 0 | 2.623 | 0 | 0.592 |
| 5 | RP1-228H13.5 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 10 | HELLS | Sponge network | 1.554 | 0 | 2.623 | 0 | 0.591 |
| 6 | KB-1572G7.2 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 10 | HELLS | Sponge network | 2.124 | 0 | 2.623 | 0 | 0.584 |
| 7 | RP11-1246C19.1 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 2.721 | 0 | 2.623 | 0 | 0.564 |
| 8 | GUSBP11 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 12 | HELLS | Sponge network | 2.066 | 0 | 2.623 | 0 | 0.541 |
| 9 | NPSR1-AS1 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | HELLS | Sponge network | 5.28 | 0 | 2.623 | 0 | 0.53 |
| 10 | CTC-459F4.3 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 10 | HELLS | Sponge network | 1.207 | 0 | 2.623 | 0 | 0.51 |
| 11 | RP5-1120P11.1 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | HELLS | Sponge network | 3.942 | 0 | 2.623 | 0 | 0.51 |
| 12 | TMCC1-AS1 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 2.298 | 0 | 2.623 | 0 | 0.51 |
| 13 | HCG18 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | HELLS | Sponge network | 1.42 | 0 | 2.623 | 0 | 0.5 |
| 14 | LINC00152 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 2.553 | 0 | 2.623 | 0 | 0.451 |
| 15 | MIR4435-1HG | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 2.541 | 0 | 2.623 | 0 | 0.441 |
| 16 | RP11-727A23.5 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 1.435 | 0 | 2.623 | 0 | 0.44 |
| 17 | RP11-540A21.2 | hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-193b-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-24-1-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | HELLS | Sponge network | 1.758 | 0 | 2.623 | 0 | 0.413 |