This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-106a-5p | AR | 1.77 | 0 | -0.44 | 0.04112 | mirMAP | -0.3 | 0 | NA | |
| 2 | hsa-miR-106b-5p | AR | 1.03 | 0 | -0.44 | 0.04112 | mirMAP | -0.24 | 0.00461 | NA | |
| 3 | hsa-miR-130a-3p | AR | -0.01 | 0.93453 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.48 | 0 | NA | |
| 4 | hsa-miR-135a-5p | AR | 1.06 | 0 | -0.44 | 0.04112 | miRNATAP | -0.37 | 0 | 26364608 | We also identified robust induction of miR-135a by androgen and strong direct binding of AR to the miR-135a locus; We further demonstrate that a negative feedback loop involving miR-135a can restore AR expression under androgen-deprivation conditions thus contributing to the upregulation of AR protein expression in castration-resistant PC |
| 5 | hsa-miR-141-3p | AR | 1.8 | 0 | -0.44 | 0.04112 | mirMAP | -0.27 | 0 | 26062412; 22314666 | miR 141 3p regulates the expression of androgen receptor by targeting its 3'UTR in prostate cancer LNCaP cells; After prostate cancer cell line LNCaP was transfected with miR-141-3p mimics expression levels of AR mRNA and protein in the LNCaP cells were detected by reverse transcription PCR and Western blotting respectively; The 3'untranslated regions 3'UTR of AR mRNA containing the binding site of miR-141-3p was amplified by PCR and inserted into pmiR-report vector a 3'downstream luciferase reporter gene; Transfection of miR-141-3p mimics decreased both mRNA and protein expression levels of AR in LNCaP cells; AR is a direct target gene of miR-141-3p;miR 141 modulates androgen receptor transcriptional activity in human prostate cancer cells through targeting the small heterodimer partner protein; Here we investigated the correlation of Shp expression with the cellular level of miR-141 and its effects on AR transcriptional activity in non-malignant and malignant human prostate epithelial cell lines; Phenethyl isothiocyanate a natural constituent of many edible cruciferous vegetables increased Shp expression downregulated miR-141 and inhibited AR transcriptional activity in LNCaP cells; Shp is a target for miR-141 and it is downregulated in cultured human PCa cells with the involvement of upregulation of miR-141 which promotes AR transcriptional activity |
| 6 | hsa-miR-146b-5p | AR | 1.7 | 0 | -0.44 | 0.04112 | mirMAP | -0.24 | 9.0E-5 | NA | |
| 7 | hsa-miR-148a-3p | AR | 1.85 | 0 | -0.44 | 0.04112 | mirMAP | -0.16 | 0.01852 | 27292025 | We found that androgen receptor is the most frequently targeted PCa oncogene and that miR-148a targets the largest number of known PCa drivers |
| 8 | hsa-miR-148b-5p | AR | 1.05 | 0 | -0.44 | 0.04112 | mirMAP | -0.17 | 0.00737 | NA | |
| 9 | hsa-miR-149-5p | AR | 0.65 | 4.0E-5 | -0.44 | 0.04112 | miRNATAP | -0.3 | 0 | NA | |
| 10 | hsa-miR-155-5p | AR | 0.66 | 0 | -0.44 | 0.04112 | mirMAP | -0.14 | 0.03211 | NA | |
| 11 | hsa-miR-15a-5p | AR | 0.92 | 0 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.52 | 0 | NA | |
| 12 | hsa-miR-16-5p | AR | 0.55 | 0 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.27 | 0.01254 | NA | |
| 13 | hsa-miR-17-5p | AR | 1.75 | 0 | -0.44 | 0.04112 | mirMAP | -0.34 | 0 | 23095762 | miR 17 5p targets the p300/CBP associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cells; Targeting of the 3'-untranslated region of PCAF mRNA by miR-17-5p caused translational suppression and RNA degradation and consequently modulation of AR transcriptional activity in PCa cells; Targeting of PCAF by miR-17-5p modulates AR transcriptional activity and cell growth in cultured PCa cells |
| 14 | hsa-miR-186-5p | AR | 0.57 | 0 | -0.44 | 0.04112 | mirMAP | -0.4 | 0.00064 | NA | |
| 15 | hsa-miR-19a-3p | AR | 1.32 | 0 | -0.44 | 0.04112 | mirMAP | -0.25 | 0 | NA | |
| 16 | hsa-miR-19b-3p | AR | 1.59 | 0 | -0.44 | 0.04112 | mirMAP | -0.41 | 0 | NA | |
| 17 | hsa-miR-20a-5p | AR | 1.86 | 0 | -0.44 | 0.04112 | mirMAP | -0.33 | 0 | NA | |
| 18 | hsa-miR-20b-5p | AR | 1.58 | 0 | -0.44 | 0.04112 | mirMAP | -0.18 | 9.0E-5 | NA | |
| 19 | hsa-miR-217 | AR | 0.65 | 0.00062 | -0.44 | 0.04112 | mirMAP | -0.17 | 0.00049 | NA | |
| 20 | hsa-miR-223-3p | AR | 0.48 | 2.0E-5 | -0.44 | 0.04112 | mirMAP | -0.21 | 0.01149 | NA | |
| 21 | hsa-miR-2355-5p | AR | 0.28 | 0.00411 | -0.44 | 0.04112 | MirTarget | -0.29 | 0.00235 | NA | |
| 22 | hsa-miR-29a-5p | AR | 0.46 | 0.00041 | -0.44 | 0.04112 | mirMAP | -0.28 | 6.0E-5 | 26052614 | A direct AR regulation of miR-22 miR-29a and miR-17-92 cluster along with their host genes was confirmed; Interestingly endogenous levels of miR-22 and miR-29a were found to be reduced in PCa cells expressing AR |
| 23 | hsa-miR-29b-2-5p | AR | 0.79 | 0 | -0.44 | 0.04112 | mirMAP | -0.31 | 0.0003 | NA | |
| 24 | hsa-miR-301a-3p | AR | 0.5 | 0.00026 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.26 | 9.0E-5 | 25940439 | Mechanism dissection revealed infiltrating pre-adipocytes might function through down-regulation of the androgen receptor AR via modulation of miR-301a and then increase PCa cell invasion via induction of TGF-β1/Smad/MMP9 signals |
| 25 | hsa-miR-3065-5p | AR | 1.05 | 0 | -0.44 | 0.04112 | miRNATAP | -0.11 | 0.02921 | NA | |
| 26 | hsa-miR-30a-5p | AR | 0.51 | 0 | -0.44 | 0.04112 | mirMAP | -0.41 | 0 | 27683042 | Results uncovered miR-30 family members as direct AR inhibitors |
| 27 | hsa-miR-30b-5p | AR | 1.17 | 0 | -0.44 | 0.04112 | mirMAP | -0.56 | 0 | 27683042 | Results uncovered miR-30 family members as direct AR inhibitors |
| 28 | hsa-miR-30c-5p | AR | 0.75 | 0 | -0.44 | 0.04112 | mirMAP | -0.84 | 0 | NA | |
| 29 | hsa-miR-320a | AR | 1 | 0 | -0.44 | 0.04112 | miRNATAP | -0.73 | 0 | 27216188 | Histone Deacetylase Inhibition in Prostate Cancer Triggers miR 320 Mediated Suppression of the Androgen Receptor; Among the upregulated miRNAs after OBP-801 treatment in the three prostate cancer cell lines miR-320a whose expression was significantly correlated with prognosis of prostate cancers P = 0.0185 was the most closely associated with AR expression; An miR-320a mimic suppressed AR protein expression together with growth suppression while anti-miR-320a oligonucleotide significantly abrogated the growth suppression by OBP-801 treatment; Our data demonstrated that OBP-801 effectively suppressed AR activity via epigenetic upregulation of miR-320a which resulted in tumor cell growth suppression of prostate cancers |
| 30 | hsa-miR-335-5p | AR | 0.96 | 0 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.23 | 0.00038 | NA | |
| 31 | hsa-miR-338-3p | AR | 0.29 | 0.08208 | -0.44 | 0.04112 | miRanda | -0.39 | 0 | NA | |
| 32 | hsa-miR-34a-5p | AR | 0.46 | 0 | -0.44 | 0.04112 | MirTarget | -0.72 | 0 | 24349627; 22347519; 21343391; 25920548; 25797256; 23145211 | In this study we found loss of miR-34a which targets AR in PCa tissue specimens especially in patients with higher Gleason grade tumors consistent with increased expression of AR; Most importantly BR-DIM intervention in PCa patients prior to radical prostatectomy showed reexpression of miR-34a which was consistent with decreased expression of AR PSA and Notch-1 in PCa tissue specimens; PCa cells treated with BR-DIM and 5-aza-dC resulted in the demethylation of miR-34a promoter concomitant with inhibition of AR and PSA expression in LNCaP and C4-2B cells; These results suggest for the first time epigenetic silencing of miR-34a in PCa which could be reversed by BR-DIM treatment and thus BR-DIM could be useful for the inactivation of AR in the treatment of PCa.This corrects the article on p;In this study we found loss of miR-34a which targets AR in PCa tissue specimens especially in patients with higher Gleason grade tumors consistent with increased expression of AR; Most importantly BR-DIM intervention in PCa patients prior to radical prostatectomy showed re-expression of miR-34a which was consistent with decreased expression of AR PSA and Notch-1 in PCa tissue specimens; PCa cells treated with BR-DIM and 5-aza-dC resulted in the demethylation of miR-34a promoter concomitant with inhibition of AR and PSA expression in LNCaP and C4-2B cells;In particular analysis of clinical prostate cancers confirmed a negative correlation of miR-34a and miR-34c expression with AR levels;To explore further the possible role of miRNAs in the AR pathway LNCaP cell line was treated with 5α-dihydrotestosterone and flutamide showing alteration in miRNAs expression especially miR-34a which was significantly underexpressed after treatment with high doses of 5α-dihydrotestosterone; Our data support a role for miRNAs especially miR-371 and miR-34a in the complex disarrangement of AR signaling pathway and in the behavior of PC;Repression of miR-34a a known AR-targeting miRNA contributes AR expression by XRN1;Inactivation of AR and Notch 1 signaling by miR 34a attenuates prostate cancer aggressiveness; We found that over-expression of miR-34a led to reduced expression of AR PSA and Notch-1; These findings suggest that the loss of miR-34a is directly linked with up-regulation of AR and Notch-1 both of which are highly expressed in PCa and thus finding innovative approaches by which miR-34a expression could be up-regulated will have a huge impact on the treatment of PCa especially for the treatment of mCRPC |
| 33 | hsa-miR-34c-5p | AR | 0.41 | 0.0147 | -0.44 | 0.04112 | MirTarget | -0.24 | 1.0E-5 | 21343391 | In particular analysis of clinical prostate cancers confirmed a negative correlation of miR-34a and miR-34c expression with AR levels |
| 34 | hsa-miR-361-5p | AR | 0.65 | 0 | -0.44 | 0.04112 | mirMAP | -0.52 | 6.0E-5 | NA | |
| 35 | hsa-miR-362-3p | AR | 0.33 | 0.03568 | -0.44 | 0.04112 | miRanda | -0.18 | 0.00211 | NA | |
| 36 | hsa-miR-374a-3p | AR | 0.61 | 0 | -0.44 | 0.04112 | mirMAP | -0.33 | 0.00096 | NA | |
| 37 | hsa-miR-452-3p | AR | -0.16 | 0.22858 | -0.44 | 0.04112 | mirMAP | -0.2 | 0.00422 | NA | |
| 38 | hsa-miR-454-3p | AR | 1.05 | 0 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.46 | 0 | NA | |
| 39 | hsa-miR-505-3p | AR | -0.01 | 0.89557 | -0.44 | 0.04112 | mirMAP | -0.29 | 0.00168 | NA | |
| 40 | hsa-miR-505-5p | AR | 0.39 | 0.0191 | -0.44 | 0.04112 | MirTarget | -0.14 | 0.01848 | NA | |
| 41 | hsa-miR-576-5p | AR | 0.96 | 0 | -0.44 | 0.04112 | mirMAP | -0.15 | 0.02787 | NA | |
| 42 | hsa-miR-582-3p | AR | 0.69 | 0 | -0.44 | 0.04112 | mirMAP | -0.35 | 0 | NA | |
| 43 | hsa-miR-590-3p | AR | 0.58 | 0 | -0.44 | 0.04112 | mirMAP; miRNATAP | -0.2 | 0.00719 | NA | |
| 44 | hsa-miR-660-5p | AR | 1.14 | 0 | -0.44 | 0.04112 | mirMAP | -0.36 | 0 | NA | |
| 45 | hsa-miR-664a-3p | AR | 0.77 | 0 | -0.44 | 0.04112 | mirMAP | -0.48 | 0 | NA | |
| 46 | hsa-miR-7-1-3p | AR | 1.46 | 0 | -0.44 | 0.04112 | mirMAP | -0.17 | 0.01062 | NA | |
| 47 | hsa-miR-9-5p | AR | 1.14 | 0 | -0.44 | 0.04112 | miRNATAP | -0.21 | 0 | NA | |
| 48 | hsa-miR-93-5p | AR | 2.03 | 0 | -0.44 | 0.04112 | mirMAP | -0.33 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 118 | 1784 | 2.459e-30 | 1.144e-26 |
| 2 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 118 | 1805 | 7.222e-30 | 1.68e-26 |
| 3 | POSITIVE REGULATION OF GENE EXPRESSION | 114 | 1733 | 5.296e-29 | 8.214e-26 |
| 4 | RESPONSE TO ENDOGENOUS STIMULUS | 101 | 1450 | 2.013e-27 | 2.342e-24 |
| 5 | REGULATION OF CELL DEATH | 101 | 1472 | 6.653e-27 | 6.191e-24 |
| 6 | CELLULAR RESPONSE TO STRESS | 102 | 1565 | 2.031e-25 | 1.575e-22 |
| 7 | REGULATION OF CELL PROLIFERATION | 98 | 1496 | 1.565e-24 | 1.04e-21 |
| 8 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 98 | 1517 | 4.451e-24 | 2.589e-21 |
| 9 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 109 | 1848 | 1.022e-23 | 5.281e-21 |
| 10 | NEGATIVE REGULATION OF GENE EXPRESSION | 96 | 1493 | 2.054e-23 | 9.555e-21 |
| 11 | RESPONSE TO HORMONE | 72 | 893 | 5.239e-23 | 2.031e-20 |
| 12 | REGULATION OF PROTEIN MODIFICATION PROCESS | 103 | 1710 | 5.104e-23 | 2.031e-20 |
| 13 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 75 | 1004 | 5.76e-22 | 2.062e-19 |
| 14 | NEGATIVE REGULATION OF CELL DEATH | 69 | 872 | 1.362e-21 | 4.528e-19 |
| 15 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 92 | 1492 | 3.855e-21 | 1.196e-18 |
| 16 | REGULATION OF RESPONSE TO STRESS | 89 | 1468 | 6.049e-20 | 1.759e-17 |
| 17 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 68 | 917 | 9.52e-20 | 2.606e-17 |
| 18 | RESPONSE TO ABIOTIC STIMULUS | 71 | 1024 | 5.4e-19 | 1.396e-16 |
| 19 | REGULATION OF BINDING | 37 | 283 | 6.542e-19 | 1.602e-16 |
| 20 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 84 | 1381 | 6.899e-19 | 1.605e-16 |
| 21 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 73 | 1087 | 9.467e-19 | 2.098e-16 |
| 22 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 69 | 1008 | 3.61e-18 | 7.635e-16 |
| 23 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 57 | 720 | 6.529e-18 | 1.321e-15 |
| 24 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 72 | 1152 | 8.241e-17 | 1.598e-14 |
| 25 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 88 | 1618 | 9.626e-17 | 1.792e-14 |
| 26 | REGULATION OF CELL CYCLE | 64 | 949 | 1.461e-16 | 2.615e-14 |
| 27 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 52 | 662 | 2.934e-16 | 4.876e-14 |
| 28 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 52 | 662 | 2.934e-16 | 4.876e-14 |
| 29 | NEGATIVE REGULATION OF CELL PROLIFERATION | 51 | 643 | 3.917e-16 | 6.284e-14 |
| 30 | REGULATION OF CELLULAR RESPONSE TO STRESS | 52 | 691 | 1.723e-15 | 2.672e-13 |
| 31 | CELL CYCLE | 75 | 1316 | 2.477e-15 | 3.717e-13 |
| 32 | RESPONSE TO STEROID HORMONE | 43 | 497 | 4.592e-15 | 6.677e-13 |
| 33 | RESPONSE TO LIPID | 59 | 888 | 5.059e-15 | 7.134e-13 |
| 34 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 68 | 1135 | 5.532e-15 | 7.57e-13 |
| 35 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 48 | 616 | 6.133e-15 | 8.153e-13 |
| 36 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 94 | 1929 | 7.068e-15 | 9.136e-13 |
| 37 | CELLULAR RESPONSE TO HORMONE STIMULUS | 45 | 552 | 9.027e-15 | 1.135e-12 |
| 38 | POSITIVE REGULATION OF CELL DEATH | 47 | 605 | 1.347e-14 | 1.649e-12 |
| 39 | CHROMATIN ORGANIZATION | 49 | 663 | 2.415e-14 | 2.881e-12 |
| 40 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 52 | 740 | 2.745e-14 | 3.194e-12 |
| 41 | CHROMOSOME ORGANIZATION | 62 | 1009 | 3.365e-14 | 3.819e-12 |
| 42 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 45 | 573 | 3.479e-14 | 3.854e-12 |
| 43 | CELL DEVELOPMENT | 76 | 1426 | 5.331e-14 | 5.768e-12 |
| 44 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 83 | 1672 | 1.463e-13 | 1.547e-11 |
| 45 | REGULATION OF TRANSFERASE ACTIVITY | 58 | 946 | 2.759e-13 | 2.853e-11 |
| 46 | REGULATION OF ORGANELLE ORGANIZATION | 66 | 1178 | 3.383e-13 | 3.422e-11 |
| 47 | AGING | 29 | 264 | 4.287e-13 | 4.244e-11 |
| 48 | NEGATIVE REGULATION OF BINDING | 21 | 131 | 5.116e-13 | 4.959e-11 |
| 49 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 62 | 1079 | 6.567e-13 | 6.236e-11 |
| 50 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 81 | 1656 | 6.717e-13 | 6.251e-11 |
| 51 | CELL CYCLE PROCESS | 62 | 1081 | 7.118e-13 | 6.494e-11 |
| 52 | CELL DEATH | 59 | 1001 | 8.879e-13 | 7.945e-11 |
| 53 | INTRACELLULAR SIGNAL TRANSDUCTION | 78 | 1572 | 9.56e-13 | 8.393e-11 |
| 54 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 38 | 465 | 1.145e-12 | 9.862e-11 |
| 55 | REGULATION OF PROTEOLYSIS | 48 | 711 | 1.27e-12 | 1.074e-10 |
| 56 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 58 | 983 | 1.36e-12 | 1.13e-10 |
| 57 | COVALENT CHROMATIN MODIFICATION | 32 | 345 | 2.67e-12 | 2.179e-10 |
| 58 | RESPONSE TO INORGANIC SUBSTANCE | 38 | 479 | 2.828e-12 | 2.268e-10 |
| 59 | CELLULAR RESPONSE TO LIPID | 37 | 457 | 3.04e-12 | 2.398e-10 |
| 60 | REGULATION OF CATABOLIC PROCESS | 48 | 731 | 3.435e-12 | 2.664e-10 |
| 61 | REGULATION OF CELL DIFFERENTIATION | 74 | 1492 | 4.21e-12 | 3.211e-10 |
| 62 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 46 | 684 | 4.435e-12 | 3.328e-10 |
| 63 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 25 | 218 | 7.088e-12 | 5.235e-10 |
| 64 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 32 | 363 | 1.035e-11 | 7.527e-10 |
| 65 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 58 | 1036 | 1.148e-11 | 8.094e-10 |
| 66 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 58 | 1036 | 1.148e-11 | 8.094e-10 |
| 67 | NEGATIVE REGULATION OF CELL CYCLE | 35 | 433 | 1.265e-11 | 8.788e-10 |
| 68 | DNA REPAIR | 37 | 480 | 1.293e-11 | 8.844e-10 |
| 69 | POSITIVE REGULATION OF CELL PROLIFERATION | 50 | 814 | 1.343e-11 | 9.057e-10 |
| 70 | RESPONSE TO EXTERNAL STIMULUS | 82 | 1821 | 3.365e-11 | 2.237e-09 |
| 71 | POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 23 | 197 | 3.448e-11 | 2.26e-09 |
| 72 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 81 | 1791 | 3.682e-11 | 2.379e-09 |
| 73 | POSITIVE REGULATION OF CELL COMMUNICATION | 73 | 1532 | 3.942e-11 | 2.512e-09 |
| 74 | DNA METABOLIC PROCESS | 47 | 758 | 4.239e-11 | 2.665e-09 |
| 75 | RESPONSE TO ALCOHOL | 31 | 362 | 4.611e-11 | 2.861e-09 |
| 76 | REGULATION OF PROTEIN BINDING | 21 | 168 | 6.843e-11 | 4.135e-09 |
| 77 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 21 | 168 | 6.843e-11 | 4.135e-09 |
| 78 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 48 | 799 | 7.68e-11 | 4.581e-09 |
| 79 | REGULATION OF PROTEIN CATABOLIC PROCESS | 32 | 393 | 8.213e-11 | 4.837e-09 |
| 80 | NEGATIVE REGULATION OF PROTEIN BINDING | 15 | 79 | 9.29e-11 | 5.382e-09 |
| 81 | CHROMATIN MODIFICATION | 38 | 539 | 9.369e-11 | 5.382e-09 |
| 82 | NEGATIVE REGULATION OF PHOSPHORYLATION | 33 | 422 | 1.194e-10 | 6.775e-09 |
| 83 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 32 | 406 | 1.888e-10 | 1.058e-08 |
| 84 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 67 | 1395 | 2.069e-10 | 1.146e-08 |
| 85 | REGULATION OF GROWTH | 41 | 633 | 2.2e-10 | 1.204e-08 |
| 86 | RESPONSE TO RADIATION | 32 | 413 | 2.912e-10 | 1.576e-08 |
| 87 | REGULATION OF KINASE ACTIVITY | 46 | 776 | 3.048e-10 | 1.627e-08 |
| 88 | REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 26 | 280 | 3.078e-10 | 1.627e-08 |
| 89 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 44 | 724 | 3.509e-10 | 1.835e-08 |
| 90 | CELL PROLIFERATION | 42 | 672 | 3.901e-10 | 2.017e-08 |
| 91 | CELLULAR CATABOLIC PROCESS | 64 | 1322 | 4.076e-10 | 2.084e-08 |
| 92 | REGULATION OF MITOTIC CELL CYCLE | 34 | 468 | 4.37e-10 | 2.21e-08 |
| 93 | APOPTOTIC SIGNALING PATHWAY | 26 | 289 | 6.115e-10 | 3.059e-08 |
| 94 | REGULATION OF PROTEIN ACETYLATION | 13 | 64 | 6.966e-10 | 3.412e-08 |
| 95 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 21 | 190 | 6.924e-10 | 3.412e-08 |
| 96 | RESPONSE TO DRUG | 32 | 431 | 8.485e-10 | 4.113e-08 |
| 97 | NEGATIVE REGULATION OF CELL COMMUNICATION | 59 | 1192 | 9.458e-10 | 4.537e-08 |
| 98 | RESPONSE TO WOUNDING | 37 | 563 | 1.18e-09 | 5.603e-08 |
| 99 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 39 | 616 | 1.192e-09 | 5.603e-08 |
| 100 | STEROID HORMONE MEDIATED SIGNALING PATHWAY | 17 | 125 | 1.218e-09 | 5.665e-08 |
| 101 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 64 | 1360 | 1.273e-09 | 5.865e-08 |
| 102 | REGULATION OF PROTEIN STABILITY | 22 | 221 | 1.998e-09 | 9.112e-08 |
| 103 | APOPTOTIC MITOCHONDRIAL CHANGES | 12 | 57 | 2.053e-09 | 9.276e-08 |
| 104 | REGULATION OF PROTEIN COMPLEX ASSEMBLY | 29 | 375 | 2.133e-09 | 9.474e-08 |
| 105 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 38 | 602 | 2.138e-09 | 9.474e-08 |
| 106 | POSITIVE REGULATION OF CELL CYCLE | 27 | 332 | 2.631e-09 | 1.155e-07 |
| 107 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 41 | 689 | 2.697e-09 | 1.173e-07 |
| 108 | TISSUE DEVELOPMENT | 68 | 1518 | 2.755e-09 | 1.187e-07 |
| 109 | RESPONSE TO METAL ION | 27 | 333 | 2.808e-09 | 1.199e-07 |
| 110 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 56 | 1142 | 3.727e-09 | 1.576e-07 |
| 111 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 18 | 152 | 3.838e-09 | 1.594e-07 |
| 112 | REGULATION OF CHROMATIN ORGANIZATION | 18 | 152 | 3.838e-09 | 1.594e-07 |
| 113 | REGULATION OF CYTOKINE PRODUCTION | 36 | 563 | 4.128e-09 | 1.693e-07 |
| 114 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 19 | 171 | 4.147e-09 | 1.693e-07 |
| 115 | REGULATION OF IMMUNE SYSTEM PROCESS | 64 | 1403 | 4.33e-09 | 1.737e-07 |
| 116 | POSITIVE REGULATION OF PROTEOLYSIS | 28 | 363 | 4.307e-09 | 1.737e-07 |
| 117 | RESPONSE TO REACTIVE OXYGEN SPECIES | 20 | 191 | 4.56e-09 | 1.813e-07 |
| 118 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 47 | 876 | 4.836e-09 | 1.891e-07 |
| 119 | REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 24 | 274 | 4.81e-09 | 1.891e-07 |
| 120 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 35 | 541 | 5.076e-09 | 1.952e-07 |
| 121 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 35 | 541 | 5.076e-09 | 1.952e-07 |
| 122 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 43 | 767 | 6.404e-09 | 2.423e-07 |
| 123 | REGULATION OF CHROMOSOME ORGANIZATION | 24 | 278 | 6.399e-09 | 2.423e-07 |
| 124 | POSITIVE REGULATION OF CATABOLIC PROCESS | 29 | 395 | 6.919e-09 | 2.575e-07 |
| 125 | GLAND DEVELOPMENT | 29 | 395 | 6.919e-09 | 2.575e-07 |
| 126 | HORMONE MEDIATED SIGNALING PATHWAY | 18 | 158 | 7.15e-09 | 2.619e-07 |
| 127 | POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 20 | 196 | 7.131e-09 | 2.619e-07 |
| 128 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 44 | 801 | 7.762e-09 | 2.822e-07 |
| 129 | REGULATION OF CELL CYCLE ARREST | 15 | 108 | 8.629e-09 | 3.113e-07 |
| 130 | RESPONSE TO OXIDATIVE STRESS | 27 | 352 | 9.226e-09 | 3.302e-07 |
| 131 | REGULATION OF DNA BINDING | 14 | 93 | 9.389e-09 | 3.335e-07 |
| 132 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 23 | 263 | 1.042e-08 | 3.672e-07 |
| 133 | REGULATION OF CELL CYCLE PROCESS | 35 | 558 | 1.117e-08 | 3.908e-07 |
| 134 | MACROMOLECULE DEACYLATION | 12 | 67 | 1.438e-08 | 4.921e-07 |
| 135 | GLOBAL GENOME NUCLEOTIDE EXCISION REPAIR | 9 | 32 | 1.418e-08 | 4.921e-07 |
| 136 | CELL AGING | 12 | 67 | 1.438e-08 | 4.921e-07 |
| 137 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 66 | 1518 | 1.579e-08 | 5.364e-07 |
| 138 | REGULATION OF IMMUNE RESPONSE | 45 | 858 | 2.082e-08 | 7.019e-07 |
| 139 | RESPONSE TO EXTRACELLULAR STIMULUS | 30 | 441 | 2.154e-08 | 7.211e-07 |
| 140 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION | 20 | 211 | 2.509e-08 | 8.338e-07 |
| 141 | RNA PROCESSING | 44 | 835 | 2.639e-08 | 8.709e-07 |
| 142 | MACROMOLECULE CATABOLIC PROCESS | 47 | 926 | 2.691e-08 | 8.819e-07 |
| 143 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 43 | 823 | 4.893e-08 | 1.592e-06 |
| 144 | CATABOLIC PROCESS | 72 | 1773 | 5.138e-08 | 1.66e-06 |
| 145 | REPRODUCTIVE SYSTEM DEVELOPMENT | 28 | 408 | 5.35e-08 | 1.717e-06 |
| 146 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 19 | 200 | 5.42e-08 | 1.727e-06 |
| 147 | REGULATION OF PROTEIN LOCALIZATION | 47 | 950 | 5.82e-08 | 1.842e-06 |
| 148 | NEUROGENESIS | 61 | 1402 | 5.869e-08 | 1.845e-06 |
| 149 | REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 18 | 181 | 6.039e-08 | 1.886e-06 |
| 150 | HOMEOSTATIC PROCESS | 59 | 1337 | 6.123e-08 | 1.899e-06 |
| 151 | REGULATION OF CELLULAR LOCALIZATION | 57 | 1277 | 7.187e-08 | 2.2e-06 |
| 152 | RESPONSE TO UV | 15 | 126 | 7.171e-08 | 2.2e-06 |
| 153 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 16 | 145 | 7.74e-08 | 2.339e-06 |
| 154 | REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 16 | 145 | 7.74e-08 | 2.339e-06 |
| 155 | PEPTIDYL AMINO ACID MODIFICATION | 43 | 841 | 8.979e-08 | 2.696e-06 |
| 156 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 13 | 96 | 1.16e-07 | 3.459e-06 |
| 157 | POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 18 | 192 | 1.487e-07 | 4.378e-06 |
| 158 | POSITIVE REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 13 | 98 | 1.486e-07 | 4.378e-06 |
| 159 | MITOTIC CELL CYCLE | 40 | 766 | 1.501e-07 | 4.391e-06 |
| 160 | RESPONSE TO NITROGEN COMPOUND | 43 | 859 | 1.614e-07 | 4.664e-06 |
| 161 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 20 | 236 | 1.606e-07 | 4.664e-06 |
| 162 | STEROID METABOLIC PROCESS | 20 | 237 | 1.72e-07 | 4.941e-06 |
| 163 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 48 | 1021 | 1.92e-07 | 5.481e-06 |
| 164 | RESPONSE TO CYTOKINE | 38 | 714 | 1.94e-07 | 5.505e-06 |
| 165 | OXIDATION REDUCTION PROCESS | 44 | 898 | 2.086e-07 | 5.882e-06 |
| 166 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 21 | 263 | 2.153e-07 | 6.035e-06 |
| 167 | CELLULAR RESPONSE TO RADIATION | 15 | 137 | 2.189e-07 | 6.1e-06 |
| 168 | RESPONSE TO TOXIC SUBSTANCE | 20 | 241 | 2.257e-07 | 6.25e-06 |
| 169 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 21 | 264 | 2.294e-07 | 6.317e-06 |
| 170 | RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 7 | 22 | 2.335e-07 | 6.354e-06 |
| 171 | HISTONE H3 DEACETYLATION | 7 | 22 | 2.335e-07 | 6.354e-06 |
| 172 | MITOTIC CELL CYCLE CHECKPOINT | 15 | 139 | 2.65e-07 | 7.168e-06 |
| 173 | INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 11 | 71 | 2.725e-07 | 7.329e-06 |
| 174 | PROTEIN CATABOLIC PROCESS | 33 | 579 | 2.775e-07 | 7.421e-06 |
| 175 | REPRODUCTION | 56 | 1297 | 2.824e-07 | 7.509e-06 |
| 176 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 25 | 365 | 2.908e-07 | 7.645e-06 |
| 177 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 32 | 552 | 2.905e-07 | 7.645e-06 |
| 178 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 34 | 609 | 2.943e-07 | 7.693e-06 |
| 179 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 16 | 162 | 3.608e-07 | 9.38e-06 |
| 180 | REGULATION OF BODY FLUID LEVELS | 30 | 506 | 4.308e-07 | 1.114e-05 |
| 181 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 41 | 829 | 4.373e-07 | 1.124e-05 |
| 182 | REGULATION OF GENE EXPRESSION EPIGENETIC | 19 | 229 | 4.545e-07 | 1.162e-05 |
| 183 | RESPONSE TO IONIZING RADIATION | 15 | 145 | 4.602e-07 | 1.164e-05 |
| 184 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 18 | 207 | 4.582e-07 | 1.164e-05 |
| 185 | EMBRYO DEVELOPMENT | 43 | 894 | 4.769e-07 | 1.199e-05 |
| 186 | MITOCHONDRION ORGANIZATION | 33 | 594 | 4.941e-07 | 1.236e-05 |
| 187 | POSITIVE REGULATION OF KINASE ACTIVITY | 29 | 482 | 4.989e-07 | 1.241e-05 |
| 188 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 14 | 127 | 5.106e-07 | 1.264e-05 |
| 189 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 24 | 351 | 5.196e-07 | 1.279e-05 |
| 190 | CELL ACTIVATION | 32 | 568 | 5.455e-07 | 1.336e-05 |
| 191 | MACROMOLECULAR COMPLEX ASSEMBLY | 58 | 1398 | 6.274e-07 | 1.528e-05 |
| 192 | RESPONSE TO NUTRIENT | 17 | 191 | 6.891e-07 | 1.67e-05 |
| 193 | NEGATIVE REGULATION OF GENE EXPRESSION EPIGENETIC | 13 | 112 | 7.189e-07 | 1.733e-05 |
| 194 | REGULATION OF NEURON APOPTOTIC PROCESS | 17 | 192 | 7.416e-07 | 1.779e-05 |
| 195 | PROTEIN STABILIZATION | 14 | 131 | 7.474e-07 | 1.783e-05 |
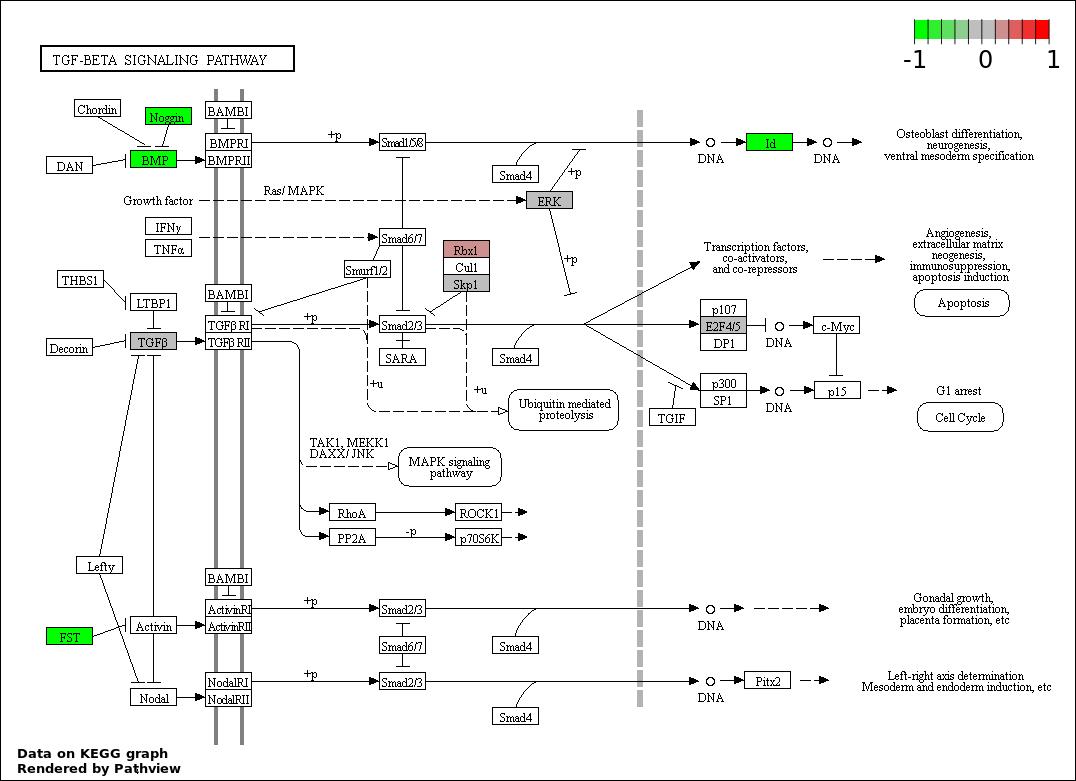
| 196 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 12 | 95 | 7.689e-07 | 1.825e-05 |
| 197 | REGULATION OF WNT SIGNALING PATHWAY | 22 | 310 | 8.361e-07 | 1.955e-05 |
| 198 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 39 | 788 | 8.341e-07 | 1.955e-05 |
| 199 | CIRCULATORY SYSTEM DEVELOPMENT | 39 | 788 | 8.341e-07 | 1.955e-05 |
| 200 | CELL CYCLE CHECKPOINT | 17 | 194 | 8.576e-07 | 1.995e-05 |
| 201 | REGULATION OF DEFENSE RESPONSE | 38 | 759 | 8.794e-07 | 2.036e-05 |
| 202 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 15 | 153 | 9.198e-07 | 2.119e-05 |
| 203 | MRNA METABOLIC PROCESS | 33 | 611 | 9.249e-07 | 2.12e-05 |
| 204 | WOUND HEALING | 28 | 470 | 9.579e-07 | 2.174e-05 |
| 205 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 28 | 470 | 9.579e-07 | 2.174e-05 |
| 206 | REGULATION OF MITOCHONDRION ORGANIZATION | 18 | 218 | 9.787e-07 | 2.211e-05 |
| 207 | CELL CYCLE ARREST | 15 | 154 | 9.996e-07 | 2.247e-05 |
| 208 | POSITIVE REGULATION OF CYTOSKELETON ORGANIZATION | 16 | 175 | 1.024e-06 | 2.281e-05 |
| 209 | HOMEOSTASIS OF NUMBER OF CELLS | 16 | 175 | 1.024e-06 | 2.281e-05 |
| 210 | REGULATION OF ORGAN MORPHOGENESIS | 19 | 242 | 1.053e-06 | 2.333e-05 |
| 211 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 11 | 81 | 1.068e-06 | 2.354e-05 |
| 212 | REGULATION OF DNA METABOLIC PROCESS | 23 | 340 | 1.081e-06 | 2.372e-05 |
| 213 | RNA SPLICING VIA TRANSESTERIFICATION REACTIONS | 20 | 267 | 1.148e-06 | 2.508e-05 |
| 214 | RESPONSE TO ETHANOL | 14 | 136 | 1.179e-06 | 2.552e-05 |
| 215 | RESPONSE TO GROWTH FACTOR | 28 | 475 | 1.179e-06 | 2.552e-05 |
| 216 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 27 | 448 | 1.204e-06 | 2.582e-05 |
| 217 | GENERATION OF PRECURSOR METABOLITES AND ENERGY | 21 | 292 | 1.2e-06 | 2.582e-05 |
| 218 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 17 | 199 | 1.223e-06 | 2.61e-05 |
| 219 | BASE EXCISION REPAIR | 8 | 39 | 1.298e-06 | 2.757e-05 |
| 220 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 12 | 100 | 1.344e-06 | 2.842e-05 |
| 221 | PHOSPHORYLATION | 52 | 1228 | 1.404e-06 | 2.956e-05 |
| 222 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 19 | 247 | 1.431e-06 | 3e-05 |
| 223 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 24 | 372 | 1.461e-06 | 3.047e-05 |
| 224 | REGULATION OF CELL CYCLE PHASE TRANSITION | 22 | 321 | 1.494e-06 | 3.104e-05 |
| 225 | REGULATION OF PROTEIN KINASE B SIGNALING | 13 | 121 | 1.748e-06 | 3.616e-05 |
| 226 | REGULATION OF MAPK CASCADE | 34 | 660 | 1.814e-06 | 3.734e-05 |
| 227 | NEGATIVE REGULATION OF TRANSPORT | 27 | 458 | 1.832e-06 | 3.756e-05 |
| 228 | NEGATIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 15 | 162 | 1.899e-06 | 3.876e-05 |
| 229 | REGULATION OF TRANSPORT | 68 | 1804 | 1.922e-06 | 3.906e-05 |
| 230 | EPITHELIUM DEVELOPMENT | 43 | 945 | 2.044e-06 | 4.134e-05 |
| 231 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 16 | 185 | 2.138e-06 | 4.307e-05 |
| 232 | CELL CYCLE PHASE TRANSITION | 19 | 255 | 2.299e-06 | 4.61e-05 |
| 233 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 14 | 144 | 2.343e-06 | 4.68e-05 |
| 234 | RESPONSE TO LIGHT STIMULUS | 20 | 280 | 2.391e-06 | 4.755e-05 |
| 235 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 26 | 437 | 2.409e-06 | 4.77e-05 |
| 236 | MYELOID CELL HOMEOSTASIS | 11 | 88 | 2.469e-06 | 4.867e-05 |
| 237 | EPITHELIAL CELL DIFFERENTIATION | 28 | 495 | 2.624e-06 | 5.153e-05 |
| 238 | PROTEIN LOCALIZATION TO ORGANELLE | 30 | 556 | 2.987e-06 | 5.84e-05 |
| 239 | POSITIVE REGULATION OF BINDING | 13 | 127 | 3.022e-06 | 5.884e-05 |
| 240 | REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION | 7 | 31 | 3.054e-06 | 5.921e-05 |
| 241 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 20 | 285 | 3.132e-06 | 6.047e-05 |
| 242 | RESPONSE TO OXYGEN LEVELS | 21 | 311 | 3.269e-06 | 6.285e-05 |
| 243 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 17 | 214 | 3.314e-06 | 6.346e-05 |
| 244 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 8 | 44 | 3.408e-06 | 6.5e-05 |
| 245 | MRNA TRANSCRIPTION | 6 | 21 | 3.581e-06 | 6.801e-05 |
| 246 | REGULATION OF HOMEOSTATIC PROCESS | 26 | 447 | 3.636e-06 | 6.877e-05 |
| 247 | POSITIVE REGULATION OF IMMUNE RESPONSE | 30 | 563 | 3.833e-06 | 7.221e-05 |
| 248 | REGULATION OF LIGASE ACTIVITY | 13 | 130 | 3.925e-06 | 7.316e-05 |
| 249 | RNA SPLICING | 23 | 367 | 3.931e-06 | 7.316e-05 |
| 250 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 40 | 872 | 3.907e-06 | 7.316e-05 |
| 251 | GLIAL CELL DEVELOPMENT | 10 | 76 | 4.426e-06 | 8.206e-05 |
| 252 | REGULATION OF RNA POLYMERASE II TRANSCRIPTIONAL PREINITIATION COMPLEX ASSEMBLY | 5 | 13 | 4.625e-06 | 8.539e-05 |
| 253 | NEGATIVE REGULATION OF WNT SIGNALING PATHWAY | 16 | 197 | 4.839e-06 | 8.899e-05 |
| 254 | NUCLEOTIDE EXCISION REPAIR | 12 | 113 | 4.937e-06 | 9.044e-05 |
| 255 | STEROID BIOSYNTHETIC PROCESS | 12 | 114 | 5.414e-06 | 9.879e-05 |
| 256 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 19 | 271 | 5.59e-06 | 0.0001016 |
| 257 | MRNA PROCESSING | 25 | 432 | 6.116e-06 | 0.0001107 |
| 258 | NUCLEOTIDE EXCISION REPAIR DNA DAMAGE RECOGNITION | 6 | 23 | 6.427e-06 | 0.0001155 |
| 259 | REGULATION OF TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 6 | 23 | 6.427e-06 | 0.0001155 |
| 260 | EMBRYONIC ORGAN DEVELOPMENT | 24 | 406 | 6.574e-06 | 0.0001176 |
| 261 | DNA TEMPLATED TRANSCRIPTION INITIATION | 16 | 202 | 6.669e-06 | 0.0001189 |
| 262 | SMALL MOLECULE METABOLIC PROCESS | 65 | 1767 | 7.483e-06 | 0.0001329 |
| 263 | REGULATION OF NEURON DEATH | 18 | 252 | 7.62e-06 | 0.0001348 |
| 264 | REGULATION OF FIBROBLAST PROLIFERATION | 10 | 81 | 7.929e-06 | 0.0001392 |
| 265 | DNA GEOMETRIC CHANGE | 10 | 81 | 7.929e-06 | 0.0001392 |
| 266 | NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 13 | 139 | 8.228e-06 | 0.0001439 |
| 267 | POSITIVE REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION | 6 | 24 | 8.417e-06 | 0.0001467 |
| 268 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 27 | 498 | 8.616e-06 | 0.0001496 |
| 269 | REGENERATION | 14 | 161 | 8.667e-06 | 0.0001499 |
| 270 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 36 | 8.85e-06 | 0.0001525 |
| 271 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 15 | 184 | 9.131e-06 | 0.0001568 |
| 272 | REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 11 | 101 | 9.587e-06 | 0.000164 |
| 273 | ORGAN MORPHOGENESIS | 38 | 841 | 9.706e-06 | 0.0001654 |
| 274 | REGULATION OF INTRACELLULAR TRANSPORT | 31 | 621 | 9.801e-06 | 0.0001664 |
| 275 | CELLULAR RESPONSE TO UV | 9 | 66 | 9.91e-06 | 0.0001677 |
| 276 | CELLULAR RESPONSE TO BIOTIC STIMULUS | 14 | 163 | 9.989e-06 | 0.0001684 |
| 277 | MRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 5 | 15 | 1.042e-05 | 0.000175 |
| 278 | PROTEIN PHOSPHORYLATION | 41 | 944 | 1.097e-05 | 0.0001837 |
| 279 | REGULATION OF CELL GROWTH | 23 | 391 | 1.107e-05 | 0.0001847 |
| 280 | REGULATION OF DNA TEMPLATED TRANSCRIPTION IN RESPONSE TO STRESS | 9 | 67 | 1.124e-05 | 0.0001867 |
| 281 | IN UTERO EMBRYONIC DEVELOPMENT | 20 | 311 | 1.151e-05 | 0.0001899 |
| 282 | HEMOSTASIS | 20 | 311 | 1.151e-05 | 0.0001899 |
| 283 | POSITIVE REGULATION OF CELL CYCLE ARREST | 10 | 85 | 1.225e-05 | 0.0002015 |
| 284 | POSITIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 8 | 52 | 1.248e-05 | 0.0002045 |
| 285 | CELLULAR HORMONE METABOLIC PROCESS | 11 | 104 | 1.271e-05 | 0.0002076 |
| 286 | IMMUNE SYSTEM PROCESS | 70 | 1984 | 1.299e-05 | 0.0002114 |
| 287 | REGULATION OF NECROTIC CELL DEATH | 6 | 26 | 1.389e-05 | 0.0002252 |
| 288 | RESPONSE TO ESTRADIOL | 13 | 146 | 1.403e-05 | 0.0002265 |
| 289 | LYMPHOCYTE ACTIVATION | 21 | 342 | 1.407e-05 | 0.0002265 |
| 290 | PROTEIN LOCALIZATION | 65 | 1805 | 1.475e-05 | 0.0002367 |
| 291 | REGULATION OF INCLUSION BODY ASSEMBLY | 5 | 16 | 1.489e-05 | 0.0002381 |
| 292 | NEGATIVE REGULATION OF ENDOCYTOSIS | 7 | 39 | 1.544e-05 | 0.0002452 |
| 293 | NUCLEOTIDE EXCISION REPAIR DNA INCISION | 7 | 39 | 1.544e-05 | 0.0002452 |
| 294 | REGULATION OF MEMBRANE PERMEABILITY | 9 | 70 | 1.616e-05 | 0.0002558 |
| 295 | REGULATION OF PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 13 | 148 | 1.625e-05 | 0.0002562 |
| 296 | ENERGY DERIVATION BY OXIDATION OF ORGANIC COMPOUNDS | 16 | 217 | 1.642e-05 | 0.0002581 |
| 297 | RESPONSE TO ACID CHEMICAL | 20 | 319 | 1.665e-05 | 0.0002609 |
| 298 | NEGATIVE REGULATION OF NEURON DEATH | 14 | 171 | 1.724e-05 | 0.0002691 |
| 299 | MUSCLE STRUCTURE DEVELOPMENT | 24 | 432 | 1.838e-05 | 0.000286 |
| 300 | MULTICELLULAR ORGANISM REPRODUCTION | 35 | 768 | 1.845e-05 | 0.0002862 |
| 301 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 38 | 867 | 1.912e-05 | 0.0002956 |
| 302 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 16 | 220 | 1.946e-05 | 0.0002999 |
| 303 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 35 | 771 | 2.001e-05 | 0.0003046 |
| 304 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 20 | 323 | 1.993e-05 | 0.0003046 |
| 305 | REGULATION OF CELL DEVELOPMENT | 37 | 836 | 2.003e-05 | 0.0003046 |
| 306 | RESPONSE TO HYDROGEN PEROXIDE | 11 | 109 | 1.99e-05 | 0.0003046 |
| 307 | NEGATIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 10 | 90 | 2.04e-05 | 0.0003081 |
| 308 | SENSORY ORGAN DEVELOPMENT | 26 | 493 | 2.039e-05 | 0.0003081 |
| 309 | POSITIVE REGULATION OF TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 5 | 17 | 2.073e-05 | 0.0003112 |
| 310 | WNT SIGNALING PATHWAY | 21 | 351 | 2.073e-05 | 0.0003112 |
| 311 | HEAD DEVELOPMENT | 33 | 709 | 2.115e-05 | 0.0003164 |
| 312 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 13 | 152 | 2.161e-05 | 0.0003223 |
| 313 | POSITIVE REGULATION OF LIGASE ACTIVITY | 11 | 110 | 2.17e-05 | 0.0003226 |
| 314 | CHEMICAL HOMEOSTASIS | 38 | 874 | 2.281e-05 | 0.0003359 |
| 315 | G1 DNA DAMAGE CHECKPOINT | 9 | 73 | 2.281e-05 | 0.0003359 |
| 316 | NEURON DIFFERENTIATION | 38 | 874 | 2.281e-05 | 0.0003359 |
| 317 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 21 | 354 | 2.351e-05 | 0.0003451 |
| 318 | POSITIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 11 | 111 | 2.364e-05 | 0.0003458 |
| 319 | VASCULATURE DEVELOPMENT | 25 | 469 | 2.464e-05 | 0.0003594 |
| 320 | NEGATIVE REGULATION OF KINASE ACTIVITY | 17 | 250 | 2.565e-05 | 0.0003718 |
| 321 | POSITIVE REGULATION OF DNA BINDING | 7 | 42 | 2.564e-05 | 0.0003718 |
| 322 | REGULATION OF INNATE IMMUNE RESPONSE | 21 | 357 | 2.662e-05 | 0.0003847 |
| 323 | NCRNA METABOLIC PROCESS | 27 | 533 | 2.868e-05 | 0.0004131 |
| 324 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 16 | 228 | 3.016e-05 | 0.0004318 |
| 325 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 43 | 3.007e-05 | 0.0004318 |
| 326 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 12 | 135 | 3.043e-05 | 0.0004344 |
| 327 | RESPONSE TO BIOTIC STIMULUS | 38 | 886 | 3.068e-05 | 0.0004365 |
| 328 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE | 21 | 361 | 3.135e-05 | 0.0004447 |
| 329 | PROTEIN DNA COMPLEX SUBUNIT ORGANIZATION | 16 | 229 | 3.181e-05 | 0.0004499 |
| 330 | CHROMATIN SILENCING | 10 | 95 | 3.28e-05 | 0.0004625 |
| 331 | REGULATION OF LEUKOCYTE PROLIFERATION | 15 | 206 | 3.476e-05 | 0.0004857 |
| 332 | RESPONSE TO KETONE | 14 | 182 | 3.459e-05 | 0.0004857 |
| 333 | FC RECEPTOR SIGNALING PATHWAY | 15 | 206 | 3.476e-05 | 0.0004857 |
| 334 | NEGATIVE REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 7 | 44 | 3.512e-05 | 0.0004892 |
| 335 | NEGATIVE REGULATION OF CHROMOSOME ORGANIZATION | 10 | 96 | 3.594e-05 | 0.0004991 |
| 336 | REGULATION OF MYELOID CELL DIFFERENTIATION | 14 | 183 | 3.675e-05 | 0.0005089 |
| 337 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 56 | 1527 | 3.781e-05 | 0.0005211 |
| 338 | PLACENTA DEVELOPMENT | 12 | 138 | 3.785e-05 | 0.0005211 |
| 339 | REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 16 | 233 | 3.923e-05 | 0.0005385 |
| 340 | LYMPHOCYTE DIFFERENTIATION | 15 | 209 | 4.108e-05 | 0.0005622 |
| 341 | REGULATION OF ATPASE ACTIVITY | 8 | 61 | 4.138e-05 | 0.0005647 |
| 342 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 11 | 118 | 4.188e-05 | 0.000569 |
| 343 | MEMBRANE ORGANIZATION | 38 | 899 | 4.195e-05 | 0.000569 |
| 344 | SKELETAL SYSTEM DEVELOPMENT | 24 | 455 | 4.226e-05 | 0.0005716 |
| 345 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 10 | 98 | 4.298e-05 | 0.0005797 |
| 346 | RESPONSE TO MECHANICAL STIMULUS | 15 | 210 | 4.34e-05 | 0.0005837 |
| 347 | ACTIVATION OF IMMUNE RESPONSE | 23 | 427 | 4.424e-05 | 0.0005932 |
| 348 | ORGANONITROGEN COMPOUND METABOLIC PROCESS | 63 | 1796 | 4.476e-05 | 0.0005978 |
| 349 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 21 | 370 | 4.484e-05 | 0.0005978 |
| 350 | NEGATIVE REGULATION OF GROWTH | 16 | 236 | 4.576e-05 | 0.0006084 |
| 351 | REGULATION OF OXIDATIVE STRESS INDUCED CELL DEATH | 7 | 46 | 4.73e-05 | 0.000627 |
| 352 | HORMONE METABOLIC PROCESS | 13 | 164 | 4.805e-05 | 0.0006352 |
| 353 | NEGATIVE REGULATION OF LOCOMOTION | 17 | 263 | 4.864e-05 | 0.0006411 |
| 354 | REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 6 | 32 | 4.911e-05 | 0.0006437 |
| 355 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 6 | 32 | 4.911e-05 | 0.0006437 |
| 356 | NEGATIVE REGULATION OF CELL CYCLE ARREST | 5 | 20 | 4.93e-05 | 0.0006443 |
| 357 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 12 | 142 | 5.015e-05 | 0.0006537 |
| 358 | IMMUNE SYSTEM DEVELOPMENT | 28 | 582 | 5.078e-05 | 0.00066 |
| 359 | MITOTIC DNA INTEGRITY CHECKPOINT | 10 | 100 | 5.117e-05 | 0.0006632 |
| 360 | RESPONSE TO PEPTIDE | 22 | 404 | 5.503e-05 | 0.0007112 |
| 361 | PROTEIN LOCALIZATION TO MEMBRANE | 21 | 376 | 5.65e-05 | 0.0007283 |
| 362 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 12 | 144 | 5.75e-05 | 0.0007391 |
| 363 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 13 | 167 | 5.797e-05 | 0.000741 |
| 364 | VIRAL LATENCY | 4 | 11 | 5.787e-05 | 0.000741 |
| 365 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 6 | 33 | 5.896e-05 | 0.0007475 |
| 366 | POSITIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 8 | 64 | 5.886e-05 | 0.0007475 |
| 367 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 19 | 321 | 5.89e-05 | 0.0007475 |
| 368 | LIPID METABOLIC PROCESS | 45 | 1158 | 6.206e-05 | 0.0007847 |
| 369 | RESPONSE TO AMINE | 7 | 48 | 6.274e-05 | 0.0007912 |
| 370 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 33 | 750 | 6.357e-05 | 0.0007974 |
| 371 | NEGATIVE REGULATION OF PROTEIN ACETYLATION | 5 | 21 | 6.358e-05 | 0.0007974 |
| 372 | ORGAN REGENERATION | 9 | 83 | 6.414e-05 | 0.0008022 |
| 373 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 12 | 146 | 6.577e-05 | 0.0008182 |
| 374 | DNA INTEGRITY CHECKPOINT | 12 | 146 | 6.577e-05 | 0.0008182 |
| 375 | NEGATIVE REGULATION OF HOMEOSTATIC PROCESS | 11 | 124 | 6.61e-05 | 0.0008202 |
| 376 | RESPONSE TO ESTROGEN | 15 | 218 | 6.652e-05 | 0.000821 |
| 377 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 15 | 218 | 6.652e-05 | 0.000821 |
| 378 | CELLULAR MACROMOLECULE LOCALIZATION | 47 | 1234 | 6.866e-05 | 0.0008451 |
| 379 | CARTILAGE DEVELOPMENT | 12 | 147 | 7.027e-05 | 0.0008627 |
| 380 | RHYTHMIC PROCESS | 18 | 298 | 7.084e-05 | 0.0008674 |
| 381 | CARDIAC CHAMBER MORPHOGENESIS | 10 | 104 | 7.159e-05 | 0.000872 |
| 382 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 10 | 104 | 7.159e-05 | 0.000872 |
| 383 | HORMONE BIOSYNTHETIC PROCESS | 7 | 49 | 7.188e-05 | 0.0008733 |
| 384 | EYE DEVELOPMENT | 19 | 326 | 7.236e-05 | 0.0008768 |
| 385 | REGULATION OF CELL DIVISION | 17 | 272 | 7.38e-05 | 0.0008919 |
| 386 | GAMETE GENERATION | 28 | 595 | 7.431e-05 | 0.0008958 |
| 387 | TISSUE MORPHOGENESIS | 26 | 533 | 7.509e-05 | 0.0009028 |
| 388 | DNA CONFORMATION CHANGE | 17 | 273 | 7.72e-05 | 0.0009258 |
| 389 | POSITIVE REGULATION OF CHROMATIN MODIFICATION | 9 | 85 | 7.741e-05 | 0.0009259 |
| 390 | LEUKOCYTE ACTIVATION | 22 | 414 | 7.878e-05 | 0.0009399 |
| 391 | REGULATION OF ANDROGEN RECEPTOR SIGNALING PATHWAY | 5 | 22 | 8.086e-05 | 0.0009549 |
| 392 | RESPONSE TO THYROID HORMONE | 5 | 22 | 8.086e-05 | 0.0009549 |
| 393 | REGULATION OF RESPONSE TO INTERFERON GAMMA | 5 | 22 | 8.086e-05 | 0.0009549 |
| 394 | NUCLEOTIDE EXCISION REPAIR DNA DUPLEX UNWINDING | 5 | 22 | 8.086e-05 | 0.0009549 |
| 395 | POSITIVE REGULATION OF NEURON DEATH | 8 | 67 | 8.208e-05 | 0.0009645 |
| 396 | RESPONSE TO GAMMA RADIATION | 7 | 50 | 8.208e-05 | 0.0009645 |
| 397 | NEGATIVE REGULATION OF RESPONSE TO OXIDATIVE STRESS | 6 | 35 | 8.337e-05 | 0.0009747 |
| 398 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO OXIDATIVE STRESS | 6 | 35 | 8.337e-05 | 0.0009747 |
| 399 | REGULATION OF FAT CELL DIFFERENTIATION | 10 | 106 | 8.415e-05 | 0.0009789 |
| 400 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 10 | 106 | 8.415e-05 | 0.0009789 |
| 401 | POSITIVE REGULATION OF CHROMOSOME ORGANIZATION | 12 | 150 | 8.541e-05 | 0.0009911 |
| 402 | SINGLE ORGANISM CELLULAR LOCALIZATION | 37 | 898 | 8.971e-05 | 0.001038 |
| 403 | PROTEIN FOLDING | 15 | 224 | 9.034e-05 | 0.001043 |
| 404 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 20 | 360 | 9.089e-05 | 0.001047 |
| 405 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 7 | 51 | 9.343e-05 | 0.001073 |
| 406 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 12 | 152 | 9.7e-05 | 0.001112 |
| 407 | POSITIVE REGULATION OF TRANSPORT | 38 | 936 | 9.771e-05 | 0.001117 |
| 408 | POSITIVE REGULATION OF PROTEIN ACETYLATION | 6 | 36 | 9.827e-05 | 0.001118 |
| 409 | REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 6 | 36 | 9.827e-05 | 0.001118 |
| 410 | RESPONSE TO CORTICOSTEROID | 13 | 176 | 9.912e-05 | 0.001125 |
| 411 | CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM | 16 | 252 | 9.949e-05 | 0.001126 |
| 412 | REGULATION OF CYTOPLASMIC TRANSPORT | 24 | 481 | 0.0001003 | 0.001132 |
| 413 | LEUKOCYTE APOPTOTIC PROCESS | 5 | 23 | 0.0001015 | 0.001142 |
| 414 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 9 | 88 | 0.0001016 | 0.001142 |
| 415 | REGULATION OF PEPTIDASE ACTIVITY | 21 | 392 | 0.0001019 | 0.001142 |
| 416 | NEURON PROJECTION DEVELOPMENT | 26 | 545 | 0.0001076 | 0.001204 |
| 417 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 15 | 229 | 0.0001156 | 0.00129 |
| 418 | ACTIVATION OF INNATE IMMUNE RESPONSE | 14 | 204 | 0.0001189 | 0.001324 |
| 419 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 7 | 53 | 0.00012 | 0.001332 |
| 420 | REGULATION OF IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE | 4 | 13 | 0.0001212 | 0.001343 |
| 421 | ORGANIC CYCLIC COMPOUND CATABOLIC PROCESS | 22 | 427 | 0.0001231 | 0.001361 |
| 422 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 8 | 71 | 0.0001244 | 0.001365 |
| 423 | CELLULAR RESPONSE TO INORGANIC SUBSTANCE | 12 | 156 | 0.0001243 | 0.001365 |
| 424 | PROTEIN LOCALIZATION TO NUCLEUS | 12 | 156 | 0.0001243 | 0.001365 |
| 425 | NEGATIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 5 | 24 | 0.000126 | 0.00138 |
| 426 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 18 | 312 | 0.0001267 | 0.00138 |
| 427 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 66 | 1977 | 0.0001266 | 0.00138 |
| 428 | CELLULAR RESPONSE TO LIGHT STIMULUS | 9 | 91 | 0.0001318 | 0.001433 |
| 429 | ESTABLISHMENT OF LOCALIZATION IN CELL | 58 | 1676 | 0.000134 | 0.001453 |
| 430 | REGULATION OF HEMOPOIESIS | 18 | 314 | 0.0001372 | 0.001484 |
| 431 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 9 | 92 | 0.0001434 | 0.001549 |
| 432 | GLIAL CELL DIFFERENTIATION | 11 | 136 | 0.0001519 | 0.001636 |
| 433 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 51 | 1423 | 0.0001527 | 0.001641 |
| 434 | C21 STEROID HORMONE METABOLIC PROCESS | 5 | 25 | 0.0001548 | 0.00166 |
| 435 | NEURON DEVELOPMENT | 30 | 687 | 0.0001562 | 0.001667 |
| 436 | NEGATIVE REGULATION OF MITOCHONDRION ORGANIZATION | 6 | 39 | 0.000156 | 0.001667 |
| 437 | VIRAL LIFE CYCLE | 17 | 290 | 0.0001604 | 0.001707 |
| 438 | POSITIVE REGULATION OF MEIOTIC CELL CYCLE | 4 | 14 | 0.0001669 | 0.001761 |
| 439 | ANDROGEN BIOSYNTHETIC PROCESS | 4 | 14 | 0.0001669 | 0.001761 |
| 440 | PEPTIDYL ARGININE METHYLATION | 4 | 14 | 0.0001669 | 0.001761 |
| 441 | REGULATION OF HISTONE H3 K9 ACETYLATION | 4 | 14 | 0.0001669 | 0.001761 |
| 442 | EPITHELIAL CELL DEVELOPMENT | 13 | 186 | 0.0001724 | 0.001815 |
| 443 | LEUKOCYTE DIFFERENTIATION | 17 | 292 | 0.000174 | 0.001828 |
| 444 | ARTERY DEVELOPMENT | 8 | 75 | 0.0001833 | 0.001917 |
| 445 | REGULATION OF DNA REPAIR | 8 | 75 | 0.0001833 | 0.001917 |
| 446 | SEX DIFFERENTIATION | 16 | 266 | 0.0001855 | 0.001935 |
| 447 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 14 | 213 | 0.000187 | 0.001946 |
| 448 | REGULATION OF CYTOSKELETON ORGANIZATION | 24 | 502 | 0.0001908 | 0.001982 |
| 449 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 13 | 188 | 0.0001917 | 0.001984 |
| 450 | MAMMARY GLAND DEVELOPMENT | 10 | 117 | 0.0001919 | 0.001984 |
| 451 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 20 | 381 | 0.0001942 | 0.002003 |
| 452 | REGULATION OF HYDROLASE ACTIVITY | 48 | 1327 | 0.0001954 | 0.00201 |
| 453 | NON CANONICAL WNT SIGNALING PATHWAY | 11 | 140 | 0.0001961 | 0.00201 |
| 454 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 11 | 140 | 0.0001961 | 0.00201 |
| 455 | POSITIVE REGULATION OF CELL DEVELOPMENT | 23 | 472 | 0.0001967 | 0.002012 |
| 456 | MYELOID CELL DIFFERENTIATION | 13 | 189 | 0.0002019 | 0.002061 |
| 457 | NEGATIVE REGULATION OF FAT CELL DIFFERENTIATION | 6 | 41 | 0.0002075 | 0.002104 |
| 458 | CELLULAR RESPONSE TO ESTROGEN STIMULUS | 6 | 41 | 0.0002075 | 0.002104 |
| 459 | POSITIVE REGULATION OF ATPASE ACTIVITY | 6 | 41 | 0.0002075 | 0.002104 |
| 460 | DOUBLE STRAND BREAK REPAIR | 12 | 165 | 0.0002106 | 0.00213 |
| 461 | NUCLEAR TRANSPORT | 19 | 355 | 0.0002185 | 0.002205 |
| 462 | REGULATION OF SYSTEM PROCESS | 24 | 507 | 0.000221 | 0.002226 |
| 463 | CELLULAR RESPONSE TO CADMIUM ION | 4 | 15 | 0.0002238 | 0.002249 |
| 464 | PROSTAGLANDIN METABOLIC PROCESS | 5 | 27 | 0.0002272 | 0.002274 |
| 465 | PROSTANOID METABOLIC PROCESS | 5 | 27 | 0.0002272 | 0.002274 |
| 466 | CELLULAR RESPIRATION | 11 | 143 | 0.0002361 | 0.002353 |
| 467 | CELLULAR RESPONSE TO OXYGEN LEVELS | 11 | 143 | 0.0002361 | 0.002353 |
| 468 | REGULATION OF INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 7 | 59 | 0.0002381 | 0.002364 |
| 469 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 20 | 387 | 0.0002383 | 0.002364 |
| 470 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 27 | 606 | 0.0002416 | 0.002392 |
| 471 | NEGATIVE REGULATION OF PROTEOLYSIS | 18 | 329 | 0.0002437 | 0.002408 |
| 472 | CARDIAC CHAMBER DEVELOPMENT | 11 | 144 | 0.0002509 | 0.002464 |
| 473 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 9 | 99 | 0.000251 | 0.002464 |
| 474 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 9 | 99 | 0.000251 | 0.002464 |
| 475 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 15 | 246 | 0.0002528 | 0.002476 |
| 476 | CONNECTIVE TISSUE DEVELOPMENT | 13 | 194 | 0.0002608 | 0.002549 |
| 477 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 7 | 60 | 0.0002647 | 0.002577 |
| 478 | LEUKOCYTE HOMEOSTASIS | 7 | 60 | 0.0002647 | 0.002577 |
| 479 | MUSCLE TISSUE DEVELOPMENT | 16 | 275 | 0.0002701 | 0.00262 |
| 480 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 24 | 514 | 0.0002703 | 0.00262 |
| 481 | NECROTIC CELL DEATH | 5 | 28 | 0.0002718 | 0.002624 |
| 482 | POSITIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 5 | 28 | 0.0002718 | 0.002624 |
| 483 | REGULATION OF MUSCLE SYSTEM PROCESS | 13 | 195 | 0.0002742 | 0.002641 |
| 484 | REGULATION OF CELL ACTIVATION | 23 | 484 | 0.0002817 | 0.002708 |
| 485 | REGULATION OF AUTOPHAGY | 15 | 249 | 0.0002878 | 0.002751 |
| 486 | ENDOCRINE SYSTEM DEVELOPMENT | 10 | 123 | 0.0002885 | 0.002751 |
| 487 | T CELL DIFFERENTIATION | 10 | 123 | 0.0002885 | 0.002751 |
| 488 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 8 | 80 | 0.0002873 | 0.002751 |
| 489 | MUSCLE ORGAN DEVELOPMENT | 16 | 277 | 0.0002928 | 0.002764 |
| 490 | POSITIVE REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 4 | 16 | 0.0002934 | 0.002764 |
| 491 | HISTONE H2A UBIQUITINATION | 4 | 16 | 0.0002934 | 0.002764 |
| 492 | REGULATION OF OXIDATIVE PHOSPHORYLATION | 4 | 16 | 0.0002934 | 0.002764 |
| 493 | APOPTOTIC PROCESS INVOLVED IN MORPHOGENESIS | 4 | 16 | 0.0002934 | 0.002764 |
| 494 | DNA LIGATION | 4 | 16 | 0.0002934 | 0.002764 |
| 495 | PROTEIN COMPLEX BIOGENESIS | 42 | 1132 | 0.0002956 | 0.002773 |
| 496 | PROTEIN COMPLEX ASSEMBLY | 42 | 1132 | 0.0002956 | 0.002773 |
| 497 | REGULATION OF MUSCLE CONTRACTION | 11 | 147 | 3e-04 | 0.002803 |
| 498 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 11 | 147 | 3e-04 | 0.002803 |
| 499 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 37 | 957 | 0.0003146 | 0.002934 |
| 500 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 11 | 148 | 0.000318 | 0.00296 |
| 501 | HEME METABOLIC PROCESS | 5 | 29 | 0.0003228 | 0.002968 |
| 502 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 29 | 0.0003228 | 0.002968 |
| 503 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 29 | 0.0003228 | 0.002968 |
| 504 | PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 5 | 29 | 0.0003228 | 0.002968 |
| 505 | NUCLEOTIDE EXCISION REPAIR PREINCISION COMPLEX ASSEMBLY | 5 | 29 | 0.0003228 | 0.002968 |
| 506 | REGULATION OF OXIDATIVE STRESS INDUCED INTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 29 | 0.0003228 | 0.002968 |
| 507 | REGULATION OF CELLULAR KETONE METABOLIC PROCESS | 12 | 173 | 0.0003258 | 0.00299 |
| 508 | REGULATION OF NEURON DIFFERENTIATION | 25 | 554 | 0.0003379 | 0.003095 |
| 509 | MULTI ORGANISM REPRODUCTIVE PROCESS | 35 | 891 | 0.0003419 | 0.003125 |
| 510 | NEGATIVE REGULATION OF CHROMATIN MODIFICATION | 6 | 45 | 0.0003501 | 0.00319 |
| 511 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 10 | 126 | 0.0003503 | 0.00319 |
| 512 | REGULATION OF LIPID METABOLIC PROCESS | 16 | 282 | 0.0003572 | 0.003246 |
| 513 | REGULATION OF CARTILAGE DEVELOPMENT | 7 | 63 | 0.0003592 | 0.003258 |
| 514 | GLIOGENESIS | 12 | 175 | 0.0003618 | 0.003269 |
| 515 | ORGANIC HYDROXY COMPOUND BIOSYNTHETIC PROCESS | 12 | 175 | 0.0003618 | 0.003269 |
| 516 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 19 | 370 | 0.0003667 | 0.003307 |
| 517 | EMBRYONIC PLACENTA DEVELOPMENT | 8 | 83 | 0.0003701 | 0.00333 |
| 518 | NEGATIVE REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 4 | 17 | 0.0003773 | 0.003383 |
| 519 | NEGATIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 4 | 17 | 0.0003773 | 0.003383 |
| 520 | BROWN FAT CELL DIFFERENTIATION | 5 | 30 | 0.0003807 | 0.003394 |
| 521 | MYD88 INDEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 5 | 30 | 0.0003807 | 0.003394 |
| 522 | HYDROGEN PEROXIDE METABOLIC PROCESS | 5 | 30 | 0.0003807 | 0.003394 |
| 523 | NEGATIVE REGULATION OF DNA BINDING | 6 | 46 | 0.0003955 | 0.003519 |
| 524 | CHROMATIN ASSEMBLY OR DISASSEMBLY | 12 | 177 | 0.0004011 | 0.003562 |
| 525 | FAT CELL DIFFERENTIATION | 9 | 106 | 0.0004184 | 0.003708 |
| 526 | REGULATION OF REPRODUCTIVE PROCESS | 10 | 129 | 0.0004228 | 0.003733 |
| 527 | REGULATION OF OSSIFICATION | 12 | 178 | 0.0004221 | 0.003733 |
| 528 | REGULATION OF RESPONSE TO OXIDATIVE STRESS | 7 | 65 | 0.000436 | 0.003842 |
| 529 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 6 | 47 | 0.0004454 | 0.003917 |
| 530 | POSITIVE REGULATION OF MAPK CASCADE | 22 | 470 | 0.0004661 | 0.004092 |
| 531 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 8 | 86 | 0.0004712 | 0.004129 |
| 532 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 10 | 131 | 0.0004778 | 0.004171 |
| 533 | LYMPHOCYTE APOPTOTIC PROCESS | 4 | 18 | 0.0004771 | 0.004171 |
| 534 | CYTOSKELETON ORGANIZATION | 33 | 838 | 0.0004828 | 0.004191 |
| 535 | MONOCARBOXYLIC ACID METABOLIC PROCESS | 23 | 503 | 0.0004828 | 0.004191 |
| 536 | NUCLEOBASE CONTAINING SMALL MOLECULE METABOLIC PROCESS | 24 | 535 | 0.000481 | 0.004191 |
| 537 | REGULATION OF MAP KINASE ACTIVITY | 17 | 319 | 0.0004859 | 0.00421 |
| 538 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 15 | 262 | 0.0004926 | 0.00426 |
| 539 | GROWTH | 20 | 410 | 0.0004995 | 0.004301 |
| 540 | SECONDARY METABOLIC PROCESS | 6 | 48 | 5e-04 | 0.004301 |
| 541 | POSITIVE REGULATION OF ACTIN FILAMENT BUNDLE ASSEMBLY | 6 | 48 | 5e-04 | 0.004301 |
| 542 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 23 | 505 | 0.0005099 | 0.004377 |
| 543 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO HYPOXIA | 5 | 32 | 0.0005196 | 0.004453 |
| 544 | POSITIVE REGULATION OF MULTI ORGANISM PROCESS | 11 | 157 | 0.0005259 | 0.004498 |
| 545 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 45 | 1275 | 0.0005272 | 0.004501 |
| 546 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO MEMBRANE | 15 | 264 | 0.0005331 | 0.004535 |
| 547 | GERM CELL DEVELOPMENT | 13 | 209 | 0.0005327 | 0.004535 |
| 548 | EMBRYONIC MORPHOGENESIS | 24 | 539 | 0.0005345 | 0.004538 |
| 549 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY | 9 | 110 | 0.0005494 | 0.004657 |
| 550 | REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 6 | 49 | 0.0005598 | 0.00471 |
| 551 | REGULATION OF STEROID BIOSYNTHETIC PROCESS | 6 | 49 | 0.0005598 | 0.00471 |
| 552 | TUBE MORPHOGENESIS | 17 | 323 | 0.0005593 | 0.00471 |
| 553 | REGULATION OF ATP METABOLIC PROCESS | 6 | 49 | 0.0005598 | 0.00471 |
| 554 | CELLULAR KETONE METABOLIC PROCESS | 7 | 68 | 0.000575 | 0.00483 |
| 555 | PROSTAGLANDIN BIOSYNTHETIC PROCESS | 4 | 19 | 0.0005943 | 0.004938 |
| 556 | PEPTIDYL ARGININE MODIFICATION | 4 | 19 | 0.0005943 | 0.004938 |
| 557 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 8 | 89 | 0.0005937 | 0.004938 |
| 558 | POSITIVE REGULATION OF PROTEIN POLYMERIZATION | 8 | 89 | 0.0005937 | 0.004938 |
| 559 | PROTEOLYSIS | 43 | 1208 | 0.0005917 | 0.004938 |
| 560 | PROSTANOID BIOSYNTHETIC PROCESS | 4 | 19 | 0.0005943 | 0.004938 |
| 561 | REGULATION OF METAL ION TRANSPORT | 17 | 325 | 0.0005995 | 0.004972 |
| 562 | CELLULAR SENESCENCE | 5 | 33 | 0.0006019 | 0.004983 |
| 563 | FATTY ACID METABOLIC PROCESS | 16 | 296 | 0.000606 | 0.005008 |
| 564 | HEART MORPHOGENESIS | 13 | 212 | 0.0006092 | 0.005026 |
| 565 | NCRNA PROCESSING | 19 | 386 | 0.0006148 | 0.005063 |
| 566 | LYMPHOCYTE HOMEOSTASIS | 6 | 50 | 0.0006249 | 0.005124 |
| 567 | FACE DEVELOPMENT | 6 | 50 | 0.0006249 | 0.005124 |
| 568 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 9 | 112 | 0.0006266 | 0.005124 |
| 569 | SINGLE ORGANISM CATABOLIC PROCESS | 36 | 957 | 0.0006266 | 0.005124 |
| 570 | MULTI MULTICELLULAR ORGANISM PROCESS | 13 | 213 | 0.0006366 | 0.005197 |
| 571 | REGULATION OF CELL MATRIX ADHESION | 8 | 90 | 0.0006398 | 0.005214 |
| 572 | NEGATIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 10 | 136 | 0.0006416 | 0.005219 |
| 573 | ACTIN FILAMENT BASED PROCESS | 21 | 450 | 0.0006481 | 0.005262 |
| 574 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 22 | 482 | 0.000652 | 0.005285 |
| 575 | PROTEIN HETEROOLIGOMERIZATION | 9 | 113 | 0.0006683 | 0.005408 |
| 576 | CD4 POSITIVE ALPHA BETA T CELL ACTIVATION | 5 | 34 | 0.0006935 | 0.005573 |
| 577 | HISTONE UBIQUITINATION | 5 | 34 | 0.0006935 | 0.005573 |
| 578 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 5 | 34 | 0.0006935 | 0.005573 |
| 579 | OLIGODENDROCYTE DEVELOPMENT | 5 | 34 | 0.0006935 | 0.005573 |
| 580 | ARTERY MORPHOGENESIS | 6 | 51 | 0.0006958 | 0.005582 |
| 581 | PROTEIN POLYUBIQUITINATION | 14 | 243 | 0.0007066 | 0.005659 |
| 582 | REGULATION OF NUCLEAR DIVISION | 11 | 163 | 0.0007196 | 0.005734 |
| 583 | CELL CYCLE G2 M PHASE TRANSITION | 10 | 138 | 0.0007189 | 0.005734 |
| 584 | MULTI ORGANISM METABOLIC PROCESS | 10 | 138 | 0.0007189 | 0.005734 |
| 585 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 15 | 272 | 0.0007251 | 0.005759 |
| 586 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 13 | 216 | 0.0007253 | 0.005759 |
| 587 | PROTEIN ADP RIBOSYLATION | 4 | 20 | 0.0007305 | 0.005781 |
| 588 | INTRACELLULAR ESTROGEN RECEPTOR SIGNALING PATHWAY | 4 | 20 | 0.0007305 | 0.005781 |
| 589 | TUBE DEVELOPMENT | 24 | 552 | 0.0007457 | 0.005891 |
| 590 | NEGATIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 7 | 71 | 0.0007473 | 0.005893 |
| 591 | NEGATIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 9 | 115 | 0.0007587 | 0.005973 |
| 592 | CELLULAR RESPONSE TO IONIZING RADIATION | 6 | 52 | 0.0007727 | 0.006073 |
| 593 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 16 | 303 | 0.0007781 | 0.006105 |
| 594 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 24 | 554 | 0.0007839 | 0.006141 |
| 595 | DEVELOPMENTAL GROWTH | 17 | 333 | 0.0007859 | 0.006146 |
| 596 | BLOOD VESSEL MORPHOGENESIS | 18 | 364 | 0.0008046 | 0.006271 |
| 597 | POSITIVE REGULATION OF DEFENSE RESPONSE | 18 | 364 | 0.0008046 | 0.006271 |
| 598 | CELLULAR COMPONENT MORPHOGENESIS | 34 | 900 | 0.0008222 | 0.006397 |
| 599 | SINGLE ORGANISM CELL ADHESION | 21 | 459 | 0.0008337 | 0.006476 |
| 600 | CELL PROJECTION ORGANIZATION | 34 | 902 | 0.0008541 | 0.006623 |
| 601 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 6 | 53 | 0.000856 | 0.006628 |
| 602 | NEGATIVE REGULATION OF CYTOPLASMIC TRANSPORT | 9 | 117 | 0.0008588 | 0.006638 |
| 603 | DEFENSE RESPONSE | 43 | 1231 | 0.0008636 | 0.006664 |
| 604 | TRANSCRIPTION COUPLED NUCLEOTIDE EXCISION REPAIR | 7 | 73 | 0.0008831 | 0.006747 |
| 605 | ERYTHROCYTE HOMEOSTASIS | 7 | 73 | 0.0008831 | 0.006747 |
| 606 | NUCLEOTIDE EXCISION REPAIR DNA INCISION 3 TO LESION | 4 | 21 | 0.0008874 | 0.006747 |
| 607 | T HELPER 1 TYPE IMMUNE RESPONSE | 4 | 21 | 0.0008874 | 0.006747 |
| 608 | APOPTOTIC PROCESS INVOLVED IN DEVELOPMENT | 4 | 21 | 0.0008874 | 0.006747 |
| 609 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 11 | 167 | 0.0008792 | 0.006747 |
| 610 | REGULATION OF ION TRANSPORT | 25 | 592 | 0.0008815 | 0.006747 |
| 611 | NEGATIVE REGULATION OF OXIDATIVE STRESS INDUCED INTRINSIC APOPTOTIC SIGNALING PATHWAY | 4 | 21 | 0.0008874 | 0.006747 |
| 612 | NUCLEOTIDE EXCISION REPAIR PREINCISION COMPLEX STABILIZATION | 4 | 21 | 0.0008874 | 0.006747 |
| 613 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 16 | 307 | 0.000894 | 0.006775 |
| 614 | REGULATION OF PROTEIN TARGETING | 16 | 307 | 0.000894 | 0.006775 |
| 615 | PLATELET ACTIVATION | 10 | 142 | 0.0008967 | 0.006784 |
| 616 | NEGATIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 5 | 36 | 0.0009076 | 0.006834 |
| 617 | PORPHYRIN CONTAINING COMPOUND METABOLIC PROCESS | 5 | 36 | 0.0009076 | 0.006834 |
| 618 | NEGATIVE REGULATION OF HISTONE MODIFICATION | 5 | 36 | 0.0009076 | 0.006834 |
| 619 | SEXUAL REPRODUCTION | 29 | 730 | 0.0009197 | 0.006913 |
| 620 | RESPONSE TO BACTERIUM | 23 | 528 | 0.0009316 | 0.006991 |
| 621 | MORPHOGENESIS OF AN EPITHELIUM | 19 | 400 | 0.0009394 | 0.007039 |
| 622 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 12 | 195 | 0.0009472 | 0.007051 |
| 623 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 6 | 54 | 0.0009461 | 0.007051 |
| 624 | REGULATION OF MITOCHONDRIAL MEMBRANE POTENTIAL | 6 | 54 | 0.0009461 | 0.007051 |
| 625 | NEGATIVE REGULATION OF INTRACELLULAR TRANSPORT | 10 | 143 | 0.0009464 | 0.007051 |
| 626 | REGULATION OF CELL ADHESION | 26 | 629 | 0.0009536 | 0.007088 |
| 627 | REGULATION OF DENDRITE MORPHOGENESIS | 7 | 74 | 0.0009579 | 0.007098 |
| 628 | REGULATION OF STEROID METABOLIC PROCESS | 7 | 74 | 0.0009579 | 0.007098 |
| 629 | POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 9 | 119 | 0.0009693 | 0.007171 |
| 630 | MAINTENANCE OF LOCATION IN CELL | 8 | 96 | 0.0009809 | 0.007221 |
| 631 | ICOSANOID METABOLIC PROCESS | 8 | 96 | 0.0009809 | 0.007221 |
| 632 | FATTY ACID DERIVATIVE METABOLIC PROCESS | 8 | 96 | 0.0009809 | 0.007221 |
| 633 | PROTEIN OLIGOMERIZATION | 20 | 434 | 0.001008 | 0.007412 |
| 634 | NEGATIVE REGULATION OF CELL GROWTH | 11 | 170 | 0.001017 | 0.007466 |
| 635 | REGULATION OF DENDRITE DEVELOPMENT | 9 | 120 | 0.001029 | 0.007539 |
| 636 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 12 | 197 | 0.001035 | 0.00757 |
| 637 | MUSCLE SYSTEM PROCESS | 15 | 282 | 0.001045 | 0.007634 |
| 638 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 8 | 97 | 0.00105 | 0.007656 |
| 639 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 19 | 404 | 0.001056 | 0.007687 |
| 640 | REGULATION OF PROTEIN HOMODIMERIZATION ACTIVITY | 4 | 22 | 0.001067 | 0.007697 |
| 641 | TISSUE HOMEOSTASIS | 11 | 171 | 0.001067 | 0.007697 |
| 642 | PEPTIDYL LYSINE MODIFICATION | 16 | 312 | 0.001059 | 0.007697 |
| 643 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 4 | 22 | 0.001067 | 0.007697 |
| 644 | ACTIVATION OF PROTEIN KINASE B ACTIVITY | 4 | 22 | 0.001067 | 0.007697 |
| 645 | AORTA MORPHOGENESIS | 4 | 22 | 0.001067 | 0.007697 |
| 646 | T CELL RECEPTOR SIGNALING PATHWAY | 10 | 146 | 0.001109 | 0.007989 |
| 647 | PROTEIN TARGETING | 19 | 406 | 0.001118 | 0.008042 |
| 648 | REGULATION OF CELLULAR RESPONSE TO HEAT | 7 | 76 | 0.001123 | 0.008048 |
| 649 | RESPONSE TO VITAMIN | 8 | 98 | 0.001122 | 0.008048 |
| 650 | LEUKOCYTE CELL CELL ADHESION | 14 | 255 | 0.001126 | 0.008058 |
| 651 | OUTFLOW TRACT MORPHOGENESIS | 6 | 56 | 0.001148 | 0.008205 |
| 652 | ANATOMICAL STRUCTURE HOMEOSTASIS | 15 | 285 | 0.001162 | 0.00829 |
| 653 | RESPONSE TO TESTOSTERONE | 5 | 38 | 0.001167 | 0.008298 |
| 654 | REGULATION OF T CELL PROLIFERATION | 10 | 147 | 0.001168 | 0.008298 |
| 655 | POSITIVE REGULATION OF DNA REPAIR | 5 | 38 | 0.001167 | 0.008298 |
| 656 | NUCLEOSIDE TRIPHOSPHATE METABOLIC PROCESS | 13 | 228 | 0.001191 | 0.008447 |
| 657 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 8 | 99 | 0.001199 | 0.008491 |
| 658 | STEROL METABOLIC PROCESS | 9 | 123 | 0.001225 | 0.008663 |
| 659 | MALE SEX DIFFERENTIATION | 10 | 148 | 0.00123 | 0.008683 |
| 660 | ADRENAL GLAND DEVELOPMENT | 4 | 23 | 0.00127 | 0.00889 |
| 661 | ALCOHOL METABOLIC PROCESS | 17 | 348 | 0.001271 | 0.00889 |
| 662 | NUCLEOBASE CONTAINING SMALL MOLECULE INTERCONVERSION | 4 | 23 | 0.00127 | 0.00889 |
| 663 | REGULATION OF SUPEROXIDE METABOLIC PROCESS | 4 | 23 | 0.00127 | 0.00889 |
| 664 | CELLULAR HOMEOSTASIS | 27 | 676 | 0.001267 | 0.00889 |
| 665 | PROTEIN INSERTION INTO MEMBRANE | 4 | 23 | 0.00127 | 0.00889 |
| 666 | REGULATION OF CELL CELL ADHESION | 18 | 380 | 0.001308 | 0.009139 |
| 667 | ERBB2 SIGNALING PATHWAY | 5 | 39 | 0.001316 | 0.009167 |
| 668 | NEGATIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 5 | 39 | 0.001316 | 0.009167 |
| 669 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 34 | 926 | 0.00133 | 0.009252 |
| 670 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 15 | 289 | 0.001334 | 0.009264 |
| 671 | CYCLOOXYGENASE PATHWAY | 3 | 11 | 0.001366 | 0.00943 |
| 672 | REGULATION OF OXIDATIVE STRESS INDUCED NEURON DEATH | 3 | 11 | 0.001366 | 0.00943 |
| 673 | REGULATION OF HISTONE H4 ACETYLATION | 3 | 11 | 0.001366 | 0.00943 |
| 674 | CHROMATIN REMODELING | 10 | 150 | 0.001361 | 0.00943 |
| 675 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 13 | 232 | 0.001393 | 0.009603 |
| 676 | REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 7 | 79 | 0.00141 | 0.009678 |
| 677 | MITOCHONDRIAL TRANSPORT | 11 | 177 | 0.00141 | 0.009678 |
| 678 | ERBB SIGNALING PATHWAY | 7 | 79 | 0.00141 | 0.009678 |
| 679 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 13 | 233 | 0.001448 | 0.009921 |
| 680 | REGULATION OF WOUND HEALING | 9 | 126 | 0.001451 | 0.009926 |
| 681 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 22 | 513 | 0.001456 | 0.009934 |
| 682 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 8 | 102 | 0.001454 | 0.009934 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | TRANSCRIPTION FACTOR BINDING | 59 | 524 | 3.278e-26 | 3.045e-23 |
| 2 | ENZYME BINDING | 101 | 1737 | 2.081e-21 | 9.666e-19 |
| 3 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 51 | 588 | 9.514e-18 | 2.946e-15 |
| 4 | MACROMOLECULAR COMPLEX BINDING | 75 | 1399 | 5.985e-14 | 1.39e-11 |
| 5 | REGULATORY REGION NUCLEIC ACID BINDING | 52 | 818 | 1.37e-12 | 2.545e-10 |
| 6 | DOUBLE STRANDED DNA BINDING | 49 | 764 | 4.699e-12 | 7.275e-10 |
| 7 | TRANSCRIPTION COACTIVATOR ACTIVITY | 29 | 296 | 7.611e-12 | 1.01e-09 |
| 8 | KINASE BINDING | 42 | 606 | 1.56e-11 | 1.812e-09 |
| 9 | RNA BINDING | 75 | 1598 | 4.118e-11 | 4.251e-09 |
| 10 | POLY A RNA BINDING | 60 | 1170 | 1.68e-10 | 1.561e-08 |
| 11 | HORMONE RECEPTOR BINDING | 20 | 168 | 4.724e-10 | 3.657e-08 |
| 12 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 25 | 264 | 4.503e-10 | 3.657e-08 |
| 13 | SEQUENCE SPECIFIC DNA BINDING | 54 | 1037 | 8.957e-10 | 6.401e-08 |
| 14 | CHROMATIN BINDING | 32 | 435 | 1.067e-09 | 7.081e-08 |
| 15 | STEROID HORMONE RECEPTOR BINDING | 14 | 81 | 1.454e-09 | 9.002e-08 |
| 16 | CORE PROMOTER BINDING | 18 | 152 | 3.838e-09 | 2.228e-07 |
| 17 | IDENTICAL PROTEIN BINDING | 58 | 1209 | 4.222e-09 | 2.307e-07 |
| 18 | DEACETYLASE ACTIVITY | 11 | 55 | 1.725e-08 | 8.904e-07 |
| 19 | PROTEIN DOMAIN SPECIFIC BINDING | 37 | 624 | 1.841e-08 | 9.003e-07 |
| 20 | NAD DEPENDENT PROTEIN DEACETYLASE ACTIVITY | 7 | 17 | 2.918e-08 | 1.356e-06 |
| 21 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 26 | 371 | 1.07e-07 | 4.735e-06 |
| 22 | PROTEIN DIMERIZATION ACTIVITY | 52 | 1149 | 1.874e-07 | 7.915e-06 |
| 23 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 35 | 629 | 2.117e-07 | 8.55e-06 |
| 24 | PROTEIN DEACETYLASE ACTIVITY | 9 | 43 | 2.32e-07 | 8.98e-06 |
| 25 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 23 | 328 | 5.8e-07 | 2.155e-05 |
| 26 | PROTEIN COMPLEX BINDING | 44 | 935 | 6.296e-07 | 2.25e-05 |
| 27 | PROTEIN HOMODIMERIZATION ACTIVITY | 37 | 722 | 7.083e-07 | 2.437e-05 |
| 28 | ANDROGEN RECEPTOR BINDING | 8 | 39 | 1.298e-06 | 4.306e-05 |
| 29 | LIGAND DEPENDENT NUCLEAR RECEPTOR TRANSCRIPTION COACTIVATOR ACTIVITY | 9 | 53 | 1.512e-06 | 4.683e-05 |
| 30 | CORE PROMOTER SEQUENCE SPECIFIC DNA BINDING | 12 | 101 | 1.496e-06 | 4.683e-05 |
| 31 | HISTONE DEACETYLASE BINDING | 12 | 105 | 2.27e-06 | 6.802e-05 |
| 32 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 50 | 1199 | 3.512e-06 | 9.888e-05 |
| 33 | RECEPTOR BINDING | 58 | 1476 | 3.486e-06 | 9.888e-05 |
| 34 | OXIDIZED NAD BINDING | 5 | 13 | 4.625e-06 | 0.0001264 |
| 35 | DAMAGED DNA BINDING | 9 | 63 | 6.701e-06 | 0.0001779 |
| 36 | CHAPERONE BINDING | 10 | 81 | 7.929e-06 | 0.0002046 |
| 37 | KINASE REGULATOR ACTIVITY | 15 | 186 | 1.04e-05 | 0.0002612 |
| 38 | HYDROLASE ACTIVITY ACTING ON CARBON NITROGEN BUT NOT PEPTIDE BONDS IN LINEAR AMIDES | 10 | 84 | 1.102e-05 | 0.0002693 |
| 39 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 20 | 315 | 1.387e-05 | 0.0003271 |
| 40 | RNA POLYMERASE II CORE PROMOTER SEQUENCE SPECIFIC DNA BINDING | 8 | 53 | 1.443e-05 | 0.0003271 |
| 41 | NAD BINDING | 8 | 53 | 1.443e-05 | 0.0003271 |
| 42 | HEAT SHOCK PROTEIN BINDING | 10 | 89 | 1.847e-05 | 0.0004086 |
| 43 | LRR DOMAIN BINDING | 5 | 17 | 2.073e-05 | 0.0004479 |
| 44 | STEROID BINDING | 10 | 91 | 2.249e-05 | 0.0004748 |
| 45 | REPRESSING TRANSCRIPTION FACTOR BINDING | 8 | 57 | 2.5e-05 | 0.0005161 |
| 46 | SINGLE STRANDED DNA BINDING | 10 | 93 | 2.723e-05 | 0.0005499 |
| 47 | OXIDOREDUCTASE ACTIVITY | 33 | 719 | 2.794e-05 | 0.0005523 |
| 48 | RIBONUCLEOTIDE BINDING | 65 | 1860 | 3.732e-05 | 0.0007224 |
| 49 | STRUCTURE SPECIFIC DNA BINDING | 11 | 118 | 4.188e-05 | 0.000794 |
| 50 | PROTEIN C TERMINUS BINDING | 14 | 186 | 4.395e-05 | 0.0008165 |
| 51 | LEUCINE ZIPPER DOMAIN BINDING | 4 | 11 | 5.787e-05 | 0.001054 |
| 52 | RNA POLYMERASE II DISTAL ENHANCER SEQUENCE SPECIFIC DNA BINDING | 8 | 65 | 6.59e-05 | 0.001177 |
| 53 | RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 10 | 104 | 7.159e-05 | 0.001255 |
| 54 | HISTONE DEACETYLASE ACTIVITY H3 K14 SPECIFIC | 4 | 12 | 8.535e-05 | 0.001468 |
| 55 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 15 | 226 | 9.979e-05 | 0.001685 |
| 56 | DEOXYRIBONUCLEASE ACTIVITY | 8 | 70 | 0.0001124 | 0.001865 |
| 57 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II DISTAL ENHANCER SEQUENCE SPECIFIC BINDING | 9 | 90 | 0.000121 | 0.001972 |
| 58 | OXIDOREDUCTASE ACTIVITY ACTING ON THE CH OH GROUP OF DONORS NAD OR NADP AS ACCEPTOR | 10 | 113 | 0.0001439 | 0.002305 |
| 59 | ENHANCER BINDING | 9 | 93 | 0.0001559 | 0.002455 |
| 60 | PROTEIN HETERODIMERIZATION ACTIVITY | 23 | 468 | 0.0001739 | 0.002693 |
| 61 | ADENYL NUCLEOTIDE BINDING | 53 | 1514 | 0.0002019 | 0.003075 |
| 62 | STEROID HORMONE RECEPTOR ACTIVITY | 7 | 59 | 0.0002381 | 0.003568 |
| 63 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 12 | 168 | 0.0002489 | 0.00367 |
| 64 | STEROID DEHYDROGENASE ACTIVITY | 5 | 28 | 0.0002718 | 0.003908 |
| 65 | TRANSCRIPTION COREPRESSOR ACTIVITY | 14 | 221 | 0.0002734 | 0.003908 |
| 66 | CHROMATIN DNA BINDING | 8 | 80 | 0.0002873 | 0.004044 |
| 67 | ALCOHOL DEHYDROGENASE NADP ACTIVITY | 4 | 16 | 0.0002934 | 0.004069 |
| 68 | UBIQUITIN LIKE PROTEIN CONJUGATING ENZYME ACTIVITY | 5 | 29 | 0.0003228 | 0.00441 |
| 69 | NUCLEOSOME BINDING | 6 | 45 | 0.0003501 | 0.004646 |
| 70 | SCAFFOLD PROTEIN BINDING | 6 | 45 | 0.0003501 | 0.004646 |
| 71 | NUCLEOSOMAL DNA BINDING | 5 | 30 | 0.0003807 | 0.004981 |
| 72 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 9 | 105 | 0.00039 | 0.005032 |
| 73 | MONOCARBOXYLIC ACID BINDING | 7 | 65 | 0.000436 | 0.005548 |
| 74 | TRANSCRIPTION FACTOR ACTIVITY DIRECT LIGAND REGULATED SEQUENCE SPECIFIC DNA BINDING | 6 | 48 | 5e-04 | 0.006277 |
| 75 | COFACTOR BINDING | 15 | 263 | 0.0005125 | 0.006348 |
| 76 | OXIDOREDUCTASE ACTIVITY ACTING ON CH OH GROUP OF DONORS | 10 | 134 | 0.0005713 | 0.006983 |
| 77 | PROTEIN KINASE C BINDING | 6 | 50 | 0.0006249 | 0.00754 |
| 78 | ENZYME REGULATOR ACTIVITY | 36 | 959 | 0.0006506 | 0.007749 |
| 79 | SNRNA BINDING | 5 | 34 | 0.0006935 | 0.008156 |
| 80 | TBP CLASS PROTEIN BINDING | 4 | 20 | 0.0007305 | 0.008483 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | TRANSCRIPTION FACTOR COMPLEX | 32 | 298 | 4.788e-14 | 2.796e-11 |
| 2 | CHROMOSOME | 54 | 880 | 1.942e-12 | 5.672e-10 |
| 3 | CHROMATIN | 35 | 441 | 2.113e-11 | 4.113e-09 |
| 4 | RNA POLYMERASE II TRANSCRIPTION FACTOR COMPLEX | 17 | 101 | 3.875e-11 | 5.658e-09 |
| 5 | NUCLEAR CHROMOSOME | 37 | 523 | 1.52e-10 | 1.776e-08 |
| 6 | NUCLEAR TRANSCRIPTION FACTOR COMPLEX | 17 | 127 | 1.563e-09 | 1.522e-07 |
| 7 | NUCLEAR CHROMATIN | 25 | 291 | 3.403e-09 | 2.839e-07 |
| 8 | NUCLEOPLASM PART | 40 | 708 | 1.81e-08 | 1.322e-06 |
| 9 | RIBONUCLEOPROTEIN COMPLEX | 40 | 721 | 2.977e-08 | 1.932e-06 |
| 10 | CATALYTIC COMPLEX | 50 | 1038 | 4.904e-08 | 2.864e-06 |
| 11 | CELL BODY | 31 | 494 | 7.851e-08 | 4.168e-06 |
| 12 | PERINUCLEAR REGION OF CYTOPLASM | 36 | 642 | 1.166e-07 | 5.239e-06 |
| 13 | MITOCHONDRION | 67 | 1633 | 1.112e-07 | 5.239e-06 |
| 14 | MYELIN SHEATH | 16 | 168 | 5.917e-07 | 2.468e-05 |
| 15 | CELL SUBSTRATE JUNCTION | 25 | 398 | 1.433e-06 | 5.578e-05 |
| 16 | NEURON PROJECTION | 43 | 942 | 1.883e-06 | 6.873e-05 |
| 17 | TRANSFERASE COMPLEX | 35 | 703 | 2.74e-06 | 9.413e-05 |
| 18 | ENVELOPE | 47 | 1090 | 2.906e-06 | 9.428e-05 |
| 19 | EXTRACELLULAR SPACE | 55 | 1376 | 3.897e-06 | 0.0001198 |
| 20 | NEURON PART | 51 | 1265 | 7.167e-06 | 0.0002093 |
| 21 | MITOCHONDRIAL ENVELOPE | 33 | 691 | 1.259e-05 | 0.00035 |
| 22 | INCLUSION BODY | 9 | 70 | 1.616e-05 | 0.000429 |
| 23 | SEX CHROMOSOME | 6 | 27 | 1.754e-05 | 0.0004454 |
| 24 | SOMATODENDRITIC COMPARTMENT | 31 | 650 | 2.377e-05 | 0.0005785 |
| 25 | NUCLEOLUS | 37 | 848 | 2.72e-05 | 0.0006354 |
| 26 | MITOCHONDRIAL PART | 40 | 953 | 3.065e-05 | 0.0006884 |
| 27 | CELL PROJECTION | 63 | 1786 | 3.794e-05 | 0.0008207 |
| 28 | ANCHORING JUNCTION | 25 | 489 | 4.88e-05 | 0.001018 |
| 29 | CELL LEADING EDGE | 20 | 350 | 6.178e-05 | 0.001244 |
| 30 | RIBONUCLEOPROTEIN GRANULE | 12 | 148 | 7.504e-05 | 0.001461 |
| 31 | HETEROCHROMATIN | 8 | 67 | 8.208e-05 | 0.001546 |
| 32 | CYTOSOLIC PART | 15 | 223 | 8.592e-05 | 0.001568 |
| 33 | CHROMOSOME TELOMERIC REGION | 12 | 162 | 0.0001774 | 0.00314 |
| 34 | ACTIN CYTOSKELETON | 22 | 444 | 0.0002138 | 0.003589 |
| 35 | PML BODY | 9 | 97 | 0.0002151 | 0.003589 |
| 36 | XY BODY | 4 | 15 | 0.0002238 | 0.00363 |
| 37 | CHROMOSOMAL REGION | 18 | 330 | 0.0002529 | 0.003991 |
| 38 | CONDENSED CHROMOSOME | 13 | 195 | 0.0002742 | 0.004214 |
| 39 | LAMELLIPODIUM | 12 | 172 | 0.000309 | 0.004627 |
| 40 | CYTOSKELETON | 63 | 1967 | 0.0005577 | 0.008142 |
| 41 | H4 HISTONE ACETYLTRANSFERASE COMPLEX | 4 | 19 | 0.0005943 | 0.008465 |
| 42 | METHYLTRANSFERASE COMPLEX | 8 | 90 | 0.0006398 | 0.008896 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04110_Cell_cycle | 17 | 128 | 1.768e-09 | 3.183e-07 | |
| 2 | hsa00140_Steroid_hormone_biosynthesis | 9 | 57 | 2.849e-06 | 0.0002564 | |
| 3 | hsa04115_p53_signaling_pathway | 9 | 69 | 1.435e-05 | 0.0008023 | |
| 4 | hsa04380_Osteoclast_differentiation | 12 | 128 | 1.783e-05 | 0.0008023 | |
| 5 | hsa04390_Hippo_signaling_pathway | 13 | 154 | 2.483e-05 | 0.0008938 | |
| 6 | hsa04120_Ubiquitin_mediated_proteolysis | 12 | 139 | 4.065e-05 | 0.00122 | |
| 7 | hsa04350_TGF.beta_signaling_pathway | 9 | 85 | 7.741e-05 | 0.001991 | |
| 8 | hsa04370_VEGF_signaling_pathway | 8 | 76 | 0.0002011 | 0.004525 | |
| 9 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 12 | 168 | 0.0002489 | 0.004977 | |
| 10 | hsa04010_MAPK_signaling_pathway | 15 | 268 | 0.0006229 | 0.009935 | |
| 11 | hsa03410_Base_excision_repair | 5 | 34 | 0.0006935 | 0.009935 | |
| 12 | hsa04114_Oocyte_meiosis | 9 | 114 | 0.0007123 | 0.009935 | |
| 13 | hsa04910_Insulin_signaling_pathway | 10 | 138 | 0.0007189 | 0.009935 | |
| 14 | hsa04150_mTOR_signaling_pathway | 6 | 52 | 0.0007727 | 0.009935 | |
| 15 | hsa04662_B_cell_receptor_signaling_pathway | 7 | 75 | 0.001038 | 0.01245 | |
| 16 | hsa04510_Focal_adhesion | 12 | 200 | 0.001179 | 0.01326 | |
| 17 | hsa04722_Neurotrophin_signaling_pathway | 9 | 127 | 0.001533 | 0.01623 | |
| 18 | hsa03010_Ribosome | 7 | 92 | 0.003375 | 0.03259 | |
| 19 | hsa04151_PI3K_AKT_signaling_pathway | 16 | 351 | 0.003469 | 0.03259 | |
| 20 | hsa04720_Long.term_potentiation | 6 | 70 | 0.003621 | 0.03259 | |
| 21 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 6 | 71 | 0.003886 | 0.03331 | |
| 22 | hsa04310_Wnt_signaling_pathway | 9 | 151 | 0.004911 | 0.04018 | |
| 23 | hsa04260_Cardiac_muscle_contraction | 6 | 77 | 0.00579 | 0.04284 | |
| 24 | hsa03040_Spliceosome | 8 | 128 | 0.005913 | 0.04284 | |
| 25 | hsa04620_Toll.like_receptor_signaling_pathway | 7 | 102 | 0.005949 | 0.04284 | |
| 26 | hsa03022_Basal_transcription_factors | 4 | 37 | 0.007507 | 0.05197 | |
| 27 | hsa04710_Circadian_rhythm_._mammal | 3 | 23 | 0.01215 | 0.08101 | |
| 28 | hsa00860_Porphyrin_and_chlorophyll_metabolism | 4 | 43 | 0.01271 | 0.08169 | |
| 29 | hsa03420_Nucleotide_excision_repair | 4 | 45 | 0.01485 | 0.09151 | |
| 30 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 6 | 95 | 0.0155 | 0.09151 | |
| 31 | hsa04810_Regulation_of_actin_cytoskeleton | 10 | 214 | 0.01604 | 0.09151 | |
| 32 | hsa03320_PPAR_signaling_pathway | 5 | 70 | 0.01627 | 0.09151 | |
| 33 | hsa04622_RIG.I.like_receptor_signaling_pathway | 5 | 71 | 0.01721 | 0.09385 | |
| 34 | hsa04912_GnRH_signaling_pathway | 6 | 101 | 0.02036 | 0.1035 | |
| 35 | hsa04916_Melanogenesis | 6 | 101 | 0.02036 | 0.1035 | |
| 36 | hsa04360_Axon_guidance | 7 | 130 | 0.0207 | 0.1035 | |
| 37 | hsa00190_Oxidative_phosphorylation | 7 | 132 | 0.0223 | 0.1085 | |
| 38 | hsa04664_Fc_epsilon_RI_signaling_pathway | 5 | 79 | 0.02601 | 0.1232 | |
| 39 | hsa04270_Vascular_smooth_muscle_contraction | 6 | 116 | 0.03679 | 0.1674 | |
| 40 | hsa04914_Progesterone.mediated_oocyte_maturation | 5 | 87 | 0.03728 | 0.1674 | |
| 41 | hsa04670_Leukocyte_transendothelial_migration | 6 | 117 | 0.03812 | 0.1674 | |
| 42 | hsa00120_Primary_bile_acid_biosynthesis | 2 | 16 | 0.04404 | 0.1887 | |
| 43 | hsa00350_Tyrosine_metabolism | 3 | 41 | 0.05545 | 0.2285 | |
| 44 | hsa04920_Adipocytokine_signaling_pathway | 4 | 68 | 0.05585 | 0.2285 | |
| 45 | hsa04530_Tight_junction | 6 | 133 | 0.06355 | 0.2542 | |
| 46 | hsa04520_Adherens_junction | 4 | 73 | 0.06888 | 0.266 | |
| 47 | hsa03050_Proteasome | 3 | 45 | 0.06946 | 0.266 | |
| 48 | hsa04330_Notch_signaling_pathway | 3 | 47 | 0.07701 | 0.2888 | |
| 49 | hsa04660_T_cell_receptor_signaling_pathway | 5 | 108 | 0.07925 | 0.2911 | |
| 50 | hsa00480_Glutathione_metabolism | 3 | 50 | 0.08897 | 0.3203 | |
| 51 | hsa04320_Dorso.ventral_axis_formation | 2 | 25 | 0.09735 | 0.3436 | |
| 52 | hsa03013_RNA_transport | 6 | 152 | 0.1041 | 0.3604 | |
| 53 | hsa04062_Chemokine_signaling_pathway | 7 | 189 | 0.1072 | 0.364 | |
| 54 | hsa04966_Collecting_duct_acid_secretion | 2 | 27 | 0.1109 | 0.369 | |
| 55 | hsa04012_ErbB_signaling_pathway | 4 | 87 | 0.1127 | 0.369 | |
| 56 | hsa04744_Phototransduction | 2 | 29 | 0.1248 | 0.4003 | |
| 57 | hsa00590_Arachidonic_acid_metabolism | 3 | 59 | 0.129 | 0.4003 | |
| 58 | hsa04621_NOD.like_receptor_signaling_pathway | 3 | 59 | 0.129 | 0.4003 | |
| 59 | hsa00020_Citrate_cycle_.TCA_cycle. | 2 | 30 | 0.1319 | 0.4026 | |
| 60 | hsa00970_Aminoacyl.tRNA_biosynthesis | 3 | 63 | 0.1485 | 0.4454 | |
| 61 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 5 | 136 | 0.1618 | 0.4776 | |
| 62 | hsa04020_Calcium_signaling_pathway | 6 | 177 | 0.1734 | 0.5034 | |
| 63 | hsa04610_Complement_and_coagulation_cascades | 3 | 69 | 0.1793 | 0.5124 | |
| 64 | hsa00982_Drug_metabolism_._cytochrome_P450 | 3 | 73 | 0.2008 | 0.556 | |
| 65 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 2 | 42 | 0.2227 | 0.5878 | |
| 66 | hsa04612_Antigen_processing_and_presentation | 3 | 78 | 0.2284 | 0.5878 | |
| 67 | hsa04630_Jak.STAT_signaling_pathway | 5 | 155 | 0.2315 | 0.5878 | |
| 68 | hsa04146_Peroxisome | 3 | 79 | 0.234 | 0.5878 | |
| 69 | hsa04014_Ras_signaling_pathway | 7 | 236 | 0.2341 | 0.5878 | |
| 70 | hsa04145_Phagosome | 5 | 156 | 0.2354 | 0.5878 | |
| 71 | hsa04962_Vasopressin.regulated_water_reabsorption | 2 | 44 | 0.2384 | 0.5878 | |
| 72 | hsa04144_Endocytosis | 6 | 203 | 0.2604 | 0.6249 | |
| 73 | hsa04672_Intestinal_immune_network_for_IgA_production | 2 | 49 | 0.2777 | 0.6578 | |
| 74 | hsa04640_Hematopoietic_cell_lineage | 3 | 88 | 0.2853 | 0.667 | |
| 75 | hsa04210_Apoptosis | 3 | 89 | 0.2911 | 0.6717 | |
| 76 | hsa00983_Drug_metabolism_._other_enzymes | 2 | 52 | 0.3013 | 0.6858 | |
| 77 | hsa04340_Hedgehog_signaling_pathway | 2 | 56 | 0.3325 | 0.7242 | |
| 78 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.3325 | 0.7242 | |
| 79 | hsa00240_Pyrimidine_metabolism | 3 | 99 | 0.349 | 0.7479 | |
| 80 | hsa00830_Retinol_metabolism | 2 | 64 | 0.3936 | 0.8334 | |
| 81 | hsa00010_Glycolysis_._Gluconeogenesis | 2 | 65 | 0.401 | 0.8393 | |
| 82 | hsa04730_Long.term_depression | 2 | 70 | 0.4376 | 0.8816 | |
| 83 | hsa03018_RNA_degradation | 2 | 71 | 0.4448 | 0.8816 | |
| 84 | hsa04976_Bile_secretion | 2 | 71 | 0.4448 | 0.8816 | |
| 85 | hsa00230_Purine_metabolism | 4 | 162 | 0.4487 | 0.8816 | |
| 86 | hsa04971_Gastric_acid_secretion | 2 | 74 | 0.466 | 0.902 | |
| 87 | hsa04142_Lysosome | 3 | 121 | 0.4732 | 0.9061 | |
| 88 | hsa03015_mRNA_surveillance_pathway | 2 | 83 | 0.5267 | 0.9765 | |
| 89 | hsa04514_Cell_adhesion_molecules_.CAMs. | 3 | 136 | 0.5514 | 0.9926 | |
| 90 | hsa04970_Salivary_secretion | 2 | 89 | 0.5645 | 1 | |
| 91 | hsa04540_Gap_junction | 2 | 90 | 0.5706 | 1 | |
| 92 | hsa04740_Olfactory_transduction | 2 | 388 | 0.9978 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-815I9.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 32 | AR | Sponge network | -0.348 | 0.02944 | -0.439 | 0.04112 | 0.8 |
| 2 | DHRS4-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 32 | AR | Sponge network | -0.343 | 0.01612 | -0.439 | 0.04112 | 0.797 |
| 3 | RP5-1047A19.4 | hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a | 12 | AR | Sponge network | -0.573 | 0.01366 | -0.439 | 0.04112 | 0.781 |
| 4 | RP11-10N23.2 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-374a-3p | 13 | AR | Sponge network | -0.227 | 0.15446 | -0.439 | 0.04112 | 0.775 |
| 5 | RP11-206L10.2 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-505-5p | 21 | AR | Sponge network | -0.272 | 0.15259 | -0.439 | 0.04112 | 0.769 |
| 6 | CTC-429P9.5 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p | 22 | AR | Sponge network | -0.62 | 0.01214 | -0.439 | 0.04112 | 0.763 |
| 7 | AC006116.24 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-93-5p | 15 | AR | Sponge network | -0.075 | 0.67001 | -0.439 | 0.04112 | 0.759 |
| 8 | CTD-3099C6.9 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 27 | AR | Sponge network | -0.721 | 0.00014 | -0.439 | 0.04112 | 0.756 |
| 9 | CTD-2620I22.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-505-3p;hsa-miR-9-5p | 14 | AR | Sponge network | -0.868 | 0.00033 | -0.439 | 0.04112 | 0.744 |
| 10 | RP11-701H24.3 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | AR | Sponge network | -0.818 | 0.05081 | -0.439 | 0.04112 | 0.726 |
| 11 | RP11-464F9.1 | hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-505-5p;hsa-miR-93-5p | 13 | AR | Sponge network | -0.222 | 0.14908 | -0.439 | 0.04112 | 0.723 |
| 12 | BX322557.10 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a | 13 | AR | Sponge network | -0.365 | 0.01273 | -0.439 | 0.04112 | 0.722 |
| 13 | AF131215.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | AR | Sponge network | -0.384 | 0.0318 | -0.439 | 0.04112 | 0.706 |
| 14 | CTC-429P9.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p | 13 | AR | Sponge network | -0.449 | 0.02664 | -0.439 | 0.04112 | 0.705 |
| 15 | FGD5-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 28 | AR | Sponge network | -0.505 | 0 | -0.439 | 0.04112 | 0.704 |
| 16 | CTD-2647L4.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-93-5p | 20 | AR | Sponge network | -0.414 | 0.12742 | -0.439 | 0.04112 | 0.698 |
| 17 | OIP5-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | AR | Sponge network | -0.421 | 0 | -0.439 | 0.04112 | 0.686 |
| 18 | RP11-34P13.13 | hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-505-3p;hsa-miR-582-3p;hsa-miR-93-5p | 15 | AR | Sponge network | 0.021 | 0.91639 | -0.439 | 0.04112 | 0.681 |
| 19 | RP11-298J20.4 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-9-5p | 10 | AR | Sponge network | -1.795 | 0.00363 | -0.439 | 0.04112 | 0.679 |
| 20 | LINC00657 | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-505-3p;hsa-miR-664a-3p | 12 | AR | Sponge network | -0.138 | 0.02318 | -0.439 | 0.04112 | 0.674 |
| 21 | CTD-2666L21.1 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-505-5p | 14 | AR | Sponge network | 0.042 | 0.82695 | -0.439 | 0.04112 | 0.669 |
| 22 | AC002064.4 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-576-5p | 13 | AR | Sponge network | -0.369 | 0.48872 | -0.439 | 0.04112 | 0.658 |
| 23 | RP11-144G6.12 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-93-5p | 17 | AR | Sponge network | -0.558 | 0.01473 | -0.439 | 0.04112 | 0.654 |
| 24 | RP11-384K6.6 | hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p | 15 | AR | Sponge network | -0.362 | 0.02261 | -0.439 | 0.04112 | 0.647 |
| 25 | RP11-182J1.1 | hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 11 | AR | Sponge network | -0.565 | 0 | -0.439 | 0.04112 | 0.643 |
| 26 | RP11-193H5.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | AR | Sponge network | -0.392 | 0.01084 | -0.439 | 0.04112 | 0.64 |
| 27 | AC093110.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-9-5p | 22 | AR | Sponge network | -1.45 | 0.00301 | -0.439 | 0.04112 | 0.635 |
| 28 | RP11-967K21.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-93-5p | 26 | AR | Sponge network | -0.764 | 0.00017 | -0.439 | 0.04112 | 0.632 |
| 29 | RP11-517P14.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | AR | Sponge network | -0.583 | 0.00227 | -0.439 | 0.04112 | 0.631 |
| 30 | RP4-614O4.11 | hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-34a-5p;hsa-miR-34c-5p | 10 | AR | Sponge network | -1.305 | 0.01011 | -0.439 | 0.04112 | 0.627 |
| 31 | RP1-122P22.2 | hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-338-3p | 11 | AR | Sponge network | -0.574 | 0.01134 | -0.439 | 0.04112 | 0.624 |
| 32 | CTD-2008P7.9 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-9-5p | 26 | AR | Sponge network | -1.427 | 0 | -0.439 | 0.04112 | 0.622 |
| 33 | FAM157C | hsa-miR-135a-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-34c-5p;hsa-miR-374a-3p;hsa-miR-505-5p | 16 | AR | Sponge network | -0.066 | 0.75133 | -0.439 | 0.04112 | 0.607 |
| 34 | RP11-282O18.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | AR | Sponge network | -0.106 | 0.38368 | -0.439 | 0.04112 | 0.6 |
| 35 | PDZRN3-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | AR | Sponge network | -0.258 | 0.2196 | -0.439 | 0.04112 | 0.598 |
| 36 | RP11-196G11.4 | hsa-miR-106a-5p;hsa-miR-135a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 11 | AR | Sponge network | -0.316 | 0.32208 | -0.439 | 0.04112 | 0.597 |
| 37 | RP1-12G14.7 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 11 | AR | Sponge network | -0.211 | 0.09329 | -0.439 | 0.04112 | 0.596 |
| 38 | FAM157A | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-505-5p | 19 | AR | Sponge network | -0.572 | 0.10413 | -0.439 | 0.04112 | 0.592 |
| 39 | RP11-835E18.5 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | AR | Sponge network | -0.401 | 0.00017 | -0.439 | 0.04112 | 0.591 |
| 40 | CTC-429P9.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-217;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-93-5p | 13 | AR | Sponge network | -0.223 | 0.08355 | -0.439 | 0.04112 | 0.582 |
| 41 | RP11-158K1.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | AR | Sponge network | -0.28 | 0.22783 | -0.439 | 0.04112 | 0.579 |
| 42 | RP11-545I5.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p;hsa-miR-93-5p | 36 | AR | Sponge network | -1.419 | 0 | -0.439 | 0.04112 | 0.578 |
| 43 | RP11-672A2.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-505-5p | 13 | AR | Sponge network | -0.591 | 0.07048 | -0.439 | 0.04112 | 0.577 |
| 44 | RP4-798P15.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-361-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | AR | Sponge network | -0.714 | 0.00168 | -0.439 | 0.04112 | 0.571 |
| 45 | RP11-283I3.6 | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-505-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | AR | Sponge network | -0.193 | 0.03292 | -0.439 | 0.04112 | 0.57 |
| 46 | RP11-473I1.9 | hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-664a-3p | 16 | AR | Sponge network | 0.097 | 0.22639 | -0.439 | 0.04112 | 0.567 |
| 47 | CTC-559E9.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-320a;hsa-miR-576-5p;hsa-miR-93-5p | 18 | AR | Sponge network | -0.679 | 0.00033 | -0.439 | 0.04112 | 0.562 |
| 48 | RP11-283G6.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-664a-3p | 13 | AR | Sponge network | -0.68 | 0.00954 | -0.439 | 0.04112 | 0.557 |
| 49 | CXADRP3 | hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-454-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 12 | AR | Sponge network | -0.66 | 0.12999 | -0.439 | 0.04112 | 0.554 |
| 50 | CTC-487M23.5 | hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p | 15 | AR | Sponge network | -0.47 | 0.0001 | -0.439 | 0.04112 | 0.552 |
| 51 | RP11-876N24.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-9-5p | 23 | AR | Sponge network | -0.331 | 0.35441 | -0.439 | 0.04112 | 0.551 |
| 52 | LINC00621 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | AR | Sponge network | -1.244 | 0.00085 | -0.439 | 0.04112 | 0.547 |
| 53 | LLNLF-65H9.1 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-454-3p | 14 | AR | Sponge network | 0.196 | 0.19403 | -0.439 | 0.04112 | 0.535 |
| 54 | RP11-497H16.9 | hsa-miR-106a-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | AR | Sponge network | -0.708 | 0.00018 | -0.439 | 0.04112 | 0.534 |
| 55 | RP5-991G20.1 | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 27 | AR | Sponge network | -0.127 | 0.40793 | -0.439 | 0.04112 | 0.532 |
| 56 | RP11-407N17.5 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 20 | AR | Sponge network | 0.03 | 0.93778 | -0.439 | 0.04112 | 0.532 |
| 57 | CTC-523E23.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-9-5p | 25 | AR | Sponge network | -0.107 | 0.75525 | -0.439 | 0.04112 | 0.512 |
| 58 | RP11-775D22.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 14 | AR | Sponge network | -0.277 | 0.42904 | -0.439 | 0.04112 | 0.501 |
| 59 | RP11-819C21.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | AR | Sponge network | -0.394 | 8.0E-5 | -0.439 | 0.04112 | 0.499 |
| 60 | RP5-994D16.9 | hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | AR | Sponge network | -0.242 | 0.00454 | -0.439 | 0.04112 | 0.497 |
| 61 | AC226118.1 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-664a-3p;hsa-miR-9-5p | 18 | AR | Sponge network | -0.745 | 0.03178 | -0.439 | 0.04112 | 0.485 |
| 62 | TMEM5-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-374a-3p | 17 | AR | Sponge network | -2.314 | 0 | -0.439 | 0.04112 | 0.481 |
| 63 | RP11-400K9.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | AR | Sponge network | -1.211 | 0 | -0.439 | 0.04112 | 0.474 |
| 64 | RP11-517B11.7 | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | AR | Sponge network | -0.065 | 0.37323 | -0.439 | 0.04112 | 0.473 |
| 65 | AC061961.2 | hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-29b-2-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-590-3p | 10 | AR | Sponge network | -4.522 | 0 | -0.439 | 0.04112 | 0.472 |
| 66 | RP11-253E3.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-5p | 22 | AR | Sponge network | -0.093 | 0.55842 | -0.439 | 0.04112 | 0.47 |
| 67 | SNHG14 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 22 | AR | Sponge network | -0.051 | 0.64448 | -0.439 | 0.04112 | 0.469 |
| 68 | LINC00094 | hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-30a-5p;hsa-miR-34a-5p | 10 | AR | Sponge network | -0.337 | 0 | -0.439 | 0.04112 | 0.454 |
| 69 | FAM87B | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | AR | Sponge network | 0.109 | 0.44393 | -0.439 | 0.04112 | 0.446 |
| 70 | LIPE-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-582-3p;hsa-miR-660-5p;hsa-miR-93-5p | 26 | AR | Sponge network | -0.697 | 0 | -0.439 | 0.04112 | 0.434 |
| 71 | CTD-2521M24.6 | hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-9-5p | 11 | AR | Sponge network | -1.5 | 6.0E-5 | -0.439 | 0.04112 | 0.434 |
| 72 | CTD-2132N18.2 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-454-3p | 13 | AR | Sponge network | -0.235 | 0.27351 | -0.439 | 0.04112 | 0.432 |
| 73 | CTD-2291D10.4 | hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-34c-5p;hsa-miR-660-5p | 12 | AR | Sponge network | -2.563 | 6.0E-5 | -0.439 | 0.04112 | 0.427 |
| 74 | RP11-428J1.5 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | AR | Sponge network | -0.166 | 0.07457 | -0.439 | 0.04112 | 0.42 |
| 75 | RP11-597D13.9 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-505-3p | 13 | AR | Sponge network | 0.874 | 0 | -0.439 | 0.04112 | 0.416 |
| 76 | RAMP2-AS1 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | AR | Sponge network | -0.492 | 0.00285 | -0.439 | 0.04112 | 0.415 |
| 77 | CTB-175E5.7 | hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-362-3p;hsa-miR-576-5p | 13 | AR | Sponge network | -0.743 | 0.02557 | -0.439 | 0.04112 | 0.414 |
| 78 | HCG11 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | AR | Sponge network | -1.393 | 0 | -0.439 | 0.04112 | 0.411 |
| 79 | RP11-269G24.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-361-5p;hsa-miR-93-5p | 13 | AR | Sponge network | -0.674 | 0.00183 | -0.439 | 0.04112 | 0.401 |
| 80 | LINC00641 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AR | Sponge network | -0.825 | 0 | -0.439 | 0.04112 | 0.398 |
| 81 | LINC00654 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | AR | Sponge network | -1.291 | 0 | -0.439 | 0.04112 | 0.394 |
| 82 | CTC-523E23.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | AR | Sponge network | 0.096 | 0.71163 | -0.439 | 0.04112 | 0.391 |
| 83 | RP11-890B15.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | AR | Sponge network | -0.374 | 0 | -0.439 | 0.04112 | 0.389 |
| 84 | A2M-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | AR | Sponge network | -1.293 | 3.0E-5 | -0.439 | 0.04112 | 0.388 |
| 85 | RP11-284N8.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | AR | Sponge network | -0.322 | 0.14616 | -0.439 | 0.04112 | 0.384 |
| 86 | LINC00987 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-335-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 14 | AR | Sponge network | -0.435 | 0 | -0.439 | 0.04112 | 0.383 |
| 87 | CTD-2554C21.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-93-5p | 23 | AR | Sponge network | -1.375 | 0 | -0.439 | 0.04112 | 0.378 |
| 88 | CTD-2260A17.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p | 13 | AR | Sponge network | -0.349 | 0.12503 | -0.439 | 0.04112 | 0.378 |
| 89 | RP5-823G15.5 | hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p | 11 | AR | Sponge network | 0.815 | 0.04831 | -0.439 | 0.04112 | 0.378 |
| 90 | RP11-44F14.2 | hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | AR | Sponge network | 0.033 | 0.89502 | -0.439 | 0.04112 | 0.375 |
| 91 | RP11-33A14.1 | hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-217;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34c-5p;hsa-miR-93-5p | 11 | AR | Sponge network | 0.148 | 0.75333 | -0.439 | 0.04112 | 0.368 |
| 92 | AC144450.2 | hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-149-5p;hsa-miR-217;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-452-3p | 12 | AR | Sponge network | 3.725 | 0 | -0.439 | 0.04112 | 0.366 |
| 93 | RP11-84G21.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | AR | Sponge network | -0.46 | 0.03275 | -0.439 | 0.04112 | 0.365 |
| 94 | RP11-57H14.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | AR | Sponge network | -0.634 | 0 | -0.439 | 0.04112 | 0.361 |
| 95 | RP11-1223D19.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | AR | Sponge network | -1 | 0.00123 | -0.439 | 0.04112 | 0.356 |
| 96 | BZRAP1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-9-5p;hsa-miR-93-5p | 22 | AR | Sponge network | -0.876 | 0 | -0.439 | 0.04112 | 0.35 |
| 97 | BOLA3-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-9-5p | 25 | AR | Sponge network | -1.462 | 0 | -0.439 | 0.04112 | 0.345 |
| 98 | U91328.21 | hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a | 11 | AR | Sponge network | -0.587 | 2.0E-5 | -0.439 | 0.04112 | 0.337 |
| 99 | TPRG1-AS1 | hsa-miR-106a-5p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-9-5p;hsa-miR-93-5p | 11 | AR | Sponge network | -0.71 | 0.06964 | -0.439 | 0.04112 | 0.335 |
| 100 | CTD-2231H16.1 | hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p | 12 | AR | Sponge network | -0.707 | 0.08398 | -0.439 | 0.04112 | 0.335 |
| 101 | CTD-2015G9.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p;hsa-miR-93-5p | 33 | AR | Sponge network | -1.112 | 0.12879 | -0.439 | 0.04112 | 0.311 |
| 102 | CTD-2035E11.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-93-5p | 13 | AR | Sponge network | -0.011 | 0.95746 | -0.439 | 0.04112 | 0.31 |
| 103 | RP11-507K2.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 10 | AR | Sponge network | -0.748 | 5.0E-5 | -0.439 | 0.04112 | 0.301 |
| 104 | RP11-686D22.8 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-590-3p | 22 | AR | Sponge network | -1.003 | 0 | -0.439 | 0.04112 | 0.298 |
| 105 | RP11-400K9.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 12 | AR | Sponge network | -1.012 | 1.0E-5 | -0.439 | 0.04112 | 0.297 |
| 106 | LINC00578 | hsa-miR-135a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 15 | AR | Sponge network | -0.497 | 0.01377 | -0.439 | 0.04112 | 0.29 |
| 107 | RP11-536C10.16 | hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 10 | AR | Sponge network | -0.384 | 0.13473 | -0.439 | 0.04112 | 0.289 |
| 108 | CTA-221G9.11 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | AR | Sponge network | -0.971 | 0 | -0.439 | 0.04112 | 0.286 |
| 109 | LINC00883 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 24 | AR | Sponge network | -1.026 | 0 | -0.439 | 0.04112 | 0.285 |
| 110 | AP000345.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 14 | AR | Sponge network | -0.595 | 0.04731 | -0.439 | 0.04112 | 0.281 |
| 111 | RP11-303E16.8 | hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-135a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 11 | AR | Sponge network | -0.034 | 0.85827 | -0.439 | 0.04112 | 0.276 |
| 112 | RP11-359B12.2 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AR | Sponge network | -0.389 | 0 | -0.439 | 0.04112 | 0.269 |
| 113 | RP11-705C15.3 | hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | AR | Sponge network | -0.62 | 0 | -0.439 | 0.04112 | 0.264 |
| 114 | AC003991.3 | hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-452-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | AR | Sponge network | 0.841 | 0 | -0.439 | 0.04112 | 0.26 |
| 115 | RP11-360F5.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-590-3p | 11 | AR | Sponge network | -0.506 | 0.01485 | -0.439 | 0.04112 | 0.26 |
| 116 | RP11-554D14.6 | hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a | 10 | AR | Sponge network | -0.245 | 0.61313 | -0.439 | 0.04112 | 0.257 |
| 117 | CTD-2201E18.3 | hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | AR | Sponge network | 0.026 | 0.8392 | -0.439 | 0.04112 | 0.255 |
| 118 | RP11-379B8.1 | hsa-miR-106a-5p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p | 16 | AR | Sponge network | 0.842 | 0.05958 | -0.439 | 0.04112 | 0.254 |
| 119 | CTD-2013N24.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | AR | Sponge network | -0.665 | 0 | -0.439 | 0.04112 | 0.254 |
| 120 | NNT-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135a-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | AR | Sponge network | -0.678 | 0 | -0.439 | 0.04112 | 0.25 |