| Num |
lncRNA |
miRNAs |
miRNAs count |
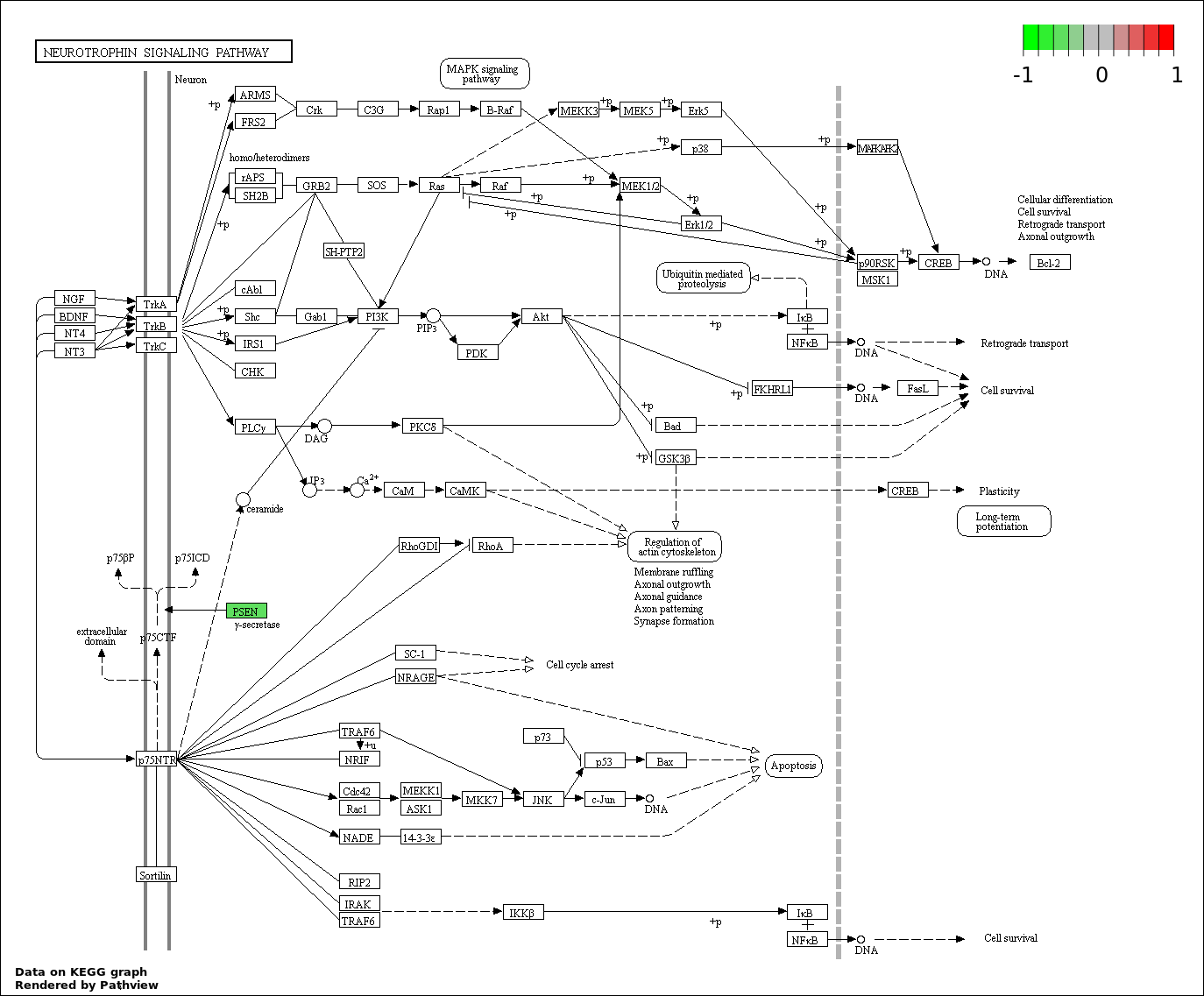
Gene |
Sponge regulatory network |
lncRNA log2FC |
lncRNA pvalue |
Gene log2FC |
Gene pvalue |
lncRNA-gene Pearson correlation |
| 1 |
RP11-10C24.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p |
11 |
KAT2B |
Sponge network |
-1.32 |
0 |
-0.701 |
0 |
0.67 |
| 2 |
DNAJC3-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.475 |
0 |
-0.701 |
0 |
0.654 |
| 3 |
FGD5-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
17 |
KAT2B |
Sponge network |
-1.192 |
0 |
-0.701 |
0 |
0.647 |
| 4 |
RP11-293M10.6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-1.903 |
0 |
-0.701 |
0 |
0.622 |
| 5 |
IQCH-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p |
18 |
KAT2B |
Sponge network |
-1.227 |
0 |
-0.701 |
0 |
0.615 |
| 6 |
RP11-337C18.8
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-0.982 |
0 |
-0.701 |
0 |
0.615 |
| 7 |
AC006116.24
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-1.468 |
0 |
-0.701 |
0 |
0.614 |
| 8 |
NNT-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-0.871 |
0 |
-0.701 |
0 |
0.608 |
| 9 |
RBM26-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.964 |
0 |
-0.701 |
0 |
0.605 |
| 10 |
LINC00472
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-2.49 |
0 |
-0.701 |
0 |
0.604 |
| 11 |
RP11-677M14.7
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.119 |
0 |
-0.701 |
0 |
0.603 |
| 12 |
KB-431C1.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-1.126 |
0 |
-0.701 |
0 |
0.595 |
| 13 |
RP11-545I5.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
20 |
KAT2B |
Sponge network |
-1.231 |
0 |
-0.701 |
0 |
0.59 |
| 14 |
CASC2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
19 |
KAT2B |
Sponge network |
-1.291 |
0 |
-0.701 |
0 |
0.59 |
| 15 |
CTD-2619J13.17
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.126 |
0 |
-0.701 |
0 |
0.585 |
| 16 |
RP1-80N2.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-2.113 |
0 |
-0.701 |
0 |
0.582 |
| 17 |
RP11-10C24.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p |
14 |
KAT2B |
Sponge network |
-1.236 |
0 |
-0.701 |
0 |
0.581 |
| 18 |
RP11-473I1.9
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-0.457 |
0 |
-0.701 |
0 |
0.581 |
| 19 |
RP11-517P14.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-1.051 |
0 |
-0.701 |
0 |
0.57 |
| 20 |
LINC00271
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-2.152 |
0 |
-0.701 |
0 |
0.565 |
| 21 |
RP11-15H20.6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p |
17 |
KAT2B |
Sponge network |
-1.192 |
0 |
-0.701 |
0 |
0.563 |
| 22 |
LINC01024
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.49 |
0 |
-0.701 |
0 |
0.561 |
| 23 |
AC005082.12
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-4.329 |
0 |
-0.701 |
0 |
0.56 |
| 24 |
RP11-362F19.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-3.705 |
0 |
-0.701 |
0 |
0.552 |
| 25 |
WDFY3-AS2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p |
16 |
KAT2B |
Sponge network |
-1.454 |
0 |
-0.701 |
0 |
0.55 |
| 26 |
RP11-890B15.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-0.759 |
0 |
-0.701 |
0 |
0.531 |
| 27 |
CTD-2366F13.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.058 |
0 |
-0.701 |
0 |
0.53 |
| 28 |
AC007246.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
18 |
KAT2B |
Sponge network |
-0.995 |
0 |
-0.701 |
0 |
0.529 |
| 29 |
DHRS4-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p |
13 |
KAT2B |
Sponge network |
-1.329 |
0 |
-0.701 |
0 |
0.525 |
| 30 |
RP11-247L20.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-1.23 |
0 |
-0.701 |
0 |
0.52 |
| 31 |
CTD-2015H6.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-1.982 |
0 |
-0.701 |
0 |
0.518 |
| 32 |
BDNF-AS
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.265 |
0 |
-0.701 |
0 |
0.514 |
| 33 |
CTD-2184D3.6
|
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.587 |
0 |
-0.701 |
0 |
0.514 |
| 34 |
PRKCQ-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p |
13 |
KAT2B |
Sponge network |
-1.703 |
0 |
-0.701 |
0 |
0.507 |
| 35 |
GAS6-AS2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-92b-3p |
14 |
KAT2B |
Sponge network |
-1.733 |
0 |
-0.701 |
0 |
0.502 |
| 36 |
DPH6-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-5p;hsa-miR-92b-3p |
14 |
KAT2B |
Sponge network |
-1.844 |
0 |
-0.701 |
0 |
0.494 |
| 37 |
LINC00865
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p |
10 |
KAT2B |
Sponge network |
-1.342 |
0 |
-0.701 |
0 |
0.493 |
| 38 |
RP11-57H14.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.628 |
0 |
-0.701 |
0 |
0.49 |
| 39 |
RP11-102N12.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.742 |
0 |
-0.701 |
0 |
0.488 |
| 40 |
RP11-16N11.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p |
10 |
KAT2B |
Sponge network |
-1.051 |
0 |
-0.701 |
0 |
0.482 |
| 41 |
MAGI2-AS3
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.733 |
0 |
-0.701 |
0 |
0.477 |
| 42 |
RP11-4O1.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p |
15 |
KAT2B |
Sponge network |
-1.18 |
0 |
-0.701 |
0 |
0.475 |
| 43 |
PWAR6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
18 |
KAT2B |
Sponge network |
-1.331 |
0 |
-0.701 |
0 |
0.475 |
| 44 |
RP1-39G22.7
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p |
10 |
KAT2B |
Sponge network |
-0.984 |
0 |
-0.701 |
0 |
0.473 |
| 45 |
RP11-502I4.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-1.257 |
0 |
-0.701 |
0 |
0.473 |
| 46 |
U91328.19
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.068 |
0 |
-0.701 |
0 |
0.472 |
| 47 |
RP11-250B2.6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.544 |
0 |
-0.701 |
0 |
0.472 |
| 48 |
AC062029.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.579 |
0 |
-0.701 |
0 |
0.47 |
| 49 |
RP11-53O19.1
|
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-1.037 |
0 |
-0.701 |
0 |
0.469 |
| 50 |
EIF3J-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.524 |
0 |
-0.701 |
0 |
0.463 |
| 51 |
RP11-110G21.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.912 |
0 |
-0.701 |
0 |
0.462 |
| 52 |
ZNF667-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-1.858 |
0 |
-0.701 |
0 |
0.459 |
| 53 |
RP11-456K23.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-1.035 |
0 |
-0.701 |
0 |
0.459 |
| 54 |
LINC00284
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-4.379 |
0 |
-0.701 |
0 |
0.455 |
| 55 |
RP11-517B11.7
|
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-0.622 |
0 |
-0.701 |
0 |
0.454 |
| 56 |
RP11-321G12.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-3.994 |
0 |
-0.701 |
0 |
0.454 |
| 57 |
TPTEP1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
17 |
KAT2B |
Sponge network |
-3.09 |
0 |
-0.701 |
0 |
0.451 |
| 58 |
ZBED5-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p |
10 |
KAT2B |
Sponge network |
-1.149 |
0 |
-0.701 |
0 |
0.449 |
| 59 |
UGDH-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-0.601 |
0 |
-0.701 |
0 |
0.448 |
| 60 |
CTD-2554C21.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-2.124 |
0 |
-0.701 |
0 |
0.448 |
| 61 |
AC074286.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-1.311 |
0 |
-0.701 |
0 |
0.447 |
| 62 |
RP11-15F12.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-2.173 |
0 |
-0.701 |
0 |
0.445 |
| 63 |
AC007255.8
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p |
10 |
KAT2B |
Sponge network |
-3.65 |
0 |
-0.701 |
0 |
0.444 |
| 64 |
RP11-44F14.8
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-5.468 |
0 |
-0.701 |
0 |
0.437 |
| 65 |
RP11-714L20.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-2.961 |
0 |
-0.701 |
0 |
0.434 |
| 66 |
RP11-834C11.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-2.522 |
0 |
-0.701 |
0 |
0.433 |
| 67 |
RP11-728F11.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-92b-3p |
12 |
KAT2B |
Sponge network |
-4.422 |
0 |
-0.701 |
0 |
0.43 |
| 68 |
LINC00675
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-92b-3p |
15 |
KAT2B |
Sponge network |
-8.255 |
0 |
-0.701 |
0 |
0.429 |
| 69 |
RP11-84G21.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.403 |
0 |
-0.701 |
0 |
0.429 |
| 70 |
AC068138.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-9.112 |
0 |
-0.701 |
0 |
0.428 |
| 71 |
CTD-2008P7.9
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p |
14 |
KAT2B |
Sponge network |
-11.092 |
0 |
-0.701 |
0 |
0.426 |
| 72 |
LINC00886
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.075 |
0 |
-0.701 |
0 |
0.425 |
| 73 |
AF131215.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-1.268 |
0 |
-0.701 |
0 |
0.421 |
| 74 |
RP11-439E19.10
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-0.995 |
0 |
-0.701 |
0 |
0.421 |
| 75 |
PSMG3-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-92b-3p |
13 |
KAT2B |
Sponge network |
-1.398 |
0 |
-0.701 |
0 |
0.421 |
| 76 |
CBR3-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-1.217 |
0 |
-0.701 |
0 |
0.42 |
| 77 |
SDCBP2-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-92b-3p |
14 |
KAT2B |
Sponge network |
-0.904 |
0 |
-0.701 |
0 |
0.419 |
| 78 |
SNHG14
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
19 |
KAT2B |
Sponge network |
-0.788 |
0 |
-0.701 |
0 |
0.419 |
| 79 |
LINC00863
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-0.878 |
0 |
-0.701 |
0 |
0.413 |
| 80 |
CTD-2243E23.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p |
10 |
KAT2B |
Sponge network |
-9.815 |
0 |
-0.701 |
0 |
0.411 |
| 81 |
RP11-789C1.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-11.363 |
0 |
-0.701 |
0 |
0.411 |
| 82 |
RP11-999E24.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p |
10 |
KAT2B |
Sponge network |
-3.92 |
0 |
-0.701 |
0 |
0.411 |
| 83 |
LINC00982
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-7.802 |
0 |
-0.701 |
0 |
0.41 |
| 84 |
LINC00864
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p |
12 |
KAT2B |
Sponge network |
-9.633 |
0 |
-0.701 |
0 |
0.408 |
| 85 |
RP11-352D13.6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-2.556 |
0 |
-0.701 |
0 |
0.408 |
| 86 |
RP11-98I9.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p |
10 |
KAT2B |
Sponge network |
-0.973 |
0 |
-0.701 |
0 |
0.407 |
| 87 |
LINC01003
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.658 |
0 |
-0.701 |
0 |
0.405 |
| 88 |
UBAC2-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-5p |
12 |
KAT2B |
Sponge network |
-1.454 |
0 |
-0.701 |
0 |
0.404 |
| 89 |
RP4-655J12.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-92b-3p |
11 |
KAT2B |
Sponge network |
-5.771 |
0 |
-0.701 |
0 |
0.403 |
| 90 |
LINC01020
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p |
14 |
KAT2B |
Sponge network |
-8.577 |
0 |
-0.701 |
0 |
0.403 |
| 91 |
FAM66C
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p |
11 |
KAT2B |
Sponge network |
-0.577 |
0 |
-0.701 |
0 |
0.403 |
| 92 |
RN7SL832P
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.834 |
0 |
-0.701 |
0 |
0.397 |
| 93 |
AC007228.11
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p |
10 |
KAT2B |
Sponge network |
-1.532 |
0 |
-0.701 |
0 |
0.393 |
| 94 |
FGF14-AS2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p |
11 |
KAT2B |
Sponge network |
-1.427 |
0 |
-0.701 |
0 |
0.392 |
| 95 |
RP11-320H14.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p |
12 |
KAT2B |
Sponge network |
-12.117 |
0 |
-0.701 |
0 |
0.391 |
| 96 |
RP11-327P2.5
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-5p |
14 |
KAT2B |
Sponge network |
-1.519 |
0 |
-0.701 |
0 |
0.39 |
| 97 |
RP11-156P1.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p |
13 |
KAT2B |
Sponge network |
-0.495 |
0 |
-0.701 |
0 |
0.389 |
| 98 |
AP000230.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-92b-3p |
11 |
KAT2B |
Sponge network |
-1.846 |
0 |
-0.701 |
0 |
0.389 |
| 99 |
RP11-742D12.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-8.559 |
0 |
-0.701 |
0 |
0.387 |
| 100 |
RP11-317M11.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-10.354 |
0 |
-0.701 |
0 |
0.386 |
| 101 |
ST3GAL6-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p |
13 |
KAT2B |
Sponge network |
-2.538 |
0 |
-0.701 |
0 |
0.382 |
| 102 |
RP11-355O1.11
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.832 |
0 |
-0.701 |
0 |
0.381 |
| 103 |
RP11-299H21.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.716 |
0 |
-0.701 |
0 |
0.381 |
| 104 |
LINC01018
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-4.305 |
0 |
-0.701 |
0 |
0.379 |
| 105 |
CHL1-AS2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p |
10 |
KAT2B |
Sponge network |
-12.213 |
0 |
-0.701 |
0 |
0.377 |
| 106 |
AC073254.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-576-5p |
12 |
KAT2B |
Sponge network |
-1.077 |
0 |
-0.701 |
0 |
0.377 |
| 107 |
SRP14-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.21 |
0 |
-0.701 |
0 |
0.376 |
| 108 |
HCG18
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-0.374 |
0 |
-0.701 |
0 |
0.375 |
| 109 |
ACVR2B-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.582 |
0 |
-0.701 |
0 |
0.373 |
| 110 |
LINC00882
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.3 |
0 |
-0.701 |
0 |
0.372 |
| 111 |
RP11-44F14.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-6.649 |
0 |
-0.701 |
0 |
0.371 |
| 112 |
CTA-221G9.11
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-1.18 |
0 |
-0.701 |
0 |
0.37 |
| 113 |
SMC5-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.187 |
0 |
-0.701 |
0 |
0.368 |
| 114 |
RP11-63A11.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-8.866 |
0 |
-0.701 |
0 |
0.364 |
| 115 |
AC144831.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-3.147 |
0 |
-0.701 |
0 |
0.363 |
| 116 |
LINC00551
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p |
12 |
KAT2B |
Sponge network |
-5.461 |
0 |
-0.701 |
0 |
0.36 |
| 117 |
RP11-6N17.4
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.433 |
0 |
-0.701 |
0 |
0.359 |
| 118 |
RP11-352D13.5
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-2.315 |
0 |
-0.701 |
0 |
0.359 |
| 119 |
RP11-297L17.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-3.465 |
0 |
-0.701 |
0 |
0.359 |
| 120 |
RP11-116O18.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-5.451 |
0 |
-0.701 |
0 |
0.359 |
| 121 |
CTA-445C9.14
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-0.872 |
0 |
-0.701 |
0 |
0.358 |
| 122 |
RP11-539I5.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-0.851 |
0 |
-0.701 |
0 |
0.358 |
| 123 |
RP11-1070N10.6
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-4.572 |
0 |
-0.701 |
0 |
0.356 |
| 124 |
CTD-2017D11.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-92b-3p |
12 |
KAT2B |
Sponge network |
-1.024 |
0 |
-0.701 |
0 |
0.356 |
| 125 |
RP11-22C11.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-7.876 |
0 |
-0.701 |
0 |
0.355 |
| 126 |
RP11-7F18.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p |
11 |
KAT2B |
Sponge network |
-0.851 |
0 |
-0.701 |
0 |
0.355 |
| 127 |
RP11-513O17.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-9.646 |
0 |
-0.701 |
0 |
0.354 |
| 128 |
LINC00323
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p |
11 |
KAT2B |
Sponge network |
-3.06 |
0 |
-0.701 |
0 |
0.354 |
| 129 |
LINC00645
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-8.028 |
0 |
-0.701 |
0 |
0.351 |
| 130 |
NR2F1-AS1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.075 |
0 |
-0.701 |
0 |
0.347 |
| 131 |
ILF3-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.012 |
0 |
-0.701 |
0 |
0.345 |
| 132 |
EMX2OS
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-1.604 |
0 |
-0.701 |
0 |
0.344 |
| 133 |
CTB-92J24.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-7.279 |
0 |
-0.701 |
0 |
0.34 |
| 134 |
RP11-30K9.6
|
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.944 |
0 |
-0.701 |
0 |
0.339 |
| 135 |
ZNF582-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-2.031 |
0 |
-0.701 |
0 |
0.332 |
| 136 |
TCL6
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-4.152 |
0 |
-0.701 |
0 |
0.329 |
| 137 |
TRHDE-AS1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.953 |
0 |
-0.701 |
0 |
0.328 |
| 138 |
TMEM9B-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.688 |
0 |
-0.701 |
0 |
0.327 |
| 139 |
CTD-2619J13.14
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-92b-3p |
11 |
KAT2B |
Sponge network |
-0.644 |
0 |
-0.701 |
0 |
0.322 |
| 140 |
SSTR5-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p |
12 |
KAT2B |
Sponge network |
-7.615 |
0 |
-0.701 |
0 |
0.321 |
| 141 |
AC093627.10
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p |
10 |
KAT2B |
Sponge network |
-0.79 |
0 |
-0.701 |
0 |
0.317 |
| 142 |
RP11-438D14.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-576-5p;hsa-miR-590-3p |
13 |
KAT2B |
Sponge network |
-7.831 |
0 |
-0.701 |
0 |
0.317 |
| 143 |
LINC00476
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-0.818 |
0 |
-0.701 |
0 |
0.317 |
| 144 |
AC008697.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-3.876 |
0 |
-0.701 |
0 |
0.317 |
| 145 |
RP11-231E6.1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-2.499 |
0 |
-0.701 |
0 |
0.316 |
| 146 |
RP11-130L8.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.646 |
0 |
-0.701 |
0 |
0.315 |
| 147 |
PART1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
19 |
KAT2B |
Sponge network |
-7.348 |
0 |
-0.701 |
0 |
0.315 |
| 148 |
RP5-952N6.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p |
10 |
KAT2B |
Sponge network |
-3.865 |
0 |
-0.701 |
0 |
0.315 |
| 149 |
RASSF8-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
16 |
KAT2B |
Sponge network |
-1.219 |
0 |
-0.701 |
0 |
0.315 |
| 150 |
AC109642.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-2.053 |
0 |
-0.701 |
0 |
0.315 |
| 151 |
RP11-1299A16.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-1.078 |
0.00018 |
-0.701 |
0 |
0.311 |
| 152 |
RP11-86H7.7
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p |
11 |
KAT2B |
Sponge network |
-1.11 |
0 |
-0.701 |
0 |
0.311 |
| 153 |
ENTPD3-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
15 |
KAT2B |
Sponge network |
-1.351 |
0 |
-0.701 |
0 |
0.31 |
| 154 |
LINC00885
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p |
11 |
KAT2B |
Sponge network |
-7.108 |
0 |
-0.701 |
0 |
0.31 |
| 155 |
CECR7
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-1.982 |
0 |
-0.701 |
0 |
0.308 |
| 156 |
RP11-384L8.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p |
10 |
KAT2B |
Sponge network |
-0.976 |
0 |
-0.701 |
0 |
0.307 |
| 157 |
ZNF503-AS2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p |
13 |
KAT2B |
Sponge network |
-2.21 |
0 |
-0.701 |
0 |
0.305 |
| 158 |
ATP6V0E2-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-1.685 |
0 |
-0.701 |
0 |
0.303 |
| 159 |
MIR22HG
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p |
17 |
KAT2B |
Sponge network |
-1.03 |
0 |
-0.701 |
0 |
0.303 |
| 160 |
CKMT2-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.603 |
0 |
-0.701 |
0 |
0.295 |
| 161 |
DANCR
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-5p |
12 |
KAT2B |
Sponge network |
-2.022 |
0 |
-0.701 |
0 |
0.292 |
| 162 |
LINC00467
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.734 |
0 |
-0.701 |
0 |
0.291 |
| 163 |
RP11-401P9.4
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-93-5p |
13 |
KAT2B |
Sponge network |
-3.455 |
0 |
-0.701 |
0 |
0.29 |
| 164 |
RP11-822E23.8
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-2.223 |
0 |
-0.701 |
0 |
0.289 |
| 165 |
WEE2-AS1
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
12 |
KAT2B |
Sponge network |
-0.803 |
0 |
-0.701 |
0 |
0.289 |
| 166 |
AF131215.8
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-590-3p |
10 |
KAT2B |
Sponge network |
-3.154 |
0 |
-0.701 |
0 |
0.287 |
| 167 |
SOCS2-AS1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-1.173 |
0 |
-0.701 |
0 |
0.287 |
| 168 |
RP11-392P7.6
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-0.981 |
0 |
-0.701 |
0 |
0.285 |
| 169 |
LINC00924
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-1.854 |
0 |
-0.701 |
0 |
0.283 |
| 170 |
AC093323.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.387 |
0 |
-0.701 |
0 |
0.283 |
| 171 |
RP11-456H18.2
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p |
11 |
KAT2B |
Sponge network |
-3.094 |
0 |
-0.701 |
0 |
0.28 |
| 172 |
RAMP2-AS1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.47 |
0.0012 |
-0.701 |
0 |
0.279 |
| 173 |
RP11-774O3.3
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p |
11 |
KAT2B |
Sponge network |
-0.742 |
0 |
-0.701 |
0 |
0.277 |
| 174 |
RP11-1260E13.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-92b-3p |
11 |
KAT2B |
Sponge network |
-1.311 |
0 |
-0.701 |
0 |
0.271 |
| 175 |
LINC00473
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-6.383 |
0 |
-0.701 |
0 |
0.268 |
| 176 |
RP11-96H17.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-3.639 |
0 |
-0.701 |
0 |
0.267 |
| 177 |
RP11-46C24.7
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-0.419 |
1.0E-5 |
-0.701 |
0 |
0.266 |
| 178 |
AC003102.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
11 |
KAT2B |
Sponge network |
-2.738 |
0 |
-0.701 |
0 |
0.264 |
| 179 |
RP1-78O14.1
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-576-5p |
10 |
KAT2B |
Sponge network |
-2.751 |
0 |
-0.701 |
0 |
0.264 |
| 180 |
FAM201A
|
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p |
14 |
KAT2B |
Sponge network |
-2.229 |
0 |
-0.701 |
0 |
0.257 |
| 181 |
AC022431.3
|
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p |
10 |
KAT2B |
Sponge network |
-4.576 |
0 |
-0.701 |
0 |
0.252 |