This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7b-5p | ABL1 | -0.23 | 0.93895 | 0.02 | 0.9921 | miRNAWalker2 validate | -0.1 | 0.00372 | NA | |
| 2 | hsa-miR-128-3p | ABL1 | -0.44 | 0.78389 | 0.02 | 0.9921 | MirTarget | -0.12 | 0.00042 | NA | |
| 3 | hsa-miR-30a-5p | ANAPC5 | 0.22 | 0.93395 | -0.16 | 0.92347 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 4 | hsa-miR-324-5p | ATM | -0.5 | 0.53742 | 0.19 | 0.86463 | miRanda | -0.12 | 0.00155 | NA | |
| 5 | hsa-miR-455-5p | ATM | -0.14 | 0.86574 | 0.19 | 0.86463 | miRanda | -0.12 | 0.00128 | NA | |
| 6 | hsa-miR-590-5p | ATM | -0.55 | 0.47274 | 0.19 | 0.86463 | mirMAP | -0.1 | 0.00142 | NA | |
| 7 | hsa-miR-139-5p | BUB1 | -0.08 | 0.92869 | -0.31 | 0.8008 | miRanda | -0.15 | 0.00112 | NA | |
| 8 | hsa-miR-199a-5p | BUB1 | 0.16 | 0.9358 | -0.31 | 0.8008 | miRanda | -0.22 | 0 | NA | |
| 9 | hsa-miR-199b-5p | BUB1 | -0.04 | 0.97717 | -0.31 | 0.8008 | miRanda | -0.15 | 0.0005 | NA | |
| 10 | hsa-miR-139-5p | BUB3 | -0.08 | 0.92869 | -0.11 | 0.94592 | miRanda | -0.17 | 0 | NA | |
| 11 | hsa-miR-130a-3p | CCNA2 | 0.09 | 0.9291 | -0.31 | 0.81453 | miRNATAP | -0.17 | 0.00109 | NA | |
| 12 | hsa-miR-199a-5p | CCNA2 | 0.16 | 0.9358 | -0.31 | 0.81453 | miRanda | -0.27 | 0 | NA | |
| 13 | hsa-miR-199b-5p | CCNA2 | -0.04 | 0.97717 | -0.31 | 0.81453 | miRanda | -0.16 | 0.00129 | NA | |
| 14 | hsa-miR-218-5p | CCNA2 | 0.23 | 0.81021 | -0.31 | 0.81453 | MirTarget | -0.29 | 0 | NA | |
| 15 | hsa-miR-29c-3p | CCNA2 | 0.16 | 0.94272 | -0.31 | 0.81453 | MirTarget | -0.17 | 0.00117 | NA | |
| 16 | hsa-miR-34c-5p | CCNA2 | -0.02 | 0.95279 | -0.31 | 0.81453 | miRanda | -0.25 | 0 | NA | |
| 17 | hsa-miR-133b | CCNB1 | 0.39 | 0.56076 | -0.28 | 0.85013 | miRanda | -0.13 | 0 | NA | |
| 18 | hsa-miR-139-5p | CCNB1 | -0.08 | 0.92869 | -0.28 | 0.85013 | miRanda | -0.18 | 0.00084 | NA | |
| 19 | hsa-let-7c-5p | CCNB2 | -0.06 | 0.9749 | -0.2 | 0.87078 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 20 | hsa-miR-23b-3p | CCNB2 | -0.11 | 0.96135 | -0.2 | 0.87078 | miRNAWalker2 validate | -0.33 | 0.00011 | NA | |
| 21 | hsa-miR-16-5p | CCND1 | 0.01 | 0.99448 | -0.25 | 0.89067 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.13 | 0.00823 | 23991964; 22922827; 18483394 | At the molecular level our results further revealed that cyclin D1 expression was negatively regulated by miR-16;CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 22 | hsa-miR-195-5p | CCND1 | 0.34 | 0.74962 | -0.25 | 0.89067 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.14 | 0.00272 | 21350001; 26631043; 25823925 | Raf-1 and Ccnd1 were identified as novel direct targets of miR-195 and miR-497 miR-195/497 expression levels in clinical specimens were found to be correlated inversely with malignancy of breast cancer;MiR 195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1; The present study was to evaluate the level of miR-195 and cyclin D1 in CC tissues and cells; We further investigated the molecular mechanisms of miR-195 and cyclin D1 in CC cell lines HeLa and SiHa; Furthermore the expression of miR-195 was inversely proportional to that of cyclin D1 mRNA or protein p = 0.013 p = 0.015 respectively; However the inhibitor of miR-195 promoted the expression of cyclin D1 and cell proliferation; In conclusion our data suggest that miR-195 may have the potential role in treatment of CC patients as well as miR-195 is a novel regulator of invasiveness and tumorigenicity in CC cells by targeting cyclin D1;MicroRNA profiling identifies MiR 195 suppresses osteosarcoma cell metastasis by targeting CCND1; Meanwhile CCND1 was identified as the target gene of miR-195 and further studied; More importantly using real-time PCR we evaluated the expression of miR-195 and CCND1 in osteosarcoma samples from 107 frozen biopsy tissues and 99 formalin- or paraformalin-fixed paraffin-embedded FFPE tissues; Results indicated lowly expressed miR-195 or highly CCND1 correlated with positive overall survival and their expression inversely related to each other; In summary our study suggests miR-195 functions as a tumor metastasis suppressor gene by down-regulating CCND1 and can be used as a potential target in the treatment of osteosarcoma |
| 23 | hsa-miR-29c-3p | CCND1 | 0.16 | 0.94272 | -0.25 | 0.89067 | mirMAP | -0.14 | 0.00181 | NA | |
| 24 | hsa-miR-125b-5p | CCNE1 | 0.04 | 0.98059 | -0.28 | 0.73651 | miRNAWalker2 validate | -0.15 | 0.00074 | NA | |
| 25 | hsa-miR-195-5p | CCNE1 | 0.34 | 0.74962 | -0.28 | 0.73651 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.18 | 0.00302 | 24402230 | Furthermore through qPCR and western blot assays we showed that overexpression of miR-195-5p reduced CCNE1 mRNA and protein levels respectively |
| 26 | hsa-miR-26a-5p | CCNE2 | 0.01 | 0.99772 | -0.15 | 0.8473 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.36 | 0.00084 | 24116110; 21901171 | The loss of miR 26a mediated post transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival; The in vitro and in vivo assays showed that overexpression of miR-26a resulted in cell cycle arrest inhibited cell proliferation and decreased tumor growth which was associated with cyclin E2 downregulation;We also show that enforced expression of miR-26a in AML cells is able to inhibit cell cycle progression by downregulating cyclin E2 expression |
| 27 | hsa-miR-30a-5p | CCNE2 | 0.22 | 0.93395 | -0.15 | 0.8473 | miRNATAP | -0.44 | 0 | NA | |
| 28 | hsa-miR-34a-5p | CCNE2 | -0.5 | 0.74203 | -0.15 | 0.8473 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.22 | 0.00018 | NA | |
| 29 | hsa-miR-34c-5p | CCNE2 | -0.02 | 0.95279 | -0.15 | 0.8473 | miRNAWalker2 validate; miRTarBase; PITA; miRanda; miRNATAP | -0.28 | 0 | NA | |
| 30 | hsa-let-7a-3p | CDC14A | -0.22 | 0.85543 | 0.02 | 0.9701 | mirMAP | -0.15 | 0.00458 | NA | |
| 31 | hsa-miR-141-3p | CDC14A | -0.32 | 0.87774 | 0.02 | 0.9701 | TargetScan; miRNATAP | -0.13 | 0.00223 | NA | |
| 32 | hsa-miR-324-5p | CDC14A | -0.5 | 0.53742 | 0.02 | 0.9701 | miRanda | -0.13 | 0.00826 | NA | |
| 33 | hsa-miR-338-3p | CDC14A | 0.49 | 0.78848 | 0.02 | 0.9701 | miRanda | -0.12 | 0.00053 | NA | |
| 34 | hsa-miR-576-5p | CDC14A | -0.51 | 0.41719 | 0.02 | 0.9701 | mirMAP | -0.13 | 0.00588 | NA | |
| 35 | hsa-miR-139-5p | CDC16 | -0.08 | 0.92869 | 0.44 | 0.76333 | miRanda | -0.11 | 0.00566 | NA | |
| 36 | hsa-miR-146b-5p | CDC16 | -0.4 | 0.83751 | 0.44 | 0.76333 | miRanda | -0.15 | 0.00069 | NA | |
| 37 | hsa-miR-23b-3p | CDC20 | -0.11 | 0.96135 | -0.26 | 0.85563 | miRNAWalker2 validate | -0.32 | 0.00074 | NA | |
| 38 | hsa-miR-30a-5p | CDC20 | 0.22 | 0.93395 | -0.26 | 0.85563 | miRNAWalker2 validate | -0.33 | 3.0E-5 | NA | |
| 39 | hsa-miR-125a-5p | CDC23 | -0 | 0.99916 | 0 | 0.99805 | miRanda | -0.1 | 0.00117 | NA | |
| 40 | hsa-miR-34c-5p | CDC23 | -0.02 | 0.95279 | 0 | 0.99805 | miRanda | -0.14 | 0 | 25064703 | In addition the levels of CDC23 an important mediator in mitotic progression were suppressed following miR-34c expression and siRNAs targeting CDC23 mimicked the effect of miR-34c on G2/M arrest |
| 41 | hsa-let-7c-5p | CDC25A | -0.06 | 0.9749 | -0.28 | 0.78493 | MirTarget | -0.22 | 0 | 25909324 | MicroRNA let 7c Inhibits Cell Proliferation and Induces Cell Cycle Arrest by Targeting CDC25A in Human Hepatocellular Carcinoma; The aim of the present study was to determine whether the cell cycle regulator CDC25A is involved in the antitumor effect of let-7c in HCC; The luciferase reporter assay showed that CDC25A was a direct target of let-7c and that let-7c inhibited the expression of CDC25A protein by directly targeting its 3' UTR; In conclusion this study indicates that let-7c suppresses HCC progression possibly by directly targeting the cell cycle regulator CDC25A and indirectly affecting its downstream target molecules |
| 42 | hsa-miR-195-5p | CDC25A | 0.34 | 0.74962 | -0.28 | 0.78493 | MirTarget; miRNATAP | -0.19 | 0.00029 | NA | |
| 43 | hsa-miR-34c-5p | CDC25A | -0.02 | 0.95279 | -0.28 | 0.78493 | miRNATAP | -0.21 | 7.0E-5 | 21321636 | Ectopic expression of miR-449b and miR-34c resulted in lowered adhesion activities by 28%-34% and in cell cycle arrests with increased cell number of 15.62% and 15.71% in G1 and with decreased cell number of 15.96% and 16.56% in S Cell cycle related proteins CDK6 and CDC25A were down-regulated; The decreases of CDK6 and CDC25A by miR-449b were 39% and 22% respecyively; 49% and 32% by miR-34c respectively |
| 44 | hsa-miR-497-5p | CDC25A | -0.01 | 0.98915 | -0.28 | 0.78493 | MirTarget; miRNATAP | -0.25 | 7.0E-5 | NA | |
| 45 | hsa-miR-497-5p | CDC27 | -0.01 | 0.98915 | -0.1 | 0.93883 | miRNATAP | -0.15 | 0 | NA | |
| 46 | hsa-miR-26a-5p | CDC6 | 0.01 | 0.99772 | -0.17 | 0.88968 | miRNAWalker2 validate | -0.39 | 0.00054 | 25100863; 27158389 | Here it is demonstrated that miR26a and miR26b inhibit replication licensing and the proliferation migration and invasion of lung cancer cells by targeting CDC6; The current study suggests that miR26a miR26b and CDC6 and factors regulating their expression represent potential cancer diagnostic and prognostic markers as well as anticancer targets;miR 26a inhibits the proliferation of ovarian cancer cells via regulating CDC6 expression; Bioinformatics analysis revealed Cdc6 was a target gene of miR-26a; dual-luciferase assay and validation assay showed miR-26a could act on the 3'UTR of Cdc6 to regulate Cdc6 expression; These findings suggest that miR-26a may act on the 3'UTR of Cdc6 to regulate Cdc6 expression which then inhibit the proliferation of ovarian cancer cells and induce their apoptosis |
| 47 | hsa-miR-199a-5p | CDC7 | 0.16 | 0.9358 | -0.5 | 0.532 | MirTarget; miRanda | -0.24 | 8.0E-5 | NA | |
| 48 | hsa-miR-199b-5p | CDC7 | -0.04 | 0.97717 | -0.5 | 0.532 | MirTarget; miRanda | -0.17 | 0.00178 | NA | |
| 49 | hsa-miR-34c-5p | CDC7 | -0.02 | 0.95279 | -0.5 | 0.532 | miRanda | -0.18 | 0.00111 | NA | |
| 50 | hsa-miR-23b-3p | CDK2 | -0.11 | 0.96135 | -0.01 | 0.99411 | miRNAWalker2 validate | -0.17 | 0.00275 | NA | |
| 51 | hsa-miR-195-5p | CDK4 | 0.34 | 0.74962 | -0.06 | 0.97429 | miRNAWalker2 validate; miRTarBase | -0.18 | 0 | NA | |
| 52 | hsa-miR-34c-5p | CDK4 | -0.02 | 0.95279 | -0.06 | 0.97429 | miRNAWalker2 validate; miRTarBase | -0.11 | 0.00308 | NA | |
| 53 | hsa-miR-21-5p | CDK6 | -0.15 | 0.97024 | 0.26 | 0.8515 | miRNAWalker2 validate; mirMAP | -0.32 | 5.0E-5 | NA | |
| 54 | hsa-miR-22-3p | CDK6 | 0.06 | 0.98656 | 0.26 | 0.8515 | miRNAWalker2 validate | -0.27 | 0.00121 | NA | |
| 55 | hsa-miR-139-5p | CDK7 | -0.08 | 0.92869 | -0.07 | 0.95086 | miRanda | -0.15 | 0.00011 | NA | |
| 56 | hsa-let-7c-5p | CDKN1A | -0.06 | 0.9749 | 0.1 | 0.95314 | MirTarget | -0.15 | 0.0033 | NA | |
| 57 | hsa-miR-101-3p | CDKN1A | 0.01 | 0.99704 | 0.1 | 0.95314 | MirTarget | -0.4 | 4.0E-5 | NA | |
| 58 | hsa-miR-708-5p | CDKN1A | -0.03 | 0.97493 | 0.1 | 0.95314 | MirTarget | -0.14 | 0.00964 | NA | |
| 59 | hsa-miR-139-5p | CDKN1B | -0.08 | 0.92869 | -0.15 | 0.91962 | miRanda | -0.13 | 0.00103 | NA | |
| 60 | hsa-miR-342-3p | CDKN1B | -0.37 | 0.77314 | -0.15 | 0.91962 | miRanda | -0.12 | 0.00143 | NA | |
| 61 | hsa-miR-582-3p | CDKN1B | -0.23 | 0.89768 | -0.15 | 0.91962 | PITA | -0.11 | 0.00639 | NA | |
| 62 | hsa-miR-107 | CDKN1C | -0.04 | 0.98836 | 0.62 | 0.38943 | miRanda | -0.48 | 0.00088 | NA | |
| 63 | hsa-miR-25-3p | CDKN1C | -0.46 | 0.87857 | 0.62 | 0.38943 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.48 | 6.0E-5 | NA | |
| 64 | hsa-miR-335-5p | CDKN1C | -0.03 | 0.97338 | 0.62 | 0.38943 | miRNAWalker2 validate | -0.21 | 0.00911 | NA | |
| 65 | hsa-miR-429 | CDKN1C | -0.46 | 0.80624 | 0.62 | 0.38943 | miRNATAP | -0.23 | 0.00064 | NA | |
| 66 | hsa-miR-590-3p | CDKN1C | -0.28 | 0.59127 | 0.62 | 0.38943 | miRanda | -0.33 | 0 | NA | |
| 67 | hsa-miR-92a-3p | CDKN1C | 0.03 | 0.99325 | 0.62 | 0.38943 | MirTarget; miRNATAP | -0.24 | 0.0066 | NA | |
| 68 | hsa-let-7g-5p | CDKN2A | -0.2 | 0.92299 | 1.38 | 0.01803 | miRNAWalker2 validate; miRTarBase | -0.54 | 0.00297 | NA | |
| 69 | hsa-miR-19a-3p | CDKN2B | -0.21 | 0.84464 | 0.62 | 0.5094 | mirMAP | -0.41 | 0 | NA | |
| 70 | hsa-miR-19b-3p | CDKN2B | -0.03 | 0.98666 | 0.62 | 0.5094 | mirMAP | -0.56 | 0 | NA | |
| 71 | hsa-miR-335-3p | CDKN2B | -0.24 | 0.8845 | 0.62 | 0.5094 | mirMAP | -0.5 | 0 | NA | |
| 72 | hsa-miR-374b-5p | CDKN2B | -0.29 | 0.8357 | 0.62 | 0.5094 | miRNAWalker2 validate | -0.57 | 0 | NA | |
| 73 | hsa-miR-429 | CDKN2B | -0.46 | 0.80624 | 0.62 | 0.5094 | miRanda | -0.34 | 0 | NA | |
| 74 | hsa-miR-450b-5p | CDKN2B | -0.01 | 0.98315 | 0.62 | 0.5094 | mirMAP | -0.29 | 0.00227 | NA | |
| 75 | hsa-miR-501-3p | CDKN2B | -0.75 | 0.55276 | 0.62 | 0.5094 | PITA | -0.29 | 0.00051 | NA | |
| 76 | hsa-miR-501-5p | CDKN2B | -0.83 | 0.05827 | 0.62 | 0.5094 | mirMAP | -0.19 | 0.00497 | NA | |
| 77 | hsa-miR-502-3p | CDKN2B | -0.48 | 0.5143 | 0.62 | 0.5094 | PITA | -0.35 | 0.00035 | NA | |
| 78 | hsa-miR-576-5p | CDKN2B | -0.51 | 0.41719 | 0.62 | 0.5094 | MirTarget | -0.37 | 4.0E-5 | NA | |
| 79 | hsa-miR-671-5p | CDKN2B | -0.37 | 0.41121 | 0.62 | 0.5094 | PITA | -0.27 | 0.00459 | NA | |
| 80 | hsa-miR-7-1-3p | CDKN2B | -0.46 | 0.6659 | 0.62 | 0.5094 | MirTarget | -0.32 | 0.00025 | NA | |
| 81 | hsa-miR-98-5p | CDKN2B | -0.17 | 0.8988 | 0.62 | 0.5094 | miRNAWalker2 validate | -0.38 | 0.00234 | NA | |
| 82 | hsa-miR-139-5p | CHEK1 | -0.08 | 0.92869 | -0.28 | 0.78554 | miRanda | -0.13 | 0.00213 | NA | |
| 83 | hsa-miR-195-5p | CHEK1 | 0.34 | 0.74962 | -0.28 | 0.78554 | MirTarget; miRNATAP | -0.16 | 0.00025 | 25840419 | MiR 195 suppresses non small cell lung cancer by targeting CHEK1; We discovered that CHEK1 was a direct target of miR-195 which decreased CHEK1 expression in lung cancer cells |
| 84 | hsa-miR-497-5p | CHEK1 | -0.01 | 0.98915 | -0.28 | 0.78554 | MirTarget; miRNATAP | -0.21 | 5.0E-5 | 24464213 | Checkpoint kinase 1 is negatively regulated by miR 497 in hepatocellular carcinoma; In silico analysis showed that CHEK1 was a candidate target of miR-497 which was previously found to be downregulated in HCC by us; To test whether miR-497 could bind to 3'untranslated region 3'UTR of CHEK1 luciferase reporter assay was conducted; The result revealed that miR-497 could bind to the 3'untranslated region 3'UTR of CHEK1 mRNA; Western blot showed that ectopic expression of miR-497 suppressed the CHEK1 expression and inhibition of miR-497 led to significant upregulation of CHEK1; Finally miR-497 expression was measured in the same 30 HCC samples and the correlation between miR-497 and CHEK1 was analyzed; The results indicated that miR-497 was downregulated in HCC and had a significant negative correlation with CHEK1; Taken together these results demonstrated that CHEK1 was negatively regulated by miR-497 and the overexpressed CHEK1 was resulted from the downregulated miR-497 in HCC which provided a potential molecular target for HCC therapy |
| 85 | hsa-miR-493-5p | CREBBP | 0.23 | 0.73198 | -0.07 | 0.96314 | miRNATAP | -0.13 | 0.00036 | NA | |
| 86 | hsa-miR-199a-5p | DBF4 | 0.16 | 0.9358 | -0.13 | 0.9016 | miRanda | -0.17 | 0.00012 | NA | |
| 87 | hsa-miR-30a-5p | DBF4 | 0.22 | 0.93395 | -0.13 | 0.9016 | MirTarget | -0.22 | 0.00031 | NA | |
| 88 | hsa-miR-23b-3p | E2F1 | -0.11 | 0.96135 | -0.09 | 0.94114 | miRNAWalker2 validate | -0.28 | 0.00275 | NA | |
| 89 | hsa-let-7c-5p | E2F2 | -0.06 | 0.9749 | -0.41 | 0.65832 | MirTarget | -0.27 | 0 | NA | |
| 90 | hsa-miR-125b-5p | E2F2 | 0.04 | 0.98059 | -0.41 | 0.65832 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.24 | 0 | 22999819 | miR 125b regulates the proliferation of glioblastoma stem cells by targeting E2F2; This study demonstrated that miR-125b plays important roles in regulating the proliferation of GSCs by directly targeting E2F2 |
| 91 | hsa-miR-30c-2-3p | E2F2 | 0.24 | 0.74693 | -0.41 | 0.65832 | MirTarget | -0.28 | 0.00019 | NA | |
| 92 | hsa-miR-365a-3p | E2F2 | 0.08 | 0.93876 | -0.41 | 0.65832 | MirTarget | -0.26 | 0.00097 | NA | |
| 93 | hsa-let-7b-5p | E2F3 | -0.23 | 0.93895 | 0.05 | 0.96837 | miRNAWalker2 validate | -0.12 | 0.0057 | NA | |
| 94 | hsa-miR-92b-3p | E2F3 | -0.57 | 0.68932 | 0.05 | 0.96837 | miRNATAP | -0.1 | 0.00023 | NA | |
| 95 | hsa-miR-23b-3p | E2F4 | -0.11 | 0.96135 | 0.08 | 0.95952 | miRNAWalker2 validate | -0.17 | 0.00092 | NA | |
| 96 | hsa-let-7e-5p | E2F5 | 0.21 | 0.92234 | -0.16 | 0.86449 | MirTarget; miRNATAP | -0.27 | 0.00167 | NA | |
| 97 | hsa-miR-1271-5p | E2F5 | -0.46 | 0.09563 | -0.16 | 0.86449 | MirTarget | -0.15 | 0.00563 | NA | |
| 98 | hsa-miR-132-3p | E2F5 | -0.24 | 0.87175 | -0.16 | 0.86449 | MirTarget | -0.24 | 0.00573 | 27186275 | miR 132 targeting E2F5 suppresses cell proliferation invasion migration in ovarian cancer cells; Mechanism investigation revealed that miR-132 inhibited the expression of transcription factor E2F5 by specifically targeting its mRNA 3'UTR; Moreover the expression level of E2F5 was significantly increased in ovarian cancer tissues than in the adjacent normal tissues and its expression was inversely correlated with miR-132 expression in clinical ovarian cancer tissues; Additionally silencing E2F5 was able to inhibit the proliferation colony formation migration and invasion of ovarian cancer cells parallel to the effect of miR-132 overexpression on the ovarian cancer cells; Meanwhile overexpression of E2F5 reversed the inhibition effect mediated by miR-132 overexpression; These results indicate that miR-132 suppresses the cell proliferation invasion migration in ovarian cancer cells by targeting E2F5 |
| 99 | hsa-miR-142-3p | E2F5 | -0.15 | 0.9461 | -0.16 | 0.86449 | miRanda | -0.17 | 0.00024 | NA | |
| 100 | hsa-miR-320b | E2F5 | -0.24 | 0.85922 | -0.16 | 0.86449 | miRanda | -0.21 | 0.00078 | NA | |
| 101 | hsa-miR-34a-5p | E2F5 | -0.5 | 0.74203 | -0.16 | 0.86449 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.24 | 7.0E-5 | 26103003 | MicroRNA 34a targets FMNL2 and E2F5 and suppresses the progression of colorectal cancer; FMNL2 and E2F5 were identified as direct targets of miR-34a; Reintroduction of FMNL2 or E2F5 without 3'UTR region reversed the inhibitory effects of miR-34a on cell proliferation and invasion; MiR-34a was down-regulated in CRC cells and inversely correlated with FMNL2 and E2F5 expressions; Our study suggests that miR-34a is an important tumor suppressor of CRC progression by targeting FMNL2 and E2F5 thus providing new insight into the molecular mechanisms underlying CRC progression and establishing a strong potential for the application of miR-34a as a novel therapeutic marker against CRC |
| 102 | hsa-miR-34c-5p | E2F5 | -0.02 | 0.95279 | -0.16 | 0.86449 | MirTarget; PITA; miRanda; miRNATAP | -0.15 | 0.00886 | NA | |
| 103 | hsa-miR-106b-5p | EP300 | -0.3 | 0.86929 | -0.28 | 0.85828 | miRNATAP | -0.13 | 0.00096 | NA | |
| 104 | hsa-miR-26b-5p | EP300 | -0.02 | 0.99038 | -0.28 | 0.85828 | miRNAWalker2 validate; miRNATAP | -0.12 | 0.00816 | NA | |
| 105 | hsa-miR-339-5p | EP300 | -0.3 | 0.71291 | -0.28 | 0.85828 | miRanda | -0.11 | 0.00013 | NA | |
| 106 | hsa-miR-369-3p | EP300 | 0.12 | 0.8323 | -0.28 | 0.85828 | MirTarget; PITA; miRNATAP | -0.15 | 1.0E-5 | NA | |
| 107 | hsa-let-7c-5p | ESPL1 | -0.06 | 0.9749 | -0.28 | 0.80065 | MirTarget | -0.16 | 2.0E-5 | NA | |
| 108 | hsa-miR-324-3p | GADD45B | -0.42 | 0.66153 | 0.52 | 0.61353 | MirTarget; miRNATAP | -0.22 | 0.00285 | NA | |
| 109 | hsa-miR-590-3p | GADD45B | -0.28 | 0.59127 | 0.52 | 0.61353 | miRanda | -0.23 | 0 | NA | |
| 110 | hsa-miR-132-3p | GSK3B | -0.24 | 0.87175 | -0.03 | 0.98417 | mirMAP; miRNATAP | -0.11 | 0.00091 | NA | |
| 111 | hsa-miR-146b-3p | GSK3B | -0.41 | 0.76252 | -0.03 | 0.98417 | miRNATAP | -0.12 | 0 | NA | |
| 112 | hsa-miR-146b-5p | GSK3B | -0.4 | 0.83751 | -0.03 | 0.98417 | miRanda | -0.14 | 0 | NA | |
| 113 | hsa-miR-155-5p | GSK3B | -0.41 | 0.82867 | -0.03 | 0.98417 | miRNAWalker2 validate; miRNATAP | -0.1 | 0 | NA | |
| 114 | hsa-miR-212-3p | GSK3B | -0.02 | 0.97323 | -0.03 | 0.98417 | mirMAP; miRNATAP | -0.11 | 2.0E-5 | NA | |
| 115 | hsa-miR-30a-5p | HDAC1 | 0.22 | 0.93395 | -0.2 | 0.91427 | miRNAWalker2 validate | -0.22 | 1.0E-5 | NA | |
| 116 | hsa-miR-34c-5p | HDAC1 | -0.02 | 0.95279 | -0.2 | 0.91427 | miRanda; miRNATAP | -0.14 | 1.0E-5 | NA | |
| 117 | hsa-miR-132-3p | HDAC2 | -0.24 | 0.87175 | -0.09 | 0.95418 | mirMAP | -0.17 | 0.00036 | NA | |
| 118 | hsa-miR-155-5p | HDAC2 | -0.41 | 0.82867 | -0.09 | 0.95418 | mirMAP | -0.11 | 0.00036 | 21946536 | Mechanistically we found that BRCA1 epigenetically represses miR-155 expression via its association with HDAC2 which deacetylates histones H2A and H3 on the miR-155 promoter |
| 119 | hsa-miR-92b-3p | HDAC2 | -0.57 | 0.68932 | -0.09 | 0.95418 | mirMAP | -0.11 | 0.00023 | NA | |
| 120 | hsa-miR-133b | MAD2L1 | 0.39 | 0.56076 | -0.32 | 0.80244 | miRanda | -0.12 | 0 | NA | |
| 121 | hsa-miR-139-5p | MAD2L1 | -0.08 | 0.92869 | -0.32 | 0.80244 | miRanda | -0.2 | 9.0E-5 | NA | |
| 122 | hsa-miR-139-5p | MCM2 | -0.08 | 0.92869 | -0.26 | 0.86599 | miRanda | -0.14 | 0.00136 | NA | |
| 123 | hsa-miR-34c-5p | MCM2 | -0.02 | 0.95279 | -0.26 | 0.86599 | miRanda | -0.13 | 0.00246 | NA | |
| 124 | hsa-miR-23b-3p | MCM4 | -0.11 | 0.96135 | -0.38 | 0.8115 | MirTarget | -0.3 | 0.00061 | NA | |
| 125 | hsa-miR-34a-5p | MCM4 | -0.5 | 0.74203 | -0.38 | 0.8115 | miRNAWalker2 validate | -0.19 | 0.00032 | NA | |
| 126 | hsa-miR-30c-2-3p | MCM6 | 0.24 | 0.74693 | -0.35 | 0.80832 | MirTarget | -0.19 | 5.0E-5 | NA | |
| 127 | hsa-miR-143-3p | MDM2 | 0.37 | 0.92734 | -0.49 | 0.72349 | miRNAWalker2 validate | -0.16 | 0.00124 | NA | |
| 128 | hsa-miR-320b | MYC | -0.24 | 0.85922 | -0.45 | 0.81149 | miRNAWalker2 validate | -0.24 | 0.00032 | 26487644 | miR 320b suppresses cell proliferation by targeting c Myc in human colorectal cancer cells; Overexpression of miR-320b in CRC cells was statistically correlated with a decrease of cell growth in vitro and in vivo while c-MYC was identified as a target gene of miR-320b in CRC; Furthermore it was found that up-regulation of c-Myc can attenuate the effects induced by miR-320b; Our identification of c-MYC as a target gene of miR-320b provides new insights into the pathophysiology of CRC proliferation and identifies miR-320b as a novel therapeutic target for the treatment of CRC |
| 129 | hsa-miR-217 | PCNA | 0.34 | 0.761 | -0.32 | 0.85329 | miRanda; miRNATAP | -0.11 | 0.00292 | 25653720 | In vitro treatment with miR-217 mimics significantly suppressed the proliferation of MCF-7 cells induced G1 phase arrest and inhibited the expression of cyclin D1; while these effects were significantly reversed by the restoration of DACH1; In MDA-MB-231 cells treatment with miR-217 inhibitors enhanced the cellular proliferation promoted cell cycle progression and upregulated the expression of cyclin D1 which were neutralized by the pre-treatment of siRNA-DACH1; In vivo inhibition of miR-217 significantly suppressed the xenografts growth and downregulated the expression of cyclin D1 |
| 130 | hsa-miR-30a-5p | PCNA | 0.22 | 0.93395 | -0.32 | 0.85329 | miRNAWalker2 validate | -0.33 | 1.0E-5 | NA | |
| 131 | hsa-miR-100-5p | PLK1 | 0.02 | 0.99282 | -0.19 | 0.89035 | miRNAWalker2 validate; miRTarBase | -0.2 | 0 | 23151088; 22246341; 23842624; 25537513; 22120675; 21636267 | MicroRNA 100 is a potential molecular marker of non small cell lung cancer and functions as a tumor suppressor by targeting polo like kinase 1; By using microRNA miR target prediction algorithms we identified miR-100 that might potentially bind the 3'-untranslated region of PLK1 transcripts; The purpose of this study was to investigate the roles of miR-100 and its association with PLK1 in NSCLC development; Finally the effects of miR-100 expression on growth apoptosis and cell cycle of NSCLC cells by posttranscriptionally regulating PLK1 expression were determined; Meanwhile miR-100 mimics could significantly inhibit PLK1 mRNA and protein expression and reduce the luciferase activity of a PLK1 3' untranslated region-based reporter construct in A549 cells; Furthermore small interfering RNA siRNA-mediated PLK1 downregulation could mimic the effects of miR-100 mimics while PLK1 overexpression could partially rescue the phenotypical changes of NSCLC cells induced by miR-100 mimics; Our findings indicate that low miR-100 may be a poor prognostic factor for NSCLC patients and functions as a tumor suppressor by posttranscriptionally regulating PLK1 expression;Together these results suggest that low miR-100 expression may be an independent poor prognostic factor and miR-100 can function as a tumor suppressor by targeting PLK1 in human EOCs;In HCC tissues miR-100 expression was inversely correlated with the expression of plk1 protein r = -0.418; P = 0.029; Therefore downregulation of miR-100 was correlated with progressive pathological feature and poor prognosis in HCC patients and miR-100 could function as a tumor suppressor by targeting plk1;Here we show that miR-100 inhibits maintenance and expansion of BrCSCs in basal-like cancer through Polo-like kinase1 Plk1 down-regulation;MiR 100 resensitizes docetaxel resistant human lung adenocarcinoma cells SPC A1 to docetaxel by targeting Plk1; Knock-down of Plk1 which was a direct target of miR-100 yielded similar effects as that of ectopic miR-100 expression; The inverse correlation between miR-100 and Plk1 expression was also detected in nude mice SPC-A1/DTX tumor xenografts and clinical lung adenocarcinoma tissues and was proved to be related with the in vivo response to docetaxel; Thus our results suggested that down-regulation of miR-100 could lead to Plk1 over-expression and eventually to docetaxel chemoresistance of human lung adenocarcinoma;Reduced miR 100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein; Through modulating miR-100 expression using miR-100 inhibitor or mimic in vitro cell growth cycle and apoptosis were tested separately by MTT or flow cytometry and meanwhile Polo-like kinase1 PLK1 mRNA and protein expressions were detected by qRT-PCR and immunoblotting; The expression of PLK1 in 125 cervical tissues was also examined by immunohistochemical staining and the correlation between miR-100 and PLK1 expression in the same tissues was analysed; The modulation of miR-100 expression remarkably influenced cell proliferation cycle and apoptosis as well as the level of PLK1 protein but not mRNA in vitro experiments; PLK1 expression was negatively correlated with miR-100 expression in CIN3 and cervical cancer tissues; The reduced miR-100 expression participates in the development of cervical cancer at least partly through loss of inhibition to target gene PLK1 which probably occurs in a relative late phase of carcinogenesis |
| 132 | hsa-let-7b-3p | RAD21 | -0.29 | 0.81216 | -0.01 | 0.99451 | miRNATAP | -0.13 | 0.00478 | NA | |
| 133 | hsa-miR-126-5p | RAD21 | 0.08 | 0.95664 | -0.01 | 0.99451 | mirMAP | -0.13 | 0.00331 | NA | |
| 134 | hsa-miR-139-5p | RAD21 | -0.08 | 0.92869 | -0.01 | 0.99451 | miRanda | -0.11 | 0.00235 | NA | |
| 135 | hsa-miR-142-5p | RAD21 | -0.12 | 0.92967 | -0.01 | 0.99451 | PITA | -0.11 | 0.00062 | NA | |
| 136 | hsa-miR-195-3p | RAD21 | 0.12 | 0.69192 | -0.01 | 0.99451 | MirTarget; miRNATAP | -0.11 | 0.0008 | NA | |
| 137 | hsa-miR-199a-5p | RAD21 | 0.16 | 0.9358 | -0.01 | 0.99451 | miRanda | -0.15 | 6.0E-5 | NA | |
| 138 | hsa-miR-199b-5p | RAD21 | -0.04 | 0.97717 | -0.01 | 0.99451 | miRanda | -0.12 | 0.00017 | NA | |
| 139 | hsa-miR-299-5p | RAD21 | -0.32 | 0.43535 | -0.01 | 0.99451 | MirTarget; PITA; miRNATAP | -0.13 | 3.0E-5 | NA | |
| 140 | hsa-miR-320a | RAD21 | -0.42 | 0.8402 | -0.01 | 0.99451 | MirTarget; PITA; miRanda; miRNATAP | -0.12 | 0.00372 | NA | |
| 141 | hsa-miR-320b | RAD21 | -0.24 | 0.85922 | -0.01 | 0.99451 | MirTarget; PITA; miRanda; miRNATAP | -0.12 | 0.00106 | NA | |
| 142 | hsa-miR-433-3p | RAD21 | -0.03 | 0.90527 | -0.01 | 0.99451 | MirTarget | -0.1 | 0.00136 | NA | |
| 143 | hsa-miR-493-5p | RAD21 | 0.23 | 0.73198 | -0.01 | 0.99451 | miRNATAP | -0.14 | 0.00113 | NA | |
| 144 | hsa-miR-126-5p | RBL2 | 0.08 | 0.95664 | -0.07 | 0.96185 | MirTarget | -0.1 | 0.00637 | NA | |
| 145 | hsa-miR-1287-5p | RBL2 | -0.39 | 0.64389 | -0.07 | 0.96185 | MirTarget | -0.11 | 0.00463 | NA | |
| 146 | hsa-miR-130a-3p | SKP1 | 0.09 | 0.9291 | 0.08 | 0.96422 | MirTarget | -0.11 | 0.00019 | NA | |
| 147 | hsa-miR-152-3p | SKP1 | 0.2 | 0.90976 | 0.08 | 0.96422 | MirTarget | -0.15 | 0.00237 | NA | |
| 148 | hsa-miR-26a-5p | SKP2 | 0.01 | 0.99772 | -0.32 | 0.72696 | MirTarget | -0.35 | 7.0E-5 | NA | |
| 149 | hsa-miR-30a-5p | SKP2 | 0.22 | 0.93395 | -0.32 | 0.72696 | MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 150 | hsa-miR-30b-5p | SKP2 | -0.01 | 0.99462 | -0.32 | 0.72696 | MirTarget; miRNATAP | -0.12 | 0.00985 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | MITOTIC CELL CYCLE | 54 | 766 | 2.784e-62 | 1.296e-58 |
| 2 | CELL CYCLE PROCESS | 58 | 1081 | 2.157e-61 | 5.019e-58 |
| 3 | CELL CYCLE | 60 | 1316 | 4.382e-60 | 6.796e-57 |
| 4 | REGULATION OF CELL CYCLE | 53 | 949 | 2.332e-55 | 2.713e-52 |
| 5 | REGULATION OF MITOTIC CELL CYCLE | 39 | 468 | 4.108e-45 | 3.823e-42 |
| 6 | REGULATION OF CELL CYCLE PHASE TRANSITION | 35 | 321 | 3.123e-44 | 2.422e-41 |
| 7 | CELL CYCLE PHASE TRANSITION | 32 | 255 | 3.316e-42 | 2.204e-39 |
| 8 | REGULATION OF CELL CYCLE PROCESS | 38 | 558 | 1.96e-40 | 1.14e-37 |
| 9 | NEGATIVE REGULATION OF CELL CYCLE | 35 | 433 | 1.511e-39 | 7.814e-37 |
| 10 | CELL CYCLE CHECKPOINT | 26 | 194 | 9.741e-35 | 4.532e-32 |
| 11 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 26 | 214 | 1.418e-33 | 5.999e-31 |
| 12 | MITOTIC CELL CYCLE CHECKPOINT | 23 | 139 | 6.935e-33 | 2.689e-30 |
| 13 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 25 | 199 | 1.256e-32 | 4.496e-30 |
| 14 | CELL CYCLE G1 S PHASE TRANSITION | 21 | 111 | 2.522e-31 | 7.825e-29 |
| 15 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 21 | 111 | 2.522e-31 | 7.825e-29 |
| 16 | CELL DIVISION | 30 | 460 | 1.117e-30 | 3.248e-28 |
| 17 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 21 | 146 | 1.22e-28 | 3.338e-26 |
| 18 | POSITIVE REGULATION OF CELL CYCLE | 25 | 332 | 6.421e-27 | 1.66e-24 |
| 19 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 23 | 247 | 7.124e-27 | 1.745e-24 |
| 20 | REGULATION OF PROTEIN MODIFICATION PROCESS | 41 | 1710 | 7.774e-26 | 1.809e-23 |
| 21 | REGULATION OF TRANSFERASE ACTIVITY | 33 | 946 | 3.63e-25 | 8.042e-23 |
| 22 | REGULATION OF CELL CYCLE ARREST | 17 | 108 | 6.175e-24 | 1.306e-21 |
| 23 | DNA INTEGRITY CHECKPOINT | 18 | 146 | 2.635e-23 | 5.331e-21 |
| 24 | MITOTIC NUCLEAR DIVISION | 23 | 361 | 4.741e-23 | 9.192e-21 |
| 25 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 32 | 1087 | 4.428e-22 | 8.242e-20 |
| 26 | POSITIVE REGULATION OF CELL CYCLE ARREST | 15 | 85 | 5.489e-22 | 9.822e-20 |
| 27 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 17 | 147 | 1.523e-21 | 2.625e-19 |
| 28 | ORGANELLE FISSION | 24 | 496 | 3.042e-21 | 5.055e-19 |
| 29 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 15 | 96 | 3.878e-21 | 6.222e-19 |
| 30 | G1 DNA DAMAGE CHECKPOINT | 14 | 73 | 4.064e-21 | 6.304e-19 |
| 31 | REGULATION OF ORGANELLE ORGANIZATION | 32 | 1178 | 5.014e-21 | 7.525e-19 |
| 32 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 15 | 98 | 5.388e-21 | 7.835e-19 |
| 33 | MITOTIC DNA INTEGRITY CHECKPOINT | 15 | 100 | 7.431e-21 | 1.048e-18 |
| 34 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 25 | 616 | 2.678e-20 | 3.664e-18 |
| 35 | REGULATION OF CELL DIVISION | 19 | 272 | 8.434e-20 | 1.121e-17 |
| 36 | REGULATION OF NUCLEAR DIVISION | 16 | 163 | 3.891e-19 | 5.029e-17 |
| 37 | REGULATION OF CELL PROLIFERATION | 33 | 1496 | 5.64e-19 | 7.093e-17 |
| 38 | CHROMOSOME ORGANIZATION | 28 | 1009 | 1.912e-18 | 2.341e-16 |
| 39 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 19 | 351 | 1.019e-17 | 1.216e-15 |
| 40 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 14 | 127 | 1.474e-17 | 1.714e-15 |
| 41 | CELL CYCLE G2 M PHASE TRANSITION | 14 | 138 | 4.868e-17 | 5.524e-15 |
| 42 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 31 | 1492 | 5.694e-17 | 6.308e-15 |
| 43 | REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 17 | 274 | 6.852e-17 | 7.414e-15 |
| 44 | REGULATION OF PROTEIN CATABOLIC PROCESS | 19 | 393 | 8.286e-17 | 8.762e-15 |
| 45 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 23 | 720 | 2.295e-16 | 2.373e-14 |
| 46 | POSITIVE REGULATION OF CELL PROLIFERATION | 24 | 814 | 2.643e-16 | 2.674e-14 |
| 47 | POSITIVE REGULATION OF PROTEOLYSIS | 18 | 363 | 3.889e-16 | 3.85e-14 |
| 48 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 27 | 1135 | 4.441e-16 | 4.305e-14 |
| 49 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 31 | 1618 | 5.537e-16 | 5.152e-14 |
| 50 | REGULATION OF SISTER CHROMATID SEGREGATION | 11 | 67 | 5.511e-16 | 5.152e-14 |
| 51 | REGULATION OF LIGASE ACTIVITY | 13 | 130 | 8.454e-16 | 7.713e-14 |
| 52 | REGULATION OF CHROMOSOME ORGANIZATION | 16 | 278 | 2.035e-15 | 1.821e-13 |
| 53 | REGULATION OF PROTEOLYSIS | 22 | 711 | 2.303e-15 | 2.022e-13 |
| 54 | POSITIVE REGULATION OF GENE EXPRESSION | 31 | 1733 | 3.724e-15 | 3.209e-13 |
| 55 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 23 | 829 | 4.748e-15 | 4.017e-13 |
| 56 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 17 | 360 | 6.427e-15 | 5.34e-13 |
| 57 | CELL CYCLE ARREST | 13 | 154 | 7.916e-15 | 6.462e-13 |
| 58 | REGULATION OF CHROMOSOME SEGREGATION | 11 | 85 | 8.706e-15 | 6.984e-13 |
| 59 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 31 | 1805 | 1.142e-14 | 9.008e-13 |
| 60 | REGULATION OF KINASE ACTIVITY | 22 | 776 | 1.386e-14 | 1.062e-12 |
| 61 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 25 | 1079 | 1.392e-14 | 1.062e-12 |
| 62 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 12 | 123 | 1.595e-14 | 1.191e-12 |
| 63 | POSITIVE REGULATION OF CELL DEATH | 20 | 605 | 1.613e-14 | 1.191e-12 |
| 64 | REGULATION OF CELL DEATH | 28 | 1472 | 3.041e-14 | 2.211e-12 |
| 65 | CHROMOSOME SEGREGATION | 15 | 272 | 3.169e-14 | 2.269e-12 |
| 66 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 18 | 470 | 3.415e-14 | 2.407e-12 |
| 67 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 11 | 97 | 3.927e-14 | 2.727e-12 |
| 68 | SISTER CHROMATID SEGREGATION | 13 | 176 | 4.523e-14 | 3.095e-12 |
| 69 | REGULATION OF CATABOLIC PROCESS | 21 | 731 | 4.882e-14 | 3.292e-12 |
| 70 | NUCLEAR CHROMOSOME SEGREGATION | 14 | 228 | 5.684e-14 | 3.778e-12 |
| 71 | REGULATION OF PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 11 | 103 | 7.744e-14 | 5.075e-12 |
| 72 | POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 13 | 192 | 1.395e-13 | 8.894e-12 |
| 73 | CELLULAR RESPONSE TO STRESS | 28 | 1565 | 1.395e-13 | 8.894e-12 |
| 74 | ANAPHASE PROMOTING COMPLEX DEPENDENT CATABOLIC PROCESS | 10 | 77 | 1.493e-13 | 9.39e-12 |
| 75 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 9 | 53 | 2.072e-13 | 1.286e-11 |
| 76 | RESPONSE TO OXYGEN LEVELS | 15 | 311 | 2.242e-13 | 1.373e-11 |
| 77 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 19 | 616 | 2.835e-13 | 1.713e-11 |
| 78 | DNA REPLICATION | 13 | 208 | 3.907e-13 | 2.331e-11 |
| 79 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 14 | 263 | 4.037e-13 | 2.365e-11 |
| 80 | RESPONSE TO ABIOTIC STIMULUS | 23 | 1024 | 4.066e-13 | 2.365e-11 |
| 81 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 8 | 36 | 4.694e-13 | 2.697e-11 |
| 82 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 18 | 552 | 5.255e-13 | 2.982e-11 |
| 83 | PROTEIN PHOSPHORYLATION | 22 | 944 | 7.24e-13 | 4.059e-11 |
| 84 | REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 14 | 280 | 9.473e-13 | 5.247e-11 |
| 85 | INTRACELLULAR SIGNAL TRANSDUCTION | 27 | 1572 | 1.189e-12 | 6.511e-11 |
| 86 | NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 11 | 139 | 2.196e-12 | 1.168e-10 |
| 87 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 8 | 43 | 2.206e-12 | 1.168e-10 |
| 88 | POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 9 | 68 | 2.209e-12 | 1.168e-10 |
| 89 | POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 12 | 196 | 4.322e-12 | 2.259e-10 |
| 90 | NEGATIVE REGULATION OF CHROMOSOME SEGREGATION | 7 | 28 | 5.852e-12 | 2.992e-10 |
| 91 | POSITIVE REGULATION OF LIGASE ACTIVITY | 10 | 110 | 5.824e-12 | 2.992e-10 |
| 92 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 29 | 7.692e-12 | 3.808e-10 |
| 93 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 29 | 7.692e-12 | 3.808e-10 |
| 94 | DNA REPLICATION INITIATION | 7 | 29 | 7.692e-12 | 3.808e-10 |
| 95 | REGULATION OF FIBROBLAST PROLIFERATION | 9 | 81 | 1.128e-11 | 5.524e-10 |
| 96 | REGULATION OF DNA METABOLIC PROCESS | 14 | 340 | 1.296e-11 | 6.28e-10 |
| 97 | RESPONSE TO LIPID | 20 | 888 | 1.89e-11 | 9.067e-10 |
| 98 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 27 | 1784 | 2.285e-11 | 1.085e-09 |
| 99 | NEGATIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 7 | 34 | 2.614e-11 | 1.229e-09 |
| 100 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 25 | 1518 | 2.738e-11 | 1.274e-09 |
| 101 | NEGATIVE REGULATION OF CELL DIVISION | 8 | 60 | 3.711e-11 | 1.709e-09 |
| 102 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 14 | 370 | 3.991e-11 | 1.821e-09 |
| 103 | PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 10 | 134 | 4.235e-11 | 1.913e-09 |
| 104 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 16 | 541 | 5.347e-11 | 2.369e-09 |
| 105 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 16 | 541 | 5.347e-11 | 2.369e-09 |
| 106 | PEPTIDYL AMINO ACID MODIFICATION | 19 | 841 | 6.388e-11 | 2.804e-09 |
| 107 | NEGATIVE REGULATION OF CELL PROLIFERATION | 17 | 643 | 6.988e-11 | 3.039e-09 |
| 108 | CELLULAR RESPONSE TO UV | 8 | 66 | 8.19e-11 | 3.529e-09 |
| 109 | POSITIVE REGULATION OF CATABOLIC PROCESS | 14 | 395 | 9.468e-11 | 4.042e-09 |
| 110 | DNA METABOLIC PROCESS | 18 | 758 | 1.005e-10 | 4.251e-09 |
| 111 | REGULATION OF PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 10 | 148 | 1.138e-10 | 4.769e-09 |
| 112 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 16 | 573 | 1.248e-10 | 5.138e-09 |
| 113 | PHOSPHORYLATION | 22 | 1228 | 1.242e-10 | 5.138e-09 |
| 114 | REGULATION OF MEMBRANE PERMEABILITY | 8 | 70 | 1.331e-10 | 5.433e-09 |
| 115 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 27 | 1929 | 1.367e-10 | 5.531e-09 |
| 116 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 26 | 1791 | 1.561e-10 | 6.263e-09 |
| 117 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 1004 | 1.684e-10 | 6.697e-09 |
| 118 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 12 | 271 | 1.891e-10 | 7.456e-09 |
| 119 | SISTER CHROMATID COHESION | 9 | 111 | 2.009e-10 | 7.854e-09 |
| 120 | NEGATIVE REGULATION OF NUCLEAR DIVISION | 7 | 46 | 2.515e-10 | 9.752e-09 |
| 121 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 21 | 1152 | 2.68e-10 | 1.031e-08 |
| 122 | SPINDLE CHECKPOINT | 6 | 25 | 2.708e-10 | 1.033e-08 |
| 123 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 15 | 514 | 2.804e-10 | 1.061e-08 |
| 124 | RESPONSE TO DRUG | 14 | 431 | 2.971e-10 | 1.115e-08 |
| 125 | REGULATION OF INTRACELLULAR TRANSPORT | 16 | 621 | 4.046e-10 | 1.506e-08 |
| 126 | POSITIVE REGULATION OF STEM CELL DIFFERENTIATION | 7 | 50 | 4.641e-10 | 1.714e-08 |
| 127 | REGULATION OF PROTEIN LOCALIZATION | 19 | 950 | 5.009e-10 | 1.835e-08 |
| 128 | RESPONSE TO UV | 9 | 126 | 6.276e-10 | 2.264e-08 |
| 129 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 9 | 126 | 6.276e-10 | 2.264e-08 |
| 130 | REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 10 | 181 | 8.227e-10 | 2.945e-08 |
| 131 | PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 18 | 873 | 9.734e-10 | 3.458e-08 |
| 132 | CELLULAR RESPONSE TO LIGHT STIMULUS | 8 | 91 | 1.13e-09 | 3.923e-08 |
| 133 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 23 | 1517 | 1.118e-09 | 3.923e-08 |
| 134 | MITOTIC SISTER CHROMATID SEGREGATION | 8 | 91 | 1.13e-09 | 3.923e-08 |
| 135 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 8 | 92 | 1.234e-09 | 4.253e-08 |
| 136 | CELL PROLIFERATION | 16 | 672 | 1.268e-09 | 4.338e-08 |
| 137 | CELLULAR RESPONSE TO RADIATION | 9 | 137 | 1.324e-09 | 4.497e-08 |
| 138 | POSITIVE REGULATION OF CELL COMMUNICATION | 23 | 1532 | 1.353e-09 | 4.563e-08 |
| 139 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 7 | 59 | 1.546e-09 | 5.174e-08 |
| 140 | RESPONSE TO STEROID HORMONE | 14 | 497 | 1.879e-09 | 6.247e-08 |
| 141 | POSITIVE REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 6 | 34 | 2.006e-09 | 6.618e-08 |
| 142 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 18 | 917 | 2.121e-09 | 6.951e-08 |
| 143 | DNA DEPENDENT DNA REPLICATION | 8 | 99 | 2.225e-09 | 7.241e-08 |
| 144 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 8 | 100 | 2.412e-09 | 7.794e-08 |
| 145 | NEGATIVE REGULATION OF PHOSPHORYLATION | 13 | 422 | 2.619e-09 | 8.403e-08 |
| 146 | NEGATIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 7 | 64 | 2.775e-09 | 8.845e-08 |
| 147 | NEGATIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 8 | 109 | 4.802e-09 | 1.52e-07 |
| 148 | RESPONSE TO ESTROGEN | 10 | 218 | 4.979e-09 | 1.565e-07 |
| 149 | RESPONSE TO ALCOHOL | 12 | 362 | 5.058e-09 | 1.58e-07 |
| 150 | REGULATION OF DNA REPLICATION | 9 | 161 | 5.512e-09 | 1.71e-07 |
| 151 | CELL DEATH | 18 | 1001 | 8.368e-09 | 2.578e-07 |
| 152 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 9 | 171 | 9.345e-09 | 2.861e-07 |
| 153 | PROTEIN CATABOLIC PROCESS | 14 | 579 | 1.31e-08 | 3.985e-07 |
| 154 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 15 | 684 | 1.359e-08 | 4.107e-07 |
| 155 | REGULATION OF MICROTUBULE BASED PROCESS | 10 | 243 | 1.405e-08 | 4.219e-07 |
| 156 | RESPONSE TO ENDOGENOUS STIMULUS | 21 | 1450 | 1.644e-08 | 4.902e-07 |
| 157 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 8 | 128 | 1.713e-08 | 5.077e-07 |
| 158 | ORGAN REGENERATION | 7 | 83 | 1.759e-08 | 5.179e-07 |
| 159 | NEGATIVE REGULATION OF KINASE ACTIVITY | 10 | 250 | 1.841e-08 | 5.387e-07 |
| 160 | RESPONSE TO RADIATION | 12 | 413 | 2.187e-08 | 6.359e-07 |
| 161 | NEGATIVE REGULATION OF GENE EXPRESSION | 21 | 1493 | 2.733e-08 | 7.898e-07 |
| 162 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 10 | 263 | 2.974e-08 | 8.542e-07 |
| 163 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 24 | 1977 | 3.499e-08 | 9.987e-07 |
| 164 | REGULATION OF TRANSCRIPTION INVOLVED IN G1 S TRANSITION OF MITOTIC CELL CYCLE | 5 | 27 | 3.712e-08 | 1.053e-06 |
| 165 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 20 | 1381 | 3.977e-08 | 1.121e-06 |
| 166 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 9 | 207 | 4.897e-08 | 1.364e-06 |
| 167 | NEGATIVE REGULATION OF CHROMOSOME ORGANIZATION | 7 | 96 | 4.868e-08 | 1.364e-06 |
| 168 | DIGESTIVE SYSTEM DEVELOPMENT | 8 | 148 | 5.335e-08 | 1.478e-06 |
| 169 | REGULATION OF BINDING | 10 | 283 | 5.926e-08 | 1.632e-06 |
| 170 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 17 | 1008 | 6.016e-08 | 1.647e-06 |
| 171 | REGULATION OF CELLULAR LOCALIZATION | 19 | 1277 | 6.211e-08 | 1.69e-06 |
| 172 | RESPONSE TO HORMONE | 16 | 893 | 6.951e-08 | 1.881e-06 |
| 173 | REPLICATIVE SENESCENCE | 4 | 12 | 7.063e-08 | 1.9e-06 |
| 174 | REGULATION OF MITOCHONDRION ORGANIZATION | 9 | 218 | 7.635e-08 | 2.03e-06 |
| 175 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 9 | 218 | 7.635e-08 | 2.03e-06 |
| 176 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 12 | 465 | 8.004e-08 | 2.116e-06 |
| 177 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 7 | 104 | 8.483e-08 | 2.23e-06 |
| 178 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 17 | 1036 | 8.93e-08 | 2.321e-06 |
| 179 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 17 | 1036 | 8.93e-08 | 2.321e-06 |
| 180 | RHYTHMIC PROCESS | 10 | 298 | 9.603e-08 | 2.482e-06 |
| 181 | RESPONSE TO GROWTH FACTOR | 12 | 475 | 1.008e-07 | 2.592e-06 |
| 182 | REGENERATION | 8 | 161 | 1.025e-07 | 2.62e-06 |
| 183 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 8 | 162 | 1.075e-07 | 2.718e-06 |
| 184 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 8 | 162 | 1.075e-07 | 2.718e-06 |
| 185 | RESPONSE TO INORGANIC SUBSTANCE | 12 | 479 | 1.104e-07 | 2.776e-06 |
| 186 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 11 | 387 | 1.118e-07 | 2.798e-06 |
| 187 | REGULATION OF CELLULAR RESPONSE TO STRESS | 14 | 691 | 1.179e-07 | 2.933e-06 |
| 188 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 8 | 167 | 1.359e-07 | 3.346e-06 |
| 189 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 6 | 67 | 1.357e-07 | 3.346e-06 |
| 190 | REGULATION OF STEM CELL DIFFERENTIATION | 7 | 113 | 1.504e-07 | 3.683e-06 |
| 191 | PROTEIN SUMOYLATION | 7 | 115 | 1.696e-07 | 4.133e-06 |
| 192 | REGULATION OF CYTOSKELETON ORGANIZATION | 12 | 502 | 1.83e-07 | 4.435e-06 |
| 193 | MITOCHONDRIAL TRANSPORT | 8 | 177 | 2.125e-07 | 5.123e-06 |
| 194 | REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION | 8 | 178 | 2.219e-07 | 5.321e-06 |
| 195 | NEGATIVE REGULATION OF PROTEOLYSIS | 10 | 329 | 2.404e-07 | 5.736e-06 |
| 196 | RESPONSE TO KETONE | 8 | 182 | 2.63e-07 | 6.244e-06 |
| 197 | PROTEIN UBIQUITINATION | 13 | 629 | 2.83e-07 | 6.685e-06 |
| 198 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 8 | 185 | 2.98e-07 | 7.003e-06 |
| 199 | MEIOTIC CELL CYCLE | 8 | 186 | 3.105e-07 | 7.261e-06 |
| 200 | NEGATIVE REGULATION OF CELL DEATH | 15 | 872 | 3.229e-07 | 7.512e-06 |
| 201 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 7 | 129 | 3.721e-07 | 8.614e-06 |
| 202 | CHROMATIN MODIFICATION | 12 | 539 | 3.914e-07 | 9.016e-06 |
| 203 | CHROMATIN ORGANIZATION | 13 | 663 | 5.143e-07 | 1.179e-05 |
| 204 | NEGATIVE REGULATION OF CATABOLIC PROCESS | 8 | 203 | 6.037e-07 | 1.377e-05 |
| 205 | RESPONSE TO LIGHT STIMULUS | 9 | 280 | 6.337e-07 | 1.438e-05 |
| 206 | MACROMOLECULE CATABOLIC PROCESS | 15 | 926 | 6.917e-07 | 1.562e-05 |
| 207 | POSITIVE REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 5 | 48 | 7.431e-07 | 1.67e-05 |
| 208 | POSITIVE REGULATION OF TRANSPORT | 15 | 936 | 7.918e-07 | 1.771e-05 |
| 209 | APOPTOTIC SIGNALING PATHWAY | 9 | 289 | 8.247e-07 | 1.827e-05 |
| 210 | REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 7 | 145 | 8.217e-07 | 1.827e-05 |
| 211 | RESPONSE TO ESTRADIOL | 7 | 146 | 8.606e-07 | 1.898e-05 |
| 212 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 21 | 1848 | 9.957e-07 | 2.185e-05 |
| 213 | POSITIVE REGULATION OF CHROMOSOME ORGANIZATION | 7 | 150 | 1.032e-06 | 2.255e-05 |
| 214 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 8 | 220 | 1.107e-06 | 2.408e-05 |
| 215 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 152 | 1.128e-06 | 2.442e-05 |
| 216 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 4 | 23 | 1.227e-06 | 2.631e-05 |
| 217 | POSITIVE REGULATION OF COLLAGEN METABOLIC PROCESS | 4 | 23 | 1.227e-06 | 2.631e-05 |
| 218 | GLAND DEVELOPMENT | 10 | 395 | 1.273e-06 | 2.717e-05 |
| 219 | NEGATIVE REGULATION OF CELLULAR CATABOLIC PROCESS | 7 | 156 | 1.343e-06 | 2.854e-05 |
| 220 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 6 | 99 | 1.394e-06 | 2.935e-05 |
| 221 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 6 | 99 | 1.394e-06 | 2.935e-05 |
| 222 | NEGATIVE REGULATION OF DNA REPLICATION | 5 | 55 | 1.481e-06 | 3.104e-05 |
| 223 | PEPTIDYL LYSINE MODIFICATION | 9 | 312 | 1.555e-06 | 3.244e-05 |
| 224 | RESPONSE TO NITROGEN COMPOUND | 14 | 859 | 1.609e-06 | 3.342e-05 |
| 225 | SMAD PROTEIN SIGNAL TRANSDUCTION | 5 | 56 | 1.622e-06 | 3.353e-05 |
| 226 | HISTONE PHOSPHORYLATION | 4 | 25 | 1.743e-06 | 3.573e-05 |
| 227 | POSITIVE REGULATION OF CHROMOSOME SEGREGATION | 4 | 25 | 1.743e-06 | 3.573e-05 |
| 228 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 13 | 740 | 1.757e-06 | 3.585e-05 |
| 229 | GROWTH | 10 | 410 | 1.779e-06 | 3.615e-05 |
| 230 | REPRODUCTION | 17 | 1297 | 2.082e-06 | 4.213e-05 |
| 231 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 5 | 60 | 2.293e-06 | 4.619e-05 |
| 232 | PROTEIN K11 LINKED UBIQUITINATION | 4 | 27 | 2.406e-06 | 4.825e-05 |
| 233 | CELLULAR RESPONSE TO HYDROGEN PEROXIDE | 5 | 61 | 2.491e-06 | 4.975e-05 |
| 234 | RESPONSE TO METAL ION | 9 | 333 | 2.654e-06 | 5.277e-05 |
| 235 | NEGATIVE REGULATION OF DNA METABOLIC PROCESS | 6 | 111 | 2.724e-06 | 5.394e-05 |
| 236 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 9 | 337 | 2.926e-06 | 5.768e-05 |
| 237 | REGULATION OF CELL DIFFERENTIATION | 18 | 1492 | 3.172e-06 | 6.195e-05 |
| 238 | NOTCH SIGNALING PATHWAY | 6 | 114 | 3.182e-06 | 6.195e-05 |
| 239 | REGULATION OF PROTEIN ACETYLATION | 5 | 64 | 3.166e-06 | 6.195e-05 |
| 240 | REGULATION OF HEART MORPHOGENESIS | 4 | 29 | 3.238e-06 | 6.278e-05 |
| 241 | NEGATIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 6 | 115 | 3.348e-06 | 6.464e-05 |
| 242 | COVALENT CHROMATIN MODIFICATION | 9 | 345 | 3.542e-06 | 6.81e-05 |
| 243 | PROTEOLYSIS | 16 | 1208 | 3.875e-06 | 7.419e-05 |
| 244 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 7 | 184 | 4.028e-06 | 7.681e-05 |
| 245 | AGING | 8 | 264 | 4.299e-06 | 8.165e-05 |
| 246 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 18 | 1527 | 4.396e-06 | 8.314e-05 |
| 247 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 6 | 121 | 4.497e-06 | 8.472e-05 |
| 248 | CELLULAR RESPONSE TO LIPID | 10 | 457 | 4.671e-06 | 8.763e-05 |
| 249 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 32 | 4.863e-06 | 9.08e-05 |
| 250 | EPITHELIUM DEVELOPMENT | 14 | 945 | 4.879e-06 | 9.08e-05 |
| 251 | RESPONSE TO REACTIVE OXYGEN SPECIES | 7 | 191 | 5.151e-06 | 9.549e-05 |
| 252 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 5 | 71 | 5.304e-06 | 9.794e-05 |
| 253 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 9 | 363 | 5.35e-06 | 9.84e-05 |
| 254 | G2 DNA DAMAGE CHECKPOINT | 4 | 33 | 5.519e-06 | 0.0001011 |
| 255 | RESPONSE TO MINERALOCORTICOID | 4 | 35 | 7.025e-06 | 0.0001277 |
| 256 | RESPONSE TO IRON ION | 4 | 35 | 7.025e-06 | 0.0001277 |
| 257 | ORGAN MORPHOGENESIS | 13 | 841 | 7.103e-06 | 0.0001286 |
| 258 | DNA REPAIR | 10 | 480 | 7.189e-06 | 0.0001297 |
| 259 | REGULATION OF CELLULAR RESPONSE TO HEAT | 5 | 76 | 7.424e-06 | 0.000133 |
| 260 | POSITIVE REGULATION OF CELL DIVISION | 6 | 132 | 7.43e-06 | 0.000133 |
| 261 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 8 | 285 | 7.537e-06 | 0.0001344 |
| 262 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 36 | 7.882e-06 | 0.00014 |
| 263 | SENSORY ORGAN DEVELOPMENT | 10 | 493 | 9.078e-06 | 0.0001605 |
| 264 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 15 | 1142 | 9.108e-06 | 0.0001605 |
| 265 | REGULATION OF MACROPHAGE CYTOKINE PRODUCTION | 3 | 12 | 9.218e-06 | 0.0001612 |
| 266 | POSITIVE REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 3 | 12 | 9.218e-06 | 0.0001612 |
| 267 | REGULATION OF CELL GROWTH | 9 | 391 | 9.722e-06 | 0.0001694 |
| 268 | MESENCHYME MORPHOGENESIS | 4 | 38 | 9.824e-06 | 0.0001699 |
| 269 | REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 4 | 38 | 9.824e-06 | 0.0001699 |
| 270 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 10 | 505 | 1.119e-05 | 0.0001928 |
| 271 | CELLULAR RESPONSE TO OXYGEN LEVELS | 6 | 143 | 1.175e-05 | 0.0002018 |
| 272 | MITOTIC CELL CYCLE ARREST | 3 | 13 | 1.195e-05 | 0.0002045 |
| 273 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 6 | 144 | 1.223e-05 | 0.0002085 |
| 274 | POSITIVE REGULATION OF CHROMATIN MODIFICATION | 5 | 85 | 1.286e-05 | 0.0002184 |
| 275 | REGULATION OF DNA DEPENDENT DNA REPLICATION | 4 | 41 | 1.337e-05 | 0.0002262 |
| 276 | POSITIVE REGULATION OF DNA REPLICATION | 5 | 86 | 1.361e-05 | 0.0002295 |
| 277 | PEPTIDYL SERINE MODIFICATION | 6 | 148 | 1.43e-05 | 0.0002402 |
| 278 | BETA CATENIN TCF COMPLEX ASSEMBLY | 4 | 43 | 1.621e-05 | 0.0002712 |
| 279 | REGULATION OF CHROMATIN ORGANIZATION | 6 | 152 | 1.664e-05 | 0.0002765 |
| 280 | MEIOTIC CELL CYCLE PROCESS | 6 | 152 | 1.664e-05 | 0.0002765 |
| 281 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 7 | 229 | 1.68e-05 | 0.0002782 |
| 282 | TISSUE DEVELOPMENT | 17 | 1518 | 1.691e-05 | 0.0002791 |
| 283 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 5 | 95 | 2.21e-05 | 0.0003633 |
| 284 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 12 | 799 | 2.228e-05 | 0.000365 |
| 285 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 12 | 801 | 2.283e-05 | 0.0003728 |
| 286 | REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 3 | 16 | 2.323e-05 | 0.0003739 |
| 287 | REGULATION OF EXIT FROM MITOSIS | 3 | 16 | 2.323e-05 | 0.0003739 |
| 288 | CELLULAR RESPONSE TO ANTIBIOTIC | 3 | 16 | 2.323e-05 | 0.0003739 |
| 289 | DEVELOPMENTAL GROWTH | 8 | 333 | 2.321e-05 | 0.0003739 |
| 290 | RESPONSE TO TOXIC SUBSTANCE | 7 | 241 | 2.334e-05 | 0.0003744 |
| 291 | REGULATION OF CELL MORPHOGENESIS | 10 | 552 | 2.406e-05 | 0.0003847 |
| 292 | PROTEIN POLYUBIQUITINATION | 7 | 243 | 2.461e-05 | 0.0003921 |
| 293 | RESPONSE TO PROGESTERONE | 4 | 50 | 2.968e-05 | 0.0004714 |
| 294 | NEGATIVE REGULATION OF CELL GROWTH | 6 | 170 | 3.131e-05 | 0.0004955 |
| 295 | MICROTUBULE CYTOSKELETON ORGANIZATION | 8 | 348 | 3.177e-05 | 0.0005011 |
| 296 | RESPONSE TO AMMONIUM ION | 4 | 51 | 3.212e-05 | 0.0005049 |
| 297 | POSITIVE REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 3 | 18 | 3.367e-05 | 0.0005257 |
| 298 | REGULATION OF UBIQUITIN PROTEIN LIGASE ACTIVITY | 3 | 18 | 3.367e-05 | 0.0005257 |
| 299 | INTRINSIC APOPTOTIC SIGNALING PATHWAY BY P53 CLASS MEDIATOR | 4 | 53 | 3.744e-05 | 0.0005826 |
| 300 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 7 | 264 | 4.173e-05 | 0.0006473 |
| 301 | RESPONSE TO HYDROGEN PEROXIDE | 5 | 109 | 4.288e-05 | 0.0006628 |
| 302 | REGULATION OF RESPONSE TO STRESS | 16 | 1468 | 4.397e-05 | 0.0006775 |
| 303 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 11 | 724 | 4.586e-05 | 0.0007043 |
| 304 | EPITHELIAL TO MESENCHYMAL TRANSITION | 4 | 56 | 4.658e-05 | 0.000713 |
| 305 | NEGATIVE REGULATION OF CELL CYCLE ARREST | 3 | 20 | 4.68e-05 | 0.000714 |
| 306 | REGULATION OF PROTEIN IMPORT | 6 | 183 | 4.728e-05 | 0.000719 |
| 307 | REGULATION OF CYTOPLASMIC TRANSPORT | 9 | 481 | 4.956e-05 | 0.0007512 |
| 308 | POSITIVE REGULATION OF KINASE ACTIVITY | 9 | 482 | 5.037e-05 | 0.0007609 |
| 309 | CELLULAR CATABOLIC PROCESS | 15 | 1322 | 5.057e-05 | 0.0007615 |
| 310 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 10 | 609 | 5.52e-05 | 0.0008285 |
| 311 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 17 | 1672 | 5.805e-05 | 0.0008686 |
| 312 | MESENCHYME DEVELOPMENT | 6 | 190 | 5.827e-05 | 0.0008689 |
| 313 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 8 | 381 | 6.019e-05 | 0.0008948 |
| 314 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 3 | 22 | 6.29e-05 | 0.0009291 |
| 315 | ENDOCARDIAL CUSHION MORPHOGENESIS | 3 | 22 | 6.29e-05 | 0.0009291 |
| 316 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 7 | 282 | 6.329e-05 | 0.000932 |
| 317 | EMBRYONIC DIGIT MORPHOGENESIS | 4 | 61 | 6.531e-05 | 0.0009586 |
| 318 | EMBRYO DEVELOPMENT | 12 | 894 | 6.639e-05 | 0.0009714 |
| 319 | RESPONSE TO INCREASED OXYGEN LEVELS | 3 | 23 | 7.215e-05 | 0.001049 |
| 320 | RESPONSE TO HYPEROXIA | 3 | 23 | 7.215e-05 | 0.001049 |
| 321 | REGULATION OF GROWTH | 10 | 633 | 7.616e-05 | 0.001104 |
| 322 | POSITIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 4 | 64 | 7.888e-05 | 0.00114 |
| 323 | POSITIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 3 | 24 | 8.225e-05 | 0.001178 |
| 324 | POSITIVE REGULATION OF G1 S TRANSITION OF MITOTIC CELL CYCLE | 3 | 24 | 8.225e-05 | 0.001178 |
| 325 | POSITIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 3 | 24 | 8.225e-05 | 0.001178 |
| 326 | POSITIVE REGULATION OF BINDING | 5 | 127 | 8.881e-05 | 0.001268 |
| 327 | MICROTUBULE BASED PROCESS | 9 | 522 | 9.277e-05 | 0.001319 |
| 328 | CELLULAR RESPONSE TO TOXIC SUBSTANCE | 3 | 25 | 9.323e-05 | 0.001319 |
| 329 | POSITIVE REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 25 | 9.323e-05 | 0.001319 |
| 330 | PROTEIN TARGETING | 8 | 406 | 9.372e-05 | 0.001321 |
| 331 | POSITIVE REGULATION OF NEURON DEATH | 4 | 67 | 9.44e-05 | 0.001323 |
| 332 | CELL AGING | 4 | 67 | 9.44e-05 | 0.001323 |
| 333 | MACROMOLECULAR COMPLEX ASSEMBLY | 15 | 1398 | 9.528e-05 | 0.001331 |
| 334 | REGULATION OF CELLULAR SENESCENCE | 3 | 26 | 0.0001051 | 0.00146 |
| 335 | RESPONSE TO CORTICOSTERONE | 3 | 26 | 0.0001051 | 0.00146 |
| 336 | MITOTIC SPINDLE ORGANIZATION | 4 | 69 | 0.0001059 | 0.001467 |
| 337 | REGULATION OF PROTEIN TARGETING | 7 | 307 | 0.0001076 | 0.001486 |
| 338 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 6 | 213 | 0.0001094 | 0.001507 |
| 339 | REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 6 | 217 | 0.0001212 | 0.001663 |
| 340 | RESPONSE TO ETHANOL | 5 | 136 | 0.0001227 | 0.001679 |
| 341 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 6 | 218 | 0.0001243 | 0.001695 |
| 342 | CIRCADIAN RHYTHM | 5 | 137 | 0.000127 | 0.001728 |
| 343 | REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 3 | 28 | 0.0001318 | 0.001788 |
| 344 | CELLULAR RESPONSE TO HORMONE STIMULUS | 9 | 552 | 0.0001415 | 0.001909 |
| 345 | TUBE DEVELOPMENT | 9 | 552 | 0.0001415 | 0.001909 |
| 346 | NEGATIVE REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 3 | 29 | 0.0001466 | 0.00196 |
| 347 | REGULATION OF EXTRACELLULAR MATRIX ORGANIZATION | 3 | 29 | 0.0001466 | 0.00196 |
| 348 | POSITIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 3 | 29 | 0.0001466 | 0.00196 |
| 349 | REGULATION OF TRANSPORT | 17 | 1804 | 0.0001483 | 0.001977 |
| 350 | PROTEIN COMPLEX BIOGENESIS | 13 | 1132 | 0.0001543 | 0.002046 |
| 351 | PROTEIN COMPLEX ASSEMBLY | 13 | 1132 | 0.0001543 | 0.002046 |
| 352 | EYE DEVELOPMENT | 7 | 326 | 0.000156 | 0.002062 |
| 353 | REGULATION OF CELL DEVELOPMENT | 11 | 836 | 0.0001647 | 0.002171 |
| 354 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 16 | 1656 | 0.0001823 | 0.002389 |
| 355 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 5 | 148 | 0.0001823 | 0.002389 |
| 356 | NEGATIVE REGULATION OF GROWTH | 6 | 236 | 0.0001913 | 0.0025 |
| 357 | HEAD DEVELOPMENT | 10 | 709 | 0.0001926 | 0.002511 |
| 358 | CHROMATIN REMODELING | 5 | 150 | 0.000194 | 0.002522 |
| 359 | SALIVARY GLAND DEVELOPMENT | 3 | 32 | 0.0001975 | 0.002553 |
| 360 | ENDOCARDIAL CUSHION DEVELOPMENT | 3 | 32 | 0.0001975 | 0.002553 |
| 361 | IMMUNE SYSTEM DEVELOPMENT | 9 | 582 | 0.0002101 | 0.002708 |
| 362 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 33 | 0.0002167 | 0.002778 |
| 363 | REGULATION OF CELL AGING | 3 | 33 | 0.0002167 | 0.002778 |
| 364 | REGULATION OF ORGAN MORPHOGENESIS | 6 | 242 | 0.000219 | 0.0028 |
| 365 | CELLULAR RESPONSE TO INORGANIC SUBSTANCE | 5 | 156 | 0.0002329 | 0.002968 |
| 366 | PROTEIN DESTABILIZATION | 3 | 34 | 0.0002371 | 0.003006 |
| 367 | HEART VALVE DEVELOPMENT | 3 | 34 | 0.0002371 | 0.003006 |
| 368 | HEART DEVELOPMENT | 8 | 466 | 0.0002406 | 0.003043 |
| 369 | MITOCHONDRION ORGANIZATION | 9 | 594 | 0.0002444 | 0.003082 |
| 370 | RESPONSE TO OXIDATIVE STRESS | 7 | 352 | 0.0002493 | 0.003136 |
| 371 | RESPONSE TO MONOAMINE | 3 | 35 | 0.0002586 | 0.003244 |
| 372 | TISSUE REMODELING | 4 | 87 | 0.0002599 | 0.003251 |
| 373 | POSITIVE REGULATION OF CELL DEVELOPMENT | 8 | 472 | 0.0002623 | 0.003272 |
| 374 | MEIOSIS I | 4 | 88 | 0.0002715 | 0.003369 |
| 375 | OVULATION CYCLE PROCESS | 4 | 88 | 0.0002715 | 0.003369 |
| 376 | POSITIVE REGULATION OF PROTEIN ACETYLATION | 3 | 36 | 0.0002814 | 0.003473 |
| 377 | HEAD MORPHOGENESIS | 3 | 36 | 0.0002814 | 0.003473 |
| 378 | MESONEPHROS DEVELOPMENT | 4 | 90 | 0.0002959 | 0.003643 |
| 379 | REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 37 | 0.0003055 | 0.00375 |
| 380 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 6 | 258 | 0.0003088 | 0.003772 |
| 381 | MEMBRANE ORGANIZATION | 11 | 899 | 0.0003083 | 0.003772 |
| 382 | RESPONSE TO CARBOHYDRATE | 5 | 168 | 0.0003279 | 0.003994 |
| 383 | REGULATION OF DNA BINDING | 4 | 93 | 0.0003355 | 0.004076 |
| 384 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 14 | 1395 | 0.0003383 | 0.004099 |
| 385 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 5 | 170 | 0.0003463 | 0.004185 |
| 386 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 7 | 372 | 0.0003482 | 0.004198 |
| 387 | REGULATION OF DNA BIOSYNTHETIC PROCESS | 4 | 94 | 0.0003494 | 0.004201 |
| 388 | CELLULAR RESPONSE TO NUTRIENT | 3 | 39 | 0.0003575 | 0.004287 |
| 389 | CELLULAR MACROMOLECULE LOCALIZATION | 13 | 1234 | 0.0003584 | 0.004287 |
| 390 | EPITHELIAL CELL DIFFERENTIATION | 8 | 495 | 0.0003608 | 0.004304 |
| 391 | REGULATION OF CELL SIZE | 5 | 172 | 0.0003654 | 0.004348 |
| 392 | RESPONSE TO CADMIUM ION | 3 | 40 | 0.0003855 | 0.004576 |
| 393 | CATABOLIC PROCESS | 16 | 1773 | 0.0003955 | 0.004683 |
| 394 | RESPONSE TO CORTICOSTEROID | 5 | 176 | 0.000406 | 0.004794 |
| 395 | HEMATOPOIETIC PROGENITOR CELL DIFFERENTIATION | 4 | 98 | 0.0004095 | 0.0048 |
| 396 | POSITIVE REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 4 | 98 | 0.0004095 | 0.0048 |
| 397 | RESPONSE TO VITAMIN | 4 | 98 | 0.0004095 | 0.0048 |
| 398 | CELL DEVELOPMENT | 14 | 1426 | 0.0004227 | 0.004942 |
| 399 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 4 | 102 | 0.0004767 | 0.005559 |
| 400 | BODY MORPHOGENESIS | 3 | 44 | 0.0005115 | 0.00595 |
| 401 | POSITIVE REGULATION OF PROTEIN IMPORT | 4 | 104 | 0.000513 | 0.005953 |
| 402 | ODONTOGENESIS | 4 | 105 | 0.0005319 | 0.006157 |
| 403 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 9 | 662 | 0.0005387 | 0.006204 |
| 404 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 9 | 662 | 0.0005387 | 0.006204 |
| 405 | EXOCRINE SYSTEM DEVELOPMENT | 3 | 45 | 0.0005467 | 0.00628 |
| 406 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 5 | 188 | 0.0005483 | 0.006284 |
| 407 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 6 | 289 | 0.0005634 | 0.00644 |
| 408 | RESPONSE TO PEPTIDE | 7 | 404 | 0.0005699 | 0.0065 |
| 409 | STEM CELL DIFFERENTIATION | 5 | 190 | 0.0005753 | 0.006529 |
| 410 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 5 | 190 | 0.0005753 | 0.006529 |
| 411 | RESPONSE TO COCAINE | 3 | 46 | 0.0005833 | 0.006604 |
| 412 | RESPONSE TO NUTRIENT | 5 | 191 | 0.0005892 | 0.006654 |
| 413 | MEMBRANE DISASSEMBLY | 3 | 47 | 0.0006215 | 0.006952 |
| 414 | RESPONSE TO ANTIBIOTIC | 3 | 47 | 0.0006215 | 0.006952 |
| 415 | NUCLEAR ENVELOPE DISASSEMBLY | 3 | 47 | 0.0006215 | 0.006952 |
| 416 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 3 | 47 | 0.0006215 | 0.006952 |
| 417 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 10 | 823 | 0.0006262 | 0.006987 |
| 418 | HOMOLOGOUS CHROMOSOME SEGREGATION | 3 | 48 | 0.0006612 | 0.007361 |
| 419 | REGULATION OF MITOTIC SPINDLE CHECKPOINT | 2 | 11 | 0.0006694 | 0.007381 |
| 420 | POSITIVE REGULATION OF RECEPTOR BIOSYNTHETIC PROCESS | 2 | 11 | 0.0006694 | 0.007381 |
| 421 | NEGATIVE REGULATION OF CELLULAR SENESCENCE | 2 | 11 | 0.0006694 | 0.007381 |
| 422 | REGULATION OF MITOTIC CELL CYCLE SPINDLE ASSEMBLY CHECKPOINT | 2 | 11 | 0.0006694 | 0.007381 |
| 423 | ESTABLISHMENT OF LOCALIZATION IN CELL | 15 | 1676 | 0.0006787 | 0.007466 |
| 424 | OVULATION CYCLE | 4 | 113 | 0.0007014 | 0.007697 |
| 425 | LYMPHOCYTE HOMEOSTASIS | 3 | 50 | 0.0007455 | 0.008143 |
| 426 | FACE DEVELOPMENT | 3 | 50 | 0.0007455 | 0.008143 |
| 427 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 8 | 554 | 0.000757 | 0.008249 |
| 428 | POSITIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 3 | 51 | 0.0007901 | 0.00859 |
| 429 | POSITIVE REGULATION OF DNA DEPENDENT DNA REPLICATION | 2 | 12 | 0.0008014 | 0.008693 |
| 430 | IN UTERO EMBRYONIC DEVELOPMENT | 6 | 311 | 0.0008262 | 0.00894 |
| 431 | REGULATION OF CYTOKINE PRODUCTION | 8 | 563 | 0.0008404 | 0.009072 |
| 432 | LYMPHOCYTE DIFFERENTIATION | 5 | 209 | 0.000884 | 0.009521 |
| 433 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 11 | 1021 | 0.0008933 | 0.009599 |
| 434 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 13 | 1360 | 0.0008968 | 0.009615 |
| 435 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 4 | 121 | 0.000906 | 0.009668 |
| 436 | REGULATION OF B CELL ACTIVATION | 4 | 121 | 0.000906 | 0.009668 |
| 437 | POSITIVE REGULATION OF PROTEIN SECRETION | 5 | 211 | 0.0009224 | 0.009822 |
| 438 | NEGATIVE REGULATION OF CELL COMMUNICATION | 12 | 1192 | 0.0009263 | 0.009841 |
| 439 | POSITIVE REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 2 | 13 | 0.000945 | 0.009859 |
| 440 | RESPONSE TO ACID CHEMICAL | 6 | 319 | 0.0009422 | 0.009859 |
| 441 | PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 2 | 13 | 0.000945 | 0.009859 |
| 442 | EYELID DEVELOPMENT IN CAMERA TYPE EYE | 2 | 13 | 0.000945 | 0.009859 |
| 443 | PROTEIN LOCALIZATION TO CHROMATIN | 2 | 13 | 0.000945 | 0.009859 |
| 444 | MITOTIC G2 DNA DAMAGE CHECKPOINT | 2 | 13 | 0.000945 | 0.009859 |
| 445 | REGULATION OF HISTONE PHOSPHORYLATION | 2 | 13 | 0.000945 | 0.009859 |
| 446 | REGULATION OF MITOCHONDRIAL MEMBRANE POTENTIAL | 3 | 54 | 0.000934 | 0.009859 |
| 447 | RESPONSE TO EXTRACELLULAR STIMULUS | 7 | 441 | 0.0009532 | 0.009922 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 9 | 28 | 3.463e-16 | 3.217e-13 |
| 2 | ENZYME BINDING | 30 | 1737 | 3.228e-14 | 1.5e-11 |
| 3 | TRANSCRIPTION FACTOR BINDING | 17 | 524 | 2.8e-12 | 8.67e-10 |
| 4 | PROTEIN KINASE ACTIVITY | 18 | 640 | 6.243e-12 | 1.45e-09 |
| 5 | KINASE BINDING | 17 | 606 | 2.774e-11 | 5.154e-09 |
| 6 | KINASE REGULATOR ACTIVITY | 11 | 186 | 5.273e-11 | 6.998e-09 |
| 7 | PROTEIN COMPLEX BINDING | 20 | 935 | 4.76e-11 | 6.998e-09 |
| 8 | KINASE ACTIVITY | 19 | 842 | 6.518e-11 | 7.57e-09 |
| 9 | MACROMOLECULAR COMPLEX BINDING | 23 | 1399 | 2.287e-10 | 2.361e-08 |
| 10 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 5 | 12 | 3.795e-10 | 3.526e-08 |
| 11 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 14 | 445 | 4.506e-10 | 3.805e-08 |
| 12 | PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 6 | 30 | 8.954e-10 | 6.932e-08 |
| 13 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 19 | 992 | 1.031e-09 | 7.365e-08 |
| 14 | CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 6 | 34 | 2.006e-09 | 1.331e-07 |
| 15 | NF KAPPAB BINDING | 5 | 30 | 6.498e-08 | 4.025e-06 |
| 16 | CYCLIN BINDING | 4 | 19 | 5.428e-07 | 3.152e-05 |
| 17 | KINASE INHIBITOR ACTIVITY | 6 | 89 | 7.434e-07 | 4.062e-05 |
| 18 | RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 6 | 104 | 1.862e-06 | 9.608e-05 |
| 19 | P53 BINDING | 5 | 67 | 3.978e-06 | 0.0001848 |
| 20 | ADENYL NUCLEOTIDE BINDING | 18 | 1514 | 3.898e-06 | 0.0001848 |
| 21 | ENZYME REGULATOR ACTIVITY | 14 | 959 | 5.776e-06 | 0.0002555 |
| 22 | CORE PROMOTER BINDING | 6 | 152 | 1.664e-05 | 0.000672 |
| 23 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 15 | 1199 | 1.625e-05 | 0.000672 |
| 24 | PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR BINDING | 3 | 15 | 1.892e-05 | 0.0007323 |
| 25 | CHROMATIN BINDING | 9 | 435 | 2.264e-05 | 0.0008415 |
| 26 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR BINDING | 4 | 50 | 2.968e-05 | 0.001061 |
| 27 | HISTONE DEACETYLASE BINDING | 5 | 105 | 3.583e-05 | 0.001233 |
| 28 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 10 | 588 | 4.111e-05 | 0.001317 |
| 29 | HISTONE KINASE ACTIVITY | 3 | 19 | 3.988e-05 | 0.001317 |
| 30 | ACTIVATING TRANSCRIPTION FACTOR BINDING | 4 | 57 | 4.997e-05 | 0.001497 |
| 31 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 8 | 371 | 4.994e-05 | 0.001497 |
| 32 | PROTEIN C TERMINUS BINDING | 6 | 186 | 5.177e-05 | 0.001503 |
| 33 | RIBONUCLEOTIDE BINDING | 18 | 1860 | 6.373e-05 | 0.001794 |
| 34 | MOLECULAR FUNCTION REGULATOR | 15 | 1353 | 6.586e-05 | 0.0018 |
| 35 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 10 | 629 | 7.226e-05 | 0.001918 |
| 36 | SMAD BINDING | 4 | 72 | 0.000125 | 0.003169 |
| 37 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 7 | 315 | 0.0001262 | 0.003169 |
| 38 | REGULATORY REGION NUCLEIC ACID BINDING | 11 | 818 | 0.0001361 | 0.003328 |
| 39 | STEROID HORMONE RECEPTOR BINDING | 4 | 81 | 0.0001974 | 0.004702 |
| 40 | ANDROGEN RECEPTOR BINDING | 3 | 39 | 0.0003575 | 0.007723 |
| 41 | DOUBLE STRANDED DNA BINDING | 10 | 764 | 0.0003499 | 0.007723 |
| 42 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 6 | 264 | 0.0003491 | 0.007723 |
| 43 | PROTEIN DOMAIN SPECIFIC BINDING | 9 | 624 | 0.000351 | 0.007723 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CHROMOSOME | 26 | 880 | 9.993e-18 | 5.836e-15 |
| 2 | CHROMOSOMAL REGION | 16 | 330 | 2.961e-14 | 8.646e-12 |
| 3 | TRANSCRIPTION FACTOR COMPLEX | 15 | 298 | 1.205e-13 | 2.345e-11 |
| 4 | NUCLEAR CHROMOSOME | 18 | 523 | 2.11e-13 | 3.08e-11 |
| 5 | CHROMATIN | 16 | 441 | 2.514e-12 | 2.936e-10 |
| 6 | TRANSFERASE COMPLEX | 18 | 703 | 2.939e-11 | 2.86e-09 |
| 7 | MICROTUBULE CYTOSKELETON | 21 | 1068 | 6.65e-11 | 5.548e-09 |
| 8 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 6 | 31 | 1.107e-09 | 8.083e-08 |
| 9 | CENTROSOME | 14 | 487 | 1.447e-09 | 9.391e-08 |
| 10 | CATALYTIC COMPLEX | 19 | 1038 | 2.183e-09 | 1.275e-07 |
| 11 | SPINDLE | 11 | 289 | 5.63e-09 | 2.989e-07 |
| 12 | CHROMOSOME CENTROMERIC REGION | 9 | 174 | 1.088e-08 | 5.294e-07 |
| 13 | ANAPHASE PROMOTING COMPLEX | 5 | 22 | 1.228e-08 | 5.515e-07 |
| 14 | PROTEIN KINASE COMPLEX | 7 | 90 | 3.103e-08 | 1.197e-06 |
| 15 | CONDENSED CHROMOSOME | 9 | 195 | 2.926e-08 | 1.197e-06 |
| 16 | MICROTUBULE ORGANIZING CENTER | 14 | 623 | 3.278e-08 | 1.197e-06 |
| 17 | CULLIN RING UBIQUITIN LIGASE COMPLEX | 8 | 150 | 5.922e-08 | 2.034e-06 |
| 18 | CYTOSKELETON | 23 | 1967 | 1.491e-07 | 4.838e-06 |
| 19 | CYTOSKELETAL PART | 19 | 1436 | 3.867e-07 | 1.129e-05 |
| 20 | NUCLEAR UBIQUITIN LIGASE COMPLEX | 5 | 42 | 3.753e-07 | 1.129e-05 |
| 21 | CONDENSED NUCLEAR CHROMOSOME | 6 | 85 | 5.658e-07 | 1.573e-05 |
| 22 | CONDENSED CHROMOSOME CENTROMERIC REGION | 6 | 102 | 1.661e-06 | 4.39e-05 |
| 23 | CHROMOSOME TELOMERIC REGION | 7 | 162 | 1.729e-06 | 4.39e-05 |
| 24 | UBIQUITIN LIGASE COMPLEX | 8 | 262 | 4.065e-06 | 9.892e-05 |
| 25 | KINETOCHORE | 6 | 120 | 4.286e-06 | 0.0001001 |
| 26 | SPINDLE POLE | 6 | 126 | 5.683e-06 | 0.0001277 |
| 27 | MCM COMPLEX | 3 | 11 | 6.931e-06 | 0.0001499 |
| 28 | CONDENSED CHROMOSOME OUTER KINETOCHORE | 3 | 12 | 9.218e-06 | 0.0001923 |
| 29 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 7 | 237 | 2.096e-05 | 0.0004221 |
| 30 | CONDENSED NUCLEAR CHROMOSOME CENTROMERIC REGION | 3 | 18 | 3.367e-05 | 0.0006555 |
| 31 | NUCLEAR CHROMATIN | 7 | 291 | 7.708e-05 | 0.001452 |
| 32 | NUCLEAR CHROMOSOME TELOMERIC REGION | 5 | 132 | 0.0001066 | 0.001945 |
| 33 | TRANSCRIPTIONAL REPRESSOR COMPLEX | 4 | 74 | 0.0001391 | 0.002461 |
| 34 | NUCLEOLUS | 11 | 848 | 0.0001864 | 0.003202 |
| 35 | CYTOPLASMIC VESICLE PART | 9 | 601 | 0.0002665 | 0.004446 |
| 36 | RNA POLYMERASE II TRANSCRIPTION FACTOR COMPLEX | 4 | 101 | 0.0004592 | 0.00745 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
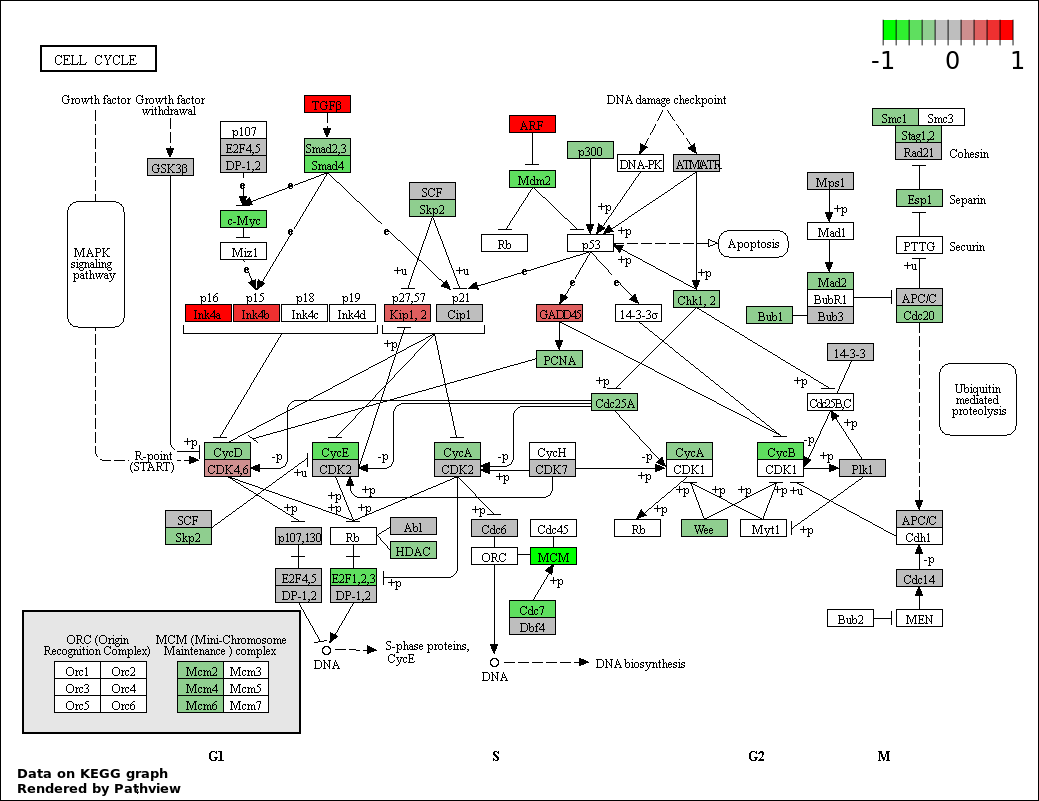
| 1 | hsa04110_Cell_cycle | 71 | 128 | 4.564e-167 | 8.215e-165 | |
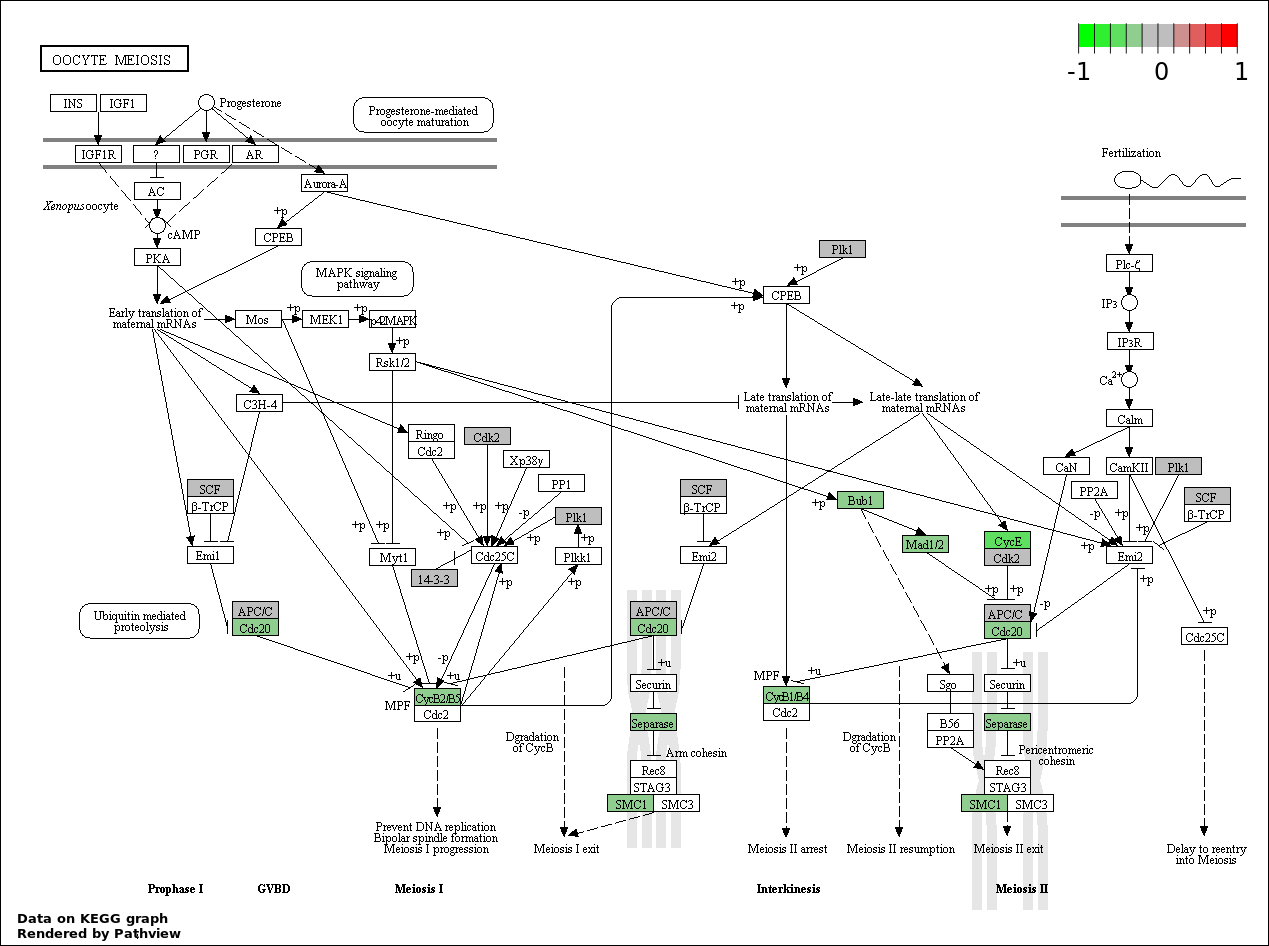
| 2 | hsa04114_Oocyte_meiosis | 21 | 114 | 4.641e-31 | 4.177e-29 | |
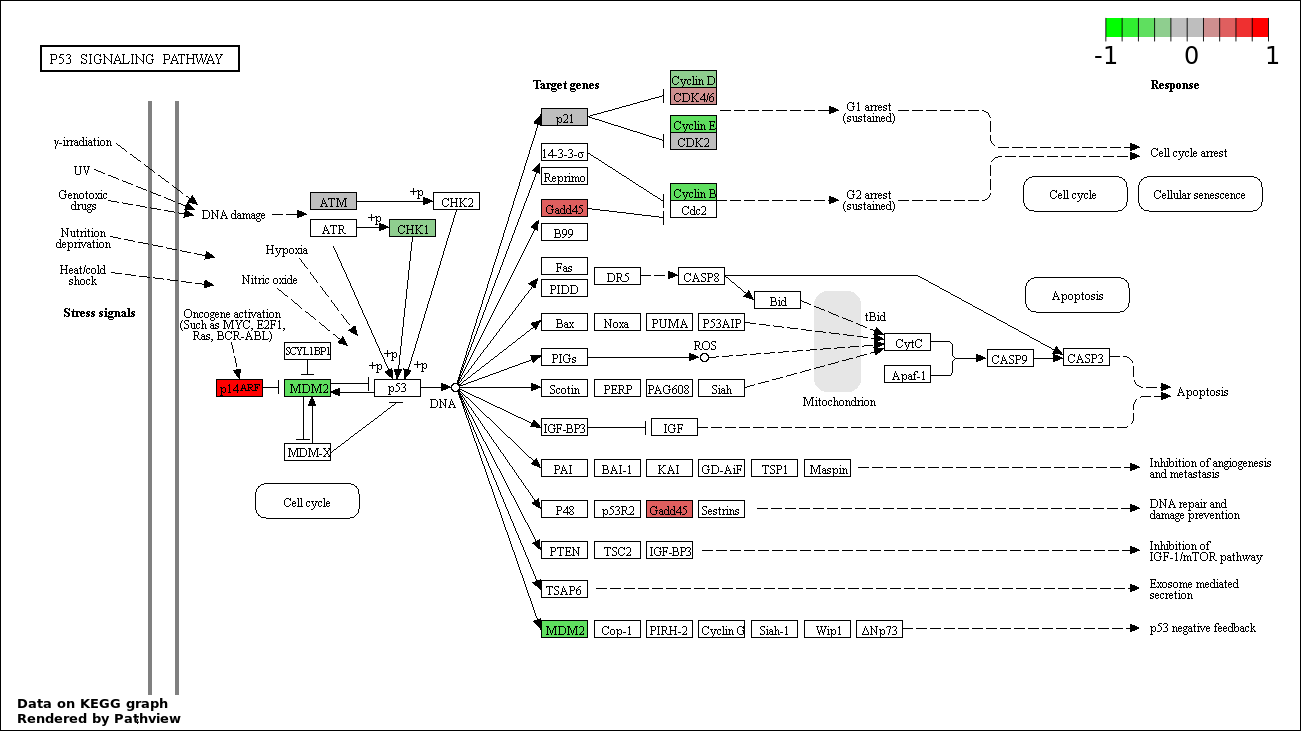
| 3 | hsa04115_p53_signaling_pathway | 14 | 69 | 1.718e-21 | 1.031e-19 | |
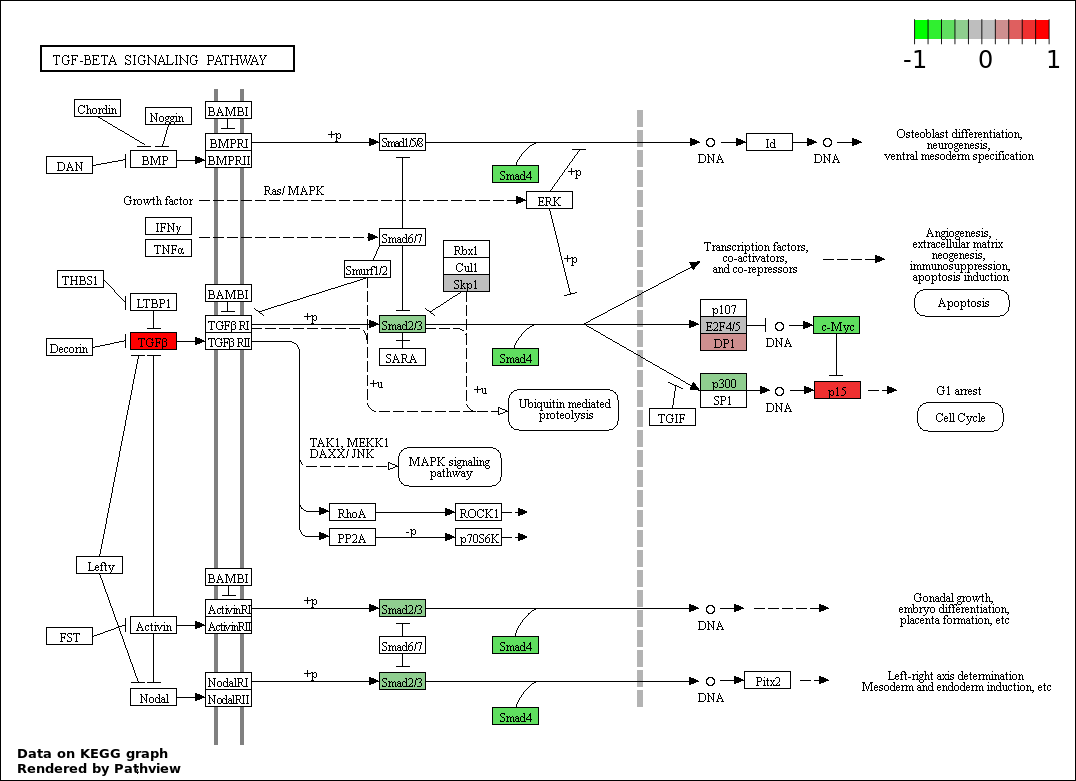
| 4 | hsa04350_TGF.beta_signaling_pathway | 14 | 85 | 4.045e-20 | 1.82e-18 | |
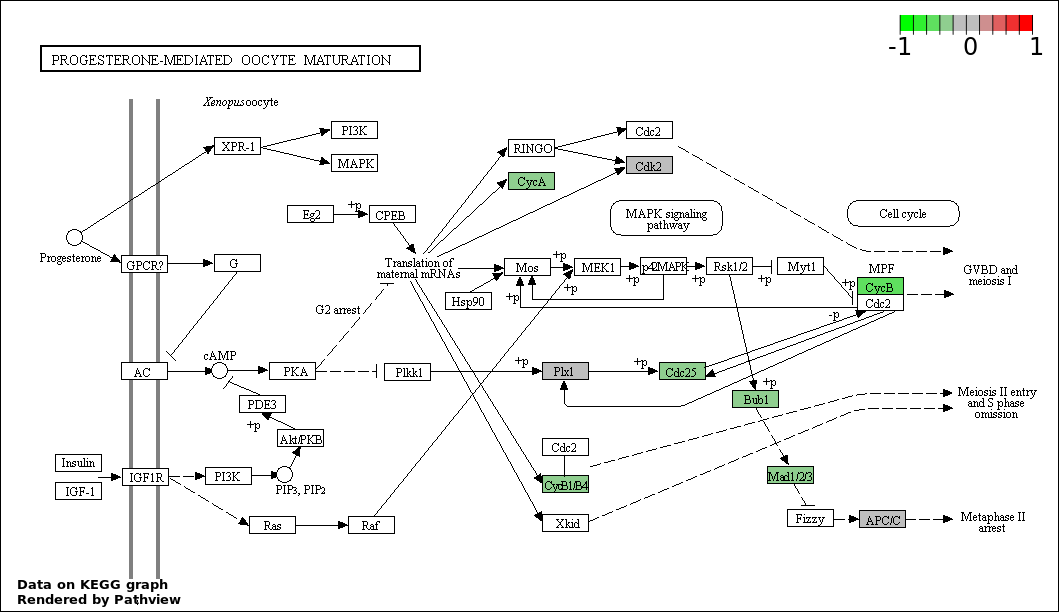
| 5 | hsa04914_Progesterone.mediated_oocyte_maturation | 12 | 87 | 2.171e-16 | 7.815e-15 | |
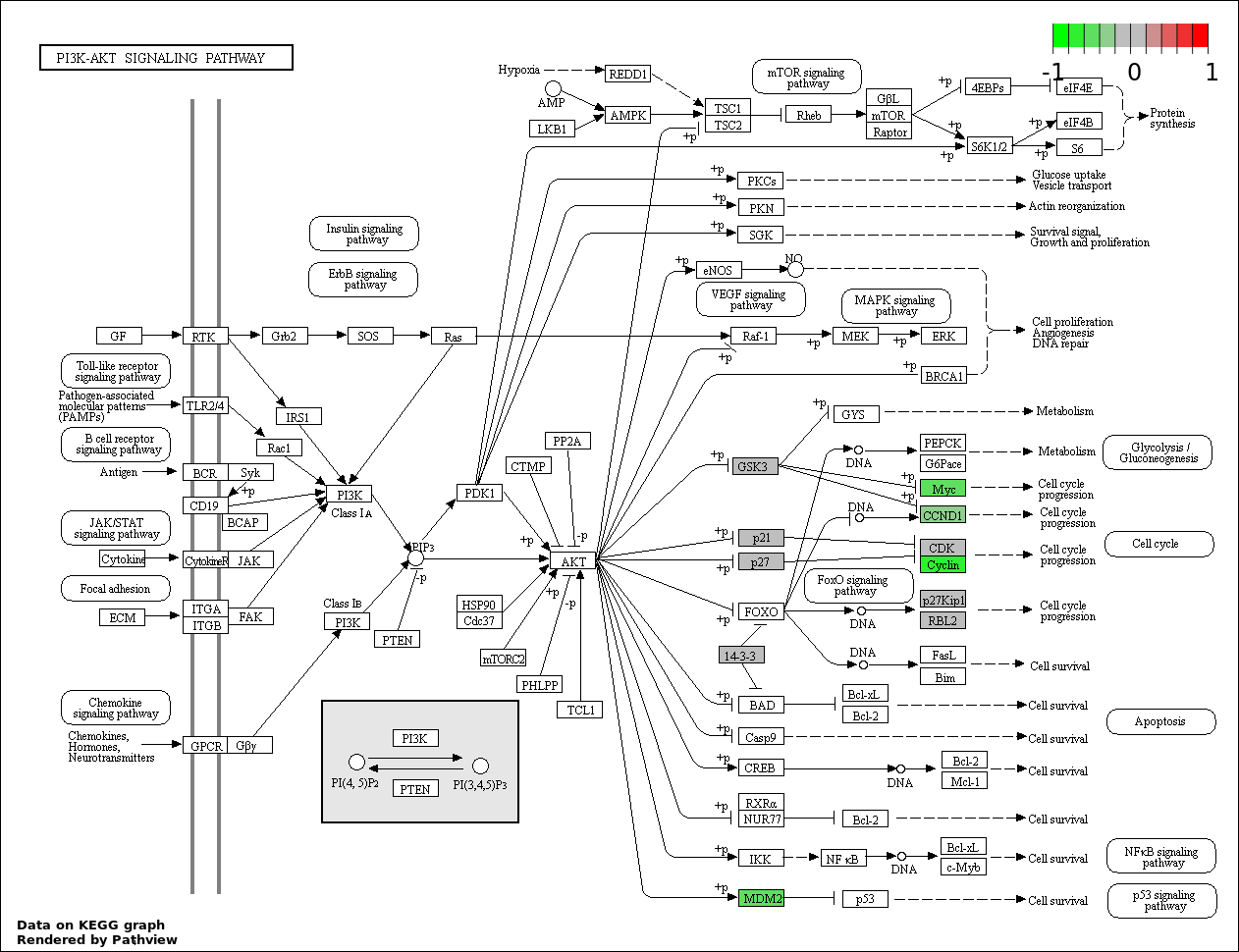
| 6 | hsa04151_PI3K_AKT_signaling_pathway | 17 | 351 | 4.235e-15 | 1.27e-13 | |
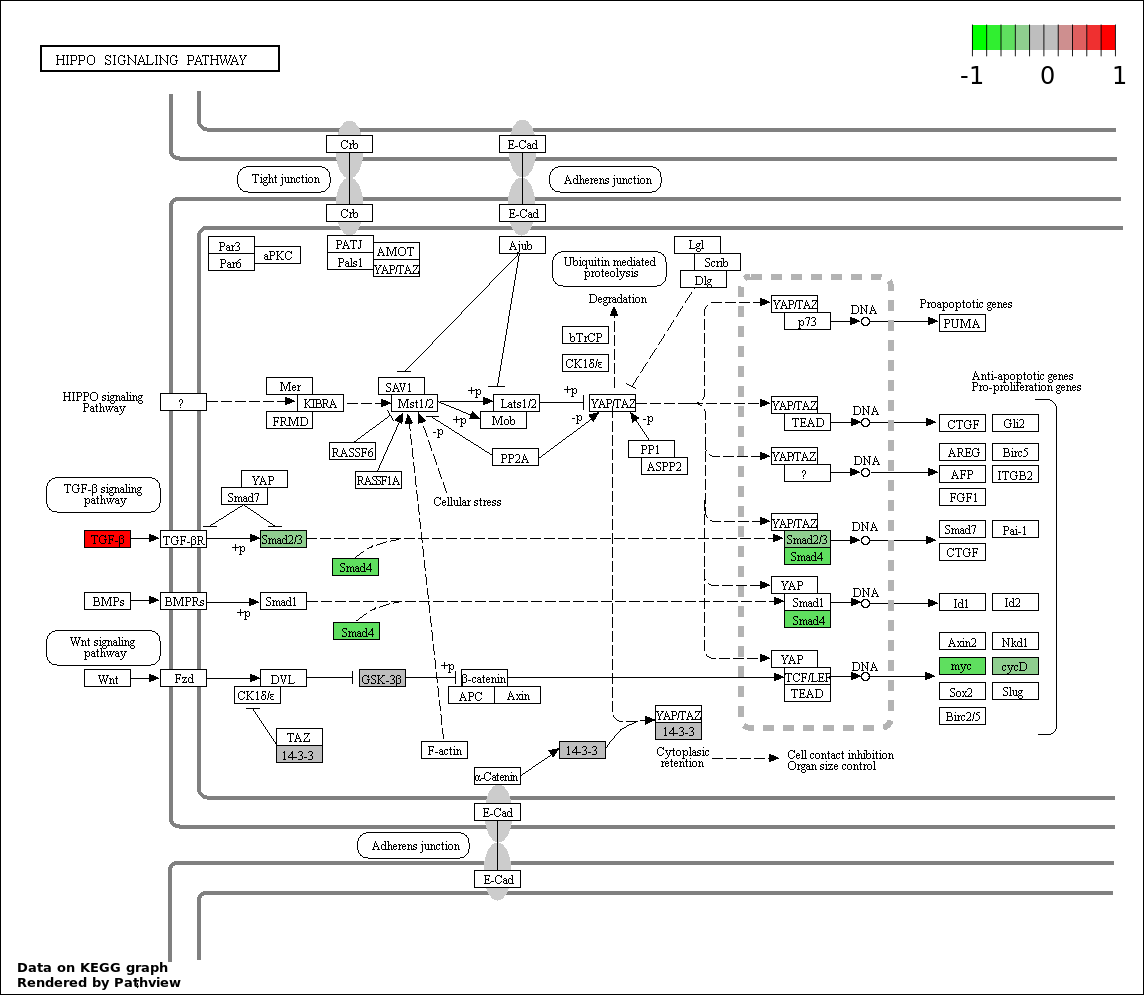
| 7 | hsa04390_Hippo_signaling_pathway | 13 | 154 | 7.916e-15 | 2.036e-13 | |
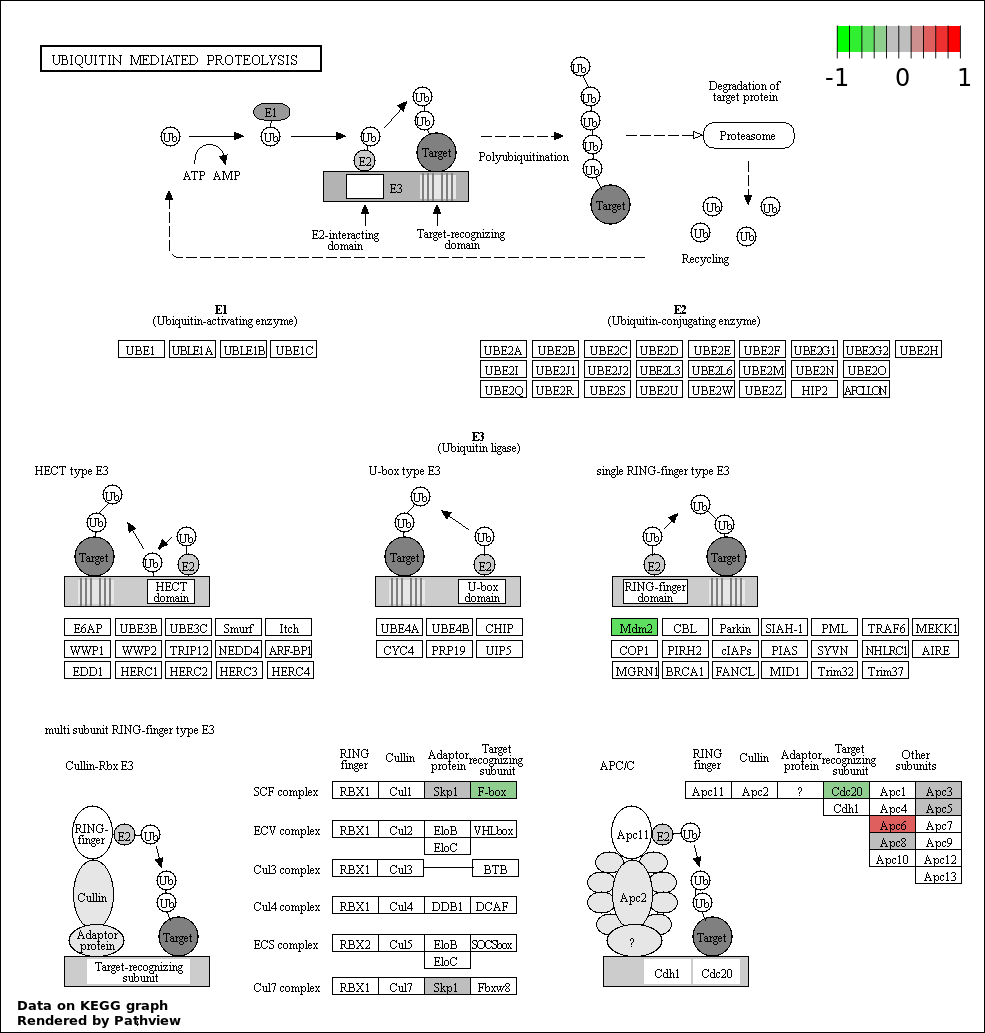
| 8 | hsa04120_Ubiquitin_mediated_proteolysis | 8 | 139 | 3.27e-08 | 7.359e-07 | |
| 9 | hsa04310_Wnt_signaling_pathway | 8 | 151 | 6.236e-08 | 1.247e-06 | |
| 10 | hsa04722_Neurotrophin_signaling_pathway | 7 | 127 | 3.345e-07 | 6.022e-06 | |
| 11 | hsa03030_DNA_replication | 4 | 36 | 7.882e-06 | 0.000129 | |
| 12 | hsa04012_ErbB_signaling_pathway | 5 | 87 | 1.441e-05 | 0.0002161 | |
| 13 | hsa04330_Notch_signaling_pathway | 4 | 47 | 2.317e-05 | 0.0003209 | |
| 14 | hsa04520_Adherens_junction | 4 | 73 | 0.0001319 | 0.001696 | |
| 15 | hsa04144_Endocytosis | 5 | 203 | 0.0007757 | 0.009308 | |
| 16 | hsa04630_Jak.STAT_signaling_pathway | 4 | 155 | 0.002254 | 0.02535 | |
| 17 | hsa04010_MAPK_signaling_pathway | 5 | 268 | 0.002637 | 0.02792 | |
| 18 | hsa04916_Melanogenesis | 3 | 101 | 0.005572 | 0.05572 | |
| 19 | hsa03420_Nucleotide_excision_repair | 2 | 45 | 0.01115 | 0.1056 | |
| 20 | hsa04720_Long.term_potentiation | 2 | 70 | 0.02569 | 0.2312 | |
| 21 | hsa04660_T_cell_receptor_signaling_pathway | 2 | 108 | 0.05643 | 0.4837 | |
| 22 | hsa04380_Osteoclast_differentiation | 2 | 128 | 0.07591 | 0.5658 | |
| 23 | hsa04360_Axon_guidance | 2 | 130 | 0.07797 | 0.5658 | |
| 24 | hsa04510_Focal_adhesion | 2 | 200 | 0.1586 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | EMX2OS |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 23 | TGFB2 | Sponge network | 1.057 | 0.31716 | 0.567 | 0.30298 | 0.36 |
| 2 | MEG3 | hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-5p | 12 | TGFB2 | Sponge network | 0.433 | 0.33816 | 0.567 | 0.30298 | 0.282 |
| 3 | CECR7 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | TGFB2 | Sponge network | 0.551 | 0.56177 | 0.567 | 0.30298 | 0.267 |