This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-139-5p | APH1A | -3.26 | 0 | 0.88 | 0 | miRanda | -0.16 | 0 | NA | |
| 2 | hsa-miR-145-5p | APH1A | -2.63 | 0 | 0.88 | 0 | miRNAWalker2 validate | -0.18 | 0 | NA | |
| 3 | hsa-miR-361-5p | CREBBP | -0.1 | 0.09543 | -0.21 | 0.00056 | PITA; miRanda; miRNATAP | -0.17 | 0 | NA | |
| 4 | hsa-miR-374a-3p | CREBBP | 0.34 | 2.0E-5 | -0.21 | 0.00056 | miRNATAP | -0.12 | 0 | NA | |
| 5 | hsa-miR-590-3p | CREBBP | 1.27 | 0 | -0.21 | 0.00056 | PITA; miRanda; mirMAP; miRNATAP | -0.1 | 0 | NA | |
| 6 | hsa-miR-10b-5p | CTBP1 | -2.11 | 0 | 0.44 | 0 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 7 | hsa-miR-1271-5p | CTBP1 | -1.08 | 0 | 0.44 | 0 | mirMAP | -0.12 | 0 | NA | |
| 8 | hsa-miR-409-3p | CTBP1 | -0.48 | 4.0E-5 | 0.44 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 9 | hsa-let-7a-3p | DLL1 | 0.47 | 0 | -0.95 | 0 | miRNATAP | -0.33 | 0 | NA | |
| 10 | hsa-miR-107 | DLL1 | 0.68 | 0 | -0.95 | 0 | PITA; miRanda; miRNATAP | -0.27 | 0 | NA | |
| 11 | hsa-miR-130b-3p | DLL1 | 1.38 | 0 | -0.95 | 0 | miRNATAP | -0.29 | 0 | NA | |
| 12 | hsa-miR-142-3p | DLL1 | 1.54 | 0 | -0.95 | 0 | miRanda | -0.19 | 0 | NA | |
| 13 | hsa-miR-149-5p | DLL1 | 0.22 | 0.26163 | -0.95 | 0 | miRNATAP | -0.11 | 0 | NA | |
| 14 | hsa-miR-15a-5p | DLL1 | 0.81 | 0 | -0.95 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 15 | hsa-miR-15b-5p | DLL1 | 0.79 | 0 | -0.95 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 16 | hsa-miR-16-5p | DLL1 | 0.47 | 0 | -0.95 | 0 | miRNATAP | -0.51 | 0 | NA | |
| 17 | hsa-miR-193a-3p | DLL1 | 0.31 | 0.00478 | -0.95 | 0 | miRanda | -0.24 | 0 | NA | |
| 18 | hsa-miR-24-3p | DLL1 | -0.12 | 0.21432 | -0.95 | 0 | miRNATAP | -0.16 | 0.00062 | NA | |
| 19 | hsa-miR-301a-3p | DLL1 | 1.32 | 0 | -0.95 | 0 | miRNATAP | -0.3 | 0 | NA | |
| 20 | hsa-miR-320b | DLL1 | 0.41 | 0.00113 | -0.95 | 0 | miRanda | -0.23 | 0 | NA | |
| 21 | hsa-miR-330-5p | DLL1 | 0.63 | 0 | -0.95 | 0 | miRanda | -0.36 | 0 | NA | |
| 22 | hsa-miR-34a-5p | DLL1 | 0.13 | 0.13326 | -0.95 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.18 | 0.0003 | 22438124 | Delta tocotrienol suppresses Notch 1 pathway by upregulating miR 34a in nonsmall cell lung cancer cells |
| 23 | hsa-miR-362-5p | DLL1 | -0.25 | 0.07261 | -0.95 | 0 | PITA; TargetScan; miRNATAP | -0.11 | 0.00039 | NA | |
| 24 | hsa-miR-423-3p | DLL1 | 0.34 | 0.00095 | -0.95 | 0 | PITA | -0.5 | 0 | NA | |
| 25 | hsa-miR-429 | DLL1 | 2.84 | 0 | -0.95 | 0 | miRanda; miRNATAP | -0.18 | 0 | NA | |
| 26 | hsa-miR-454-3p | DLL1 | 1.28 | 0 | -0.95 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 27 | hsa-miR-576-5p | DLL1 | 0.06 | 0.58879 | -0.95 | 0 | PITA | -0.23 | 0 | NA | |
| 28 | hsa-miR-589-5p | DLL1 | 0.37 | 2.0E-5 | -0.95 | 0 | miRNATAP | -0.2 | 8.0E-5 | NA | |
| 29 | hsa-miR-590-3p | DLL1 | 1.27 | 0 | -0.95 | 0 | PITA; miRanda; mirMAP | -0.33 | 0 | NA | |
| 30 | hsa-let-7a-3p | DLL4 | 0.47 | 0 | -0.84 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 31 | hsa-miR-15a-5p | DLL4 | 0.81 | 0 | -0.84 | 0 | miRNATAP | -0.11 | 0.004 | NA | |
| 32 | hsa-miR-15b-5p | DLL4 | 0.79 | 0 | -0.84 | 0 | miRNATAP | -0.21 | 0 | NA | |
| 33 | hsa-miR-16-5p | DLL4 | 0.47 | 0 | -0.84 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 34 | hsa-miR-29a-5p | DLL4 | 0.33 | 0.00445 | -0.84 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 35 | hsa-miR-30c-5p | DLL4 | -0.39 | 8.0E-5 | -0.84 | 0 | miRNATAP | -0.11 | 0.00152 | NA | |
| 36 | hsa-miR-30d-5p | DLL4 | -0.25 | 0.00407 | -0.84 | 0 | miRNATAP | -0.15 | 8.0E-5 | NA | |
| 37 | hsa-miR-424-5p | DLL4 | 0.42 | 0.0048 | -0.84 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 38 | hsa-miR-429 | DLL4 | 2.84 | 0 | -0.84 | 0 | miRanda | -0.2 | 0 | NA | |
| 39 | hsa-miR-484 | DLL4 | 0.35 | 0.00154 | -0.84 | 0 | miRNATAP | -0.21 | 0 | NA | |
| 40 | hsa-miR-324-5p | DTX1 | 0.21 | 0.13941 | -2.34 | 0 | PITA; miRanda | -0.17 | 1.0E-5 | NA | |
| 41 | hsa-miR-331-3p | DTX1 | 0.74 | 0 | -2.34 | 0 | PITA; miRNATAP | -0.32 | 0 | NA | |
| 42 | hsa-miR-429 | DTX1 | 2.84 | 0 | -2.34 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 43 | hsa-miR-484 | DTX1 | 0.35 | 0.00154 | -2.34 | 0 | miRNATAP | -0.31 | 0 | NA | |
| 44 | hsa-let-7a-5p | DTX2 | -0.21 | 0.00725 | 0.9 | 0 | TargetScan; miRNATAP | -0.27 | 0 | NA | |
| 45 | hsa-let-7b-5p | DTX2 | -0.54 | 0 | 0.9 | 0 | miRNATAP | -0.3 | 0 | NA | |
| 46 | hsa-let-7e-5p | DTX2 | 0.26 | 0.00191 | 0.9 | 0 | miRNATAP | -0.11 | 0.00184 | NA | |
| 47 | hsa-let-7g-5p | DTX2 | 0.02 | 0.79164 | 0.9 | 0 | miRNATAP | -0.11 | 0.00699 | NA | |
| 48 | hsa-miR-146b-5p | DTX3 | 0.63 | 0 | 0.41 | 1.0E-5 | miRanda; miRNATAP | -0.13 | 0 | NA | |
| 49 | hsa-miR-185-5p | DTX3 | 0.48 | 0 | 0.41 | 1.0E-5 | MirTarget | -0.11 | 0.0052 | NA | |
| 50 | hsa-miR-10b-3p | DTX3L | -1.58 | 0 | 0.66 | 0 | mirMAP | -0.12 | 0 | NA | |
| 51 | hsa-miR-139-5p | DTX3L | -3.26 | 0 | 0.66 | 0 | miRanda | -0.15 | 0 | NA | |
| 52 | hsa-miR-495-3p | DTX3L | -1.68 | 0 | 0.66 | 0 | mirMAP | -0.11 | 0 | NA | |
| 53 | hsa-let-7a-5p | DTX4 | -0.21 | 0.00725 | -0.81 | 0 | TargetScan; miRNATAP | -0.27 | 1.0E-5 | NA | |
| 54 | hsa-let-7b-5p | DTX4 | -0.54 | 0 | -0.81 | 0 | miRNATAP | -0.14 | 0.00426 | NA | |
| 55 | hsa-let-7e-5p | DTX4 | 0.26 | 0.00191 | -0.81 | 0 | miRNATAP | -0.22 | 0.00011 | NA | |
| 56 | hsa-miR-29b-3p | DTX4 | 0.44 | 6.0E-5 | -0.81 | 0 | miRNATAP | -0.13 | 0.00275 | NA | |
| 57 | hsa-miR-29c-3p | DTX4 | -0.07 | 0.56377 | -0.81 | 0 | miRNATAP | -0.3 | 0 | NA | |
| 58 | hsa-miR-3065-3p | DTX4 | 1.63 | 0 | -0.81 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 59 | hsa-miR-330-3p | DTX4 | 0.43 | 0.00041 | -0.81 | 0 | PITA; mirMAP; miRNATAP | -0.16 | 5.0E-5 | NA | |
| 60 | hsa-miR-429 | DTX4 | 2.84 | 0 | -0.81 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 61 | hsa-miR-10a-5p | DVL1 | -0.8 | 0 | 0.59 | 0 | miRNAWalker2 validate | -0.11 | 1.0E-5 | NA | |
| 62 | hsa-miR-30b-3p | DVL1 | -0.54 | 2.0E-5 | 0.59 | 0 | MirTarget | -0.12 | 2.0E-5 | NA | |
| 63 | hsa-miR-339-5p | DVL1 | -0.21 | 0.06859 | 0.59 | 0 | miRanda | -0.11 | 0.00022 | NA | |
| 64 | hsa-let-7a-5p | DVL3 | -0.21 | 0.00725 | 0.46 | 0 | TargetScan | -0.1 | 0 | NA | |
| 65 | hsa-miR-106b-5p | EP300 | 1.22 | 0 | -0.38 | 1.0E-5 | miRNATAP | -0.19 | 0 | NA | |
| 66 | hsa-miR-181b-5p | EP300 | 1.23 | 0 | -0.38 | 1.0E-5 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 67 | hsa-miR-20a-5p | EP300 | 0.45 | 0.0006 | -0.38 | 1.0E-5 | miRNATAP | -0.15 | 0 | NA | |
| 68 | hsa-miR-25-3p | EP300 | -0.01 | 0.94406 | -0.38 | 1.0E-5 | miRNAWalker2 validate | -0.1 | 0.00585 | NA | |
| 69 | hsa-miR-361-3p | EP300 | -0.04 | 0.6249 | -0.38 | 1.0E-5 | PITA | -0.14 | 0.00025 | NA | |
| 70 | hsa-miR-195-3p | HDAC2 | -1.5 | 0 | 0.41 | 0 | mirMAP | -0.17 | 0 | NA | |
| 71 | hsa-miR-30a-5p | HDAC2 | -0.39 | 0.00955 | 0.41 | 0 | mirMAP | -0.11 | 0 | NA | |
| 72 | hsa-miR-222-3p | HES1 | -0.43 | 0.00104 | 0.37 | 0.0001 | miRNAWalker2 validate | -0.1 | 7.0E-5 | NA | |
| 73 | hsa-miR-23a-3p | HES1 | -0.22 | 0.00963 | 0.37 | 0.0001 | miRNAWalker2 validate; miRTarBase | -0.24 | 0 | NA | |
| 74 | hsa-miR-324-5p | HES1 | 0.21 | 0.13941 | 0.37 | 0.0001 | miRanda | -0.13 | 0 | NA | |
| 75 | hsa-miR-92a-3p | HES1 | -0.34 | 0.00073 | 0.37 | 0.0001 | miRNAWalker2 validate | -0.15 | 1.0E-5 | NA | |
| 76 | hsa-miR-128-3p | JAG1 | 0.9 | 0 | -0.64 | 0 | MirTarget | -0.29 | 0 | NA | |
| 77 | hsa-miR-141-3p | JAG1 | 3 | 0 | -0.64 | 0 | TargetScan; miRNATAP | -0.18 | 0 | NA | |
| 78 | hsa-miR-16-1-3p | JAG1 | 1.3 | 0 | -0.64 | 0 | MirTarget | -0.15 | 0 | NA | |
| 79 | hsa-miR-200a-3p | JAG1 | 2.55 | 0 | -0.64 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 80 | hsa-miR-200c-3p | JAG1 | 2.07 | 0 | -0.64 | 0 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 81 | hsa-miR-21-5p | JAG1 | 2.32 | 0 | -0.64 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.13 | 1.0E-5 | NA | |
| 82 | hsa-miR-34a-5p | JAG1 | 0.13 | 0.13326 | -0.64 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.11 | 0.00421 | NA | |
| 83 | hsa-miR-361-3p | JAG1 | -0.04 | 0.6249 | -0.64 | 0 | miRNATAP | -0.14 | 0.00172 | NA | |
| 84 | hsa-miR-429 | JAG1 | 2.84 | 0 | -0.64 | 0 | miRanda | -0.18 | 0 | NA | |
| 85 | hsa-miR-590-5p | JAG1 | 0.75 | 0 | -0.64 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.14 | 0 | NA | |
| 86 | hsa-miR-93-3p | JAG1 | 0.63 | 0 | -0.64 | 0 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 87 | hsa-miR-98-5p | JAG1 | 0.66 | 0 | -0.64 | 0 | miRNAWalker2 validate | -0.24 | 0 | NA | |
| 88 | hsa-let-7a-3p | JAG2 | 0.47 | 0 | -0.62 | 0 | miRNATAP | -0.14 | 0.00018 | NA | |
| 89 | hsa-let-7b-3p | JAG2 | -0.78 | 0 | -0.62 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 90 | hsa-miR-3607-3p | JAG2 | 0.01 | 0.97207 | -0.62 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 91 | hsa-miR-484 | JAG2 | 0.35 | 0.00154 | -0.62 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 92 | hsa-miR-106b-5p | KAT2B | 1.22 | 0 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.37 | 0 | NA | |
| 93 | hsa-miR-17-5p | KAT2B | 0.6 | 1.0E-5 | -1.59 | 0 | MirTarget; TargetScan | -0.19 | 0 | 23095762 | miR 17 5p targets the p300/CBP associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cells; Targeting of PCAF by miR-17-5p was evaluated using the luciferase reporter assay; Expression of PCAF in PCa cells was associated with the downregulation of miR-17-5p; Targeting of the 3'-untranslated region of PCAF mRNA by miR-17-5p caused translational suppression and RNA degradation and consequently modulation of AR transcriptional activity in PCa cells; PCAF is upregulated in cultured PCa cells and upregulation of PCAF is associated with the downregulation of miR-17-5p; Targeting of PCAF by miR-17-5p modulates AR transcriptional activity and cell growth in cultured PCa cells |
| 94 | hsa-miR-181a-5p | KAT2B | 0.64 | 0 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.33 | 0 | NA | |
| 95 | hsa-miR-181b-5p | KAT2B | 1.23 | 0 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.35 | 0 | NA | |
| 96 | hsa-miR-181c-5p | KAT2B | 0.57 | 0 | -1.59 | 0 | MirTarget | -0.28 | 0 | NA | |
| 97 | hsa-miR-181d-5p | KAT2B | 0.78 | 0 | -1.59 | 0 | MirTarget | -0.28 | 0 | NA | |
| 98 | hsa-miR-192-5p | KAT2B | 1.33 | 0 | -1.59 | 0 | miRNAWalker2 validate | -0.13 | 1.0E-5 | NA | |
| 99 | hsa-miR-19a-3p | KAT2B | 0.82 | 0 | -1.59 | 0 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 100 | hsa-miR-19b-3p | KAT2B | 0.14 | 0.25566 | -1.59 | 0 | miRNAWalker2 validate | -0.12 | 4.0E-5 | NA | |
| 101 | hsa-miR-20a-5p | KAT2B | 0.45 | 0.0006 | -1.59 | 0 | MirTarget | -0.25 | 0 | NA | |
| 102 | hsa-miR-25-3p | KAT2B | -0.01 | 0.94406 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.21 | 1.0E-5 | NA | |
| 103 | hsa-miR-32-5p | KAT2B | 1.39 | 0 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.16 | 0 | NA | |
| 104 | hsa-miR-361-5p | KAT2B | -0.1 | 0.09543 | -1.59 | 0 | miRanda | -0.2 | 0.00106 | NA | |
| 105 | hsa-miR-374a-3p | KAT2B | 0.34 | 2.0E-5 | -1.59 | 0 | MirTarget | -0.12 | 0.00632 | NA | |
| 106 | hsa-miR-429 | KAT2B | 2.84 | 0 | -1.59 | 0 | miRanda | -0.25 | 0 | NA | |
| 107 | hsa-miR-450b-5p | KAT2B | 0.78 | 0 | -1.59 | 0 | MirTarget; miRNATAP | -0.11 | 0.00028 | NA | |
| 108 | hsa-miR-542-3p | KAT2B | 0.57 | 0 | -1.59 | 0 | miRanda | -0.15 | 0 | NA | |
| 109 | hsa-miR-590-3p | KAT2B | 1.27 | 0 | -1.59 | 0 | MirTarget; miRanda; mirMAP; miRNATAP | -0.12 | 4.0E-5 | NA | |
| 110 | hsa-miR-590-5p | KAT2B | 0.75 | 0 | -1.59 | 0 | miRanda | -0.11 | 7.0E-5 | NA | |
| 111 | hsa-miR-92a-3p | KAT2B | -0.34 | 0.00073 | -1.59 | 0 | miRNAWalker2 validate; MirTarget | -0.12 | 0.00059 | NA | |
| 112 | hsa-miR-92b-3p | KAT2B | 1.3 | 0 | -1.59 | 0 | MirTarget | -0.21 | 0 | NA | |
| 113 | hsa-miR-93-5p | KAT2B | 1.02 | 0 | -1.59 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.4 | 0 | NA | |
| 114 | hsa-miR-125b-5p | LFNG | -1.94 | 0 | 0.52 | 0.00059 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 115 | hsa-miR-132-3p | LFNG | -0.04 | 0.6686 | 0.52 | 0.00059 | miRNAWalker2 validate | -0.18 | 0.00316 | NA | |
| 116 | hsa-miR-146b-5p | LFNG | 0.63 | 0 | 0.52 | 0.00059 | PITA; miRanda; miRNATAP | -0.32 | 0 | NA | |
| 117 | hsa-miR-3127-5p | LFNG | 0.9 | 0 | 0.52 | 0.00059 | MirTarget | -0.14 | 0.00073 | NA | |
| 118 | hsa-miR-324-5p | LFNG | 0.21 | 0.13941 | 0.52 | 0.00059 | miRanda | -0.22 | 0 | NA | |
| 119 | hsa-miR-590-3p | LFNG | 1.27 | 0 | 0.52 | 0.00059 | miRanda | -0.15 | 0.00025 | NA | |
| 120 | hsa-miR-429 | MAML2 | 2.84 | 0 | -2.51 | 0 | miRanda | -0.33 | 0 | NA | |
| 121 | hsa-miR-664a-3p | MAML2 | -0.24 | 0.01971 | -2.51 | 0 | mirMAP | -0.2 | 0.00077 | NA | |
| 122 | hsa-miR-125a-3p | MAML3 | -0.18 | 0.10108 | -0.67 | 0 | miRanda | -0.12 | 0.00035 | NA | |
| 123 | hsa-miR-330-5p | MAML3 | 0.63 | 0 | -0.67 | 0 | miRanda | -0.23 | 0 | NA | |
| 124 | hsa-miR-339-5p | MAML3 | -0.21 | 0.06859 | -0.67 | 0 | miRanda | -0.11 | 0.0004 | NA | |
| 125 | hsa-miR-361-5p | MAML3 | -0.1 | 0.09543 | -0.67 | 0 | miRanda | -0.33 | 0 | NA | |
| 126 | hsa-miR-590-3p | MAML3 | 1.27 | 0 | -0.67 | 0 | miRanda | -0.29 | 0 | NA | |
| 127 | hsa-miR-590-3p | MFNG | 1.27 | 0 | -1.05 | 0 | miRanda | -0.15 | 0 | NA | |
| 128 | hsa-miR-16-5p | NCOR2 | 0.47 | 0 | 0.25 | 7.0E-5 | miRNAWalker2 validate; miRTarBase | -0.11 | 0 | NA | |
| 129 | hsa-miR-29a-3p | NCOR2 | -0.7 | 0 | 0.25 | 7.0E-5 | miRNATAP | -0.11 | 0 | NA | |
| 130 | hsa-miR-491-5p | NCOR2 | -1.04 | 0 | 0.25 | 7.0E-5 | miRanda | -0.1 | 0 | NA | |
| 131 | hsa-miR-199b-5p | NCSTN | -1.37 | 0 | 0.38 | 0 | miRNATAP | -0.11 | 0 | NA | |
| 132 | hsa-miR-200c-3p | NOTCH1 | 2.07 | 0 | -0.55 | 0 | MirTarget | -0.14 | 0 | NA | |
| 133 | hsa-miR-103a-3p | NOTCH2 | 0.61 | 0 | -0.57 | 0 | MirTarget | -0.39 | 0 | NA | |
| 134 | hsa-miR-107 | NOTCH2 | 0.68 | 0 | -0.57 | 0 | miRNAWalker2 validate; MirTarget; miRanda; miRNATAP | -0.3 | 0 | NA | |
| 135 | hsa-miR-15a-5p | NOTCH2 | 0.81 | 0 | -0.57 | 0 | MirTarget | -0.34 | 0 | NA | |
| 136 | hsa-miR-16-1-3p | NOTCH2 | 1.3 | 0 | -0.57 | 0 | mirMAP | -0.12 | 2.0E-5 | NA | |
| 137 | hsa-miR-181a-5p | NOTCH2 | 0.64 | 0 | -0.57 | 0 | miRNAWalker2 validate; MirTarget; mirMAP | -0.13 | 4.0E-5 | NA | |
| 138 | hsa-miR-181b-5p | NOTCH2 | 1.23 | 0 | -0.57 | 0 | MirTarget; mirMAP | -0.13 | 0 | NA | |
| 139 | hsa-miR-181c-5p | NOTCH2 | 0.57 | 0 | -0.57 | 0 | miRNAWalker2 validate; MirTarget | -0.14 | 0 | 26983574; 25494473 | This is supported by the depletion of CTCF in glioblastoma cells affecting the expression levels of NOTCH2 as a target of miR-181c;Finally investigation of the mechanism defined Notch2 a key molecular of Notch signaling as the functional downstream target of miR-181c; An inverse correlation was found between miR-181c and Notch2 in glioma cells and verified in fresh glioma samples; miR-181c acts as a tumor suppressor that attenuates proliferation invasion and self-renewal capacities by downregulation of Notch2 in glioma cells |
| 140 | hsa-miR-181d-5p | NOTCH2 | 0.78 | 0 | -0.57 | 0 | MirTarget | -0.11 | 0 | NA | |
| 141 | hsa-miR-185-5p | NOTCH2 | 0.48 | 0 | -0.57 | 0 | MirTarget | -0.26 | 0 | NA | |
| 142 | hsa-miR-25-3p | NOTCH2 | -0.01 | 0.94406 | -0.57 | 0 | miRNAWalker2 validate | -0.16 | 0.00012 | NA | |
| 143 | hsa-miR-29c-3p | NOTCH2 | -0.07 | 0.56377 | -0.57 | 0 | MirTarget | -0.21 | 0 | NA | |
| 144 | hsa-miR-340-5p | NOTCH2 | 1.09 | 0 | -0.57 | 0 | mirMAP | -0.2 | 0 | NA | |
| 145 | hsa-miR-34a-5p | NOTCH2 | 0.13 | 0.13326 | -0.57 | 0 | miRNAWalker2 validate; miRNATAP | -0.11 | 0.00302 | NA | |
| 146 | hsa-miR-423-5p | NOTCH2 | -0.06 | 0.46228 | -0.57 | 0 | MirTarget | -0.13 | 0.0005 | NA | |
| 147 | hsa-miR-125a-3p | NOTCH4 | -0.18 | 0.10108 | -1.51 | 0 | miRanda | -0.12 | 0.00052 | NA | |
| 148 | hsa-miR-146b-5p | NOTCH4 | 0.63 | 0 | -1.51 | 0 | miRanda | -0.13 | 3.0E-5 | NA | |
| 149 | hsa-miR-181c-5p | NOTCH4 | 0.57 | 0 | -1.51 | 0 | miRNAWalker2 validate; miRTarBase | -0.16 | 0 | NA | |
| 150 | hsa-miR-193a-3p | NOTCH4 | 0.31 | 0.00478 | -1.51 | 0 | miRanda | -0.16 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NOTCH SIGNALING PATHWAY | 21 | 114 | 4.495e-41 | 2.091e-37 |
| 2 | NOTCH RECEPTOR PROCESSING | 8 | 16 | 1.588e-19 | 3.695e-16 |
| 3 | REGULATION OF NOTCH SIGNALING PATHWAY | 10 | 67 | 3.699e-18 | 5.738e-15 |
| 4 | POSITIVE REGULATION OF NOTCH SIGNALING PATHWAY | 8 | 34 | 2.2e-16 | 2.559e-13 |
| 5 | CELL FATE COMMITMENT | 10 | 227 | 1.047e-12 | 9.747e-10 |
| 6 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 14 | 750 | 1.416e-12 | 1.098e-09 |
| 7 | EPITHELIUM DEVELOPMENT | 15 | 945 | 1.73e-12 | 1.15e-09 |
| 8 | ORGAN MORPHOGENESIS | 14 | 841 | 6.615e-12 | 3.848e-09 |
| 9 | MORPHOGENESIS OF AN EPITHELIUM | 11 | 400 | 1.056e-11 | 5.461e-09 |
| 10 | EMBRYO DEVELOPMENT | 14 | 894 | 1.5e-11 | 6.424e-09 |
| 11 | TUBE DEVELOPMENT | 12 | 552 | 1.519e-11 | 6.424e-09 |
| 12 | EPITHELIAL CELL FATE COMMITMENT | 5 | 15 | 1.894e-11 | 7.343e-09 |
| 13 | HEART MORPHOGENESIS | 9 | 212 | 2.347e-11 | 8.402e-09 |
| 14 | IMMUNE SYSTEM DEVELOPMENT | 12 | 582 | 2.807e-11 | 8.708e-09 |
| 15 | CELL SURFACE RECEPTOR SIGNALING PATHWAY INVOLVED IN HEART DEVELOPMENT | 5 | 16 | 2.752e-11 | 8.708e-09 |
| 16 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 17 | 1672 | 3.751e-11 | 1.091e-08 |
| 17 | HEART DEVELOPMENT | 11 | 466 | 5.435e-11 | 1.488e-08 |
| 18 | POSITIVE REGULATION OF GENE EXPRESSION | 17 | 1733 | 6.621e-11 | 1.542e-08 |
| 19 | LYMPHOCYTE ACTIVATION | 10 | 342 | 6.047e-11 | 1.542e-08 |
| 20 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 8 | 153 | 6.63e-11 | 1.542e-08 |
| 21 | REGULATION OF CELL DEVELOPMENT | 13 | 836 | 1.113e-10 | 2.388e-08 |
| 22 | TISSUE DEVELOPMENT | 16 | 1518 | 1.129e-10 | 2.388e-08 |
| 23 | CARDIAC CHAMBER MORPHOGENESIS | 7 | 104 | 1.988e-10 | 4.022e-08 |
| 24 | TISSUE MORPHOGENESIS | 11 | 533 | 2.271e-10 | 4.404e-08 |
| 25 | REGULATION OF NEURON DIFFERENTIATION | 11 | 554 | 3.42e-10 | 6.366e-08 |
| 26 | LEUKOCYTE ACTIVATION | 10 | 414 | 3.9e-10 | 6.98e-08 |
| 27 | LEUKOCYTE DIFFERENTIATION | 9 | 292 | 4.055e-10 | 6.988e-08 |
| 28 | PATTERN SPECIFICATION PROCESS | 10 | 418 | 4.282e-10 | 7.115e-08 |
| 29 | EAR DEVELOPMENT | 8 | 195 | 4.604e-10 | 7.199e-08 |
| 30 | NEUROGENESIS | 15 | 1402 | 4.642e-10 | 7.199e-08 |
| 31 | AUDITORY RECEPTOR CELL DIFFERENTIATION | 5 | 28 | 6.111e-10 | 8.886e-08 |
| 32 | DNA TEMPLATED TRANSCRIPTION INITIATION | 8 | 202 | 6.092e-10 | 8.886e-08 |
| 33 | NEURON FATE COMMITMENT | 6 | 67 | 7.748e-10 | 1.092e-07 |
| 34 | LYMPHOCYTE DIFFERENTIATION | 8 | 209 | 7.981e-10 | 1.092e-07 |
| 35 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 12 | 788 | 9.116e-10 | 1.178e-07 |
| 36 | CIRCULATORY SYSTEM DEVELOPMENT | 12 | 788 | 9.116e-10 | 1.178e-07 |
| 37 | TUBE MORPHOGENESIS | 9 | 323 | 9.865e-10 | 1.241e-07 |
| 38 | REGULATION OF CELL DIFFERENTIATION | 15 | 1492 | 1.103e-09 | 1.351e-07 |
| 39 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 16 | 1784 | 1.236e-09 | 1.474e-07 |
| 40 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 13 | 1021 | 1.296e-09 | 1.507e-07 |
| 41 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 16 | 1805 | 1.467e-09 | 1.665e-07 |
| 42 | POSITIVE REGULATION OF CELL COMMUNICATION | 15 | 1532 | 1.592e-09 | 1.763e-07 |
| 43 | CARDIAC CHAMBER DEVELOPMENT | 7 | 144 | 1.971e-09 | 2.118e-07 |
| 44 | HAIR CELL DIFFERENTIATION | 5 | 35 | 2.003e-09 | 2.118e-07 |
| 45 | SENSORY ORGAN DEVELOPMENT | 10 | 493 | 2.108e-09 | 2.18e-07 |
| 46 | REGULATION OF TIMING OF CELL DIFFERENTIATION | 4 | 12 | 2.317e-09 | 2.294e-07 |
| 47 | REGULATION OF DEVELOPMENT HETEROCHRONIC | 4 | 12 | 2.317e-09 | 2.294e-07 |
| 48 | NEURON DIFFERENTIATION | 12 | 874 | 2.946e-09 | 2.856e-07 |
| 49 | MATURE B CELL DIFFERENTIATION INVOLVED IN IMMUNE RESPONSE | 4 | 13 | 3.343e-09 | 3.174e-07 |
| 50 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 16 | 1929 | 3.878e-09 | 3.609e-07 |
| 51 | B CELL DIFFERENTIATION | 6 | 89 | 4.406e-09 | 4.02e-07 |
| 52 | ENDOTHELIUM DEVELOPMENT | 6 | 90 | 4.716e-09 | 4.22e-07 |
| 53 | EMBRYONIC MORPHOGENESIS | 10 | 539 | 4.958e-09 | 4.353e-07 |
| 54 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 11 | 724 | 5.667e-09 | 4.883e-07 |
| 55 | CELL FATE DETERMINATION | 5 | 43 | 5.889e-09 | 4.982e-07 |
| 56 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 10 | 554 | 6.444e-09 | 5.354e-07 |
| 57 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 12 | 957 | 8.168e-09 | 6.558e-07 |
| 58 | CELL ACTIVATION | 10 | 568 | 8.175e-09 | 6.558e-07 |
| 59 | REGULATION OF BINDING | 8 | 283 | 8.661e-09 | 6.831e-07 |
| 60 | MATURE B CELL DIFFERENTIATION | 4 | 17 | 1.108e-08 | 8.592e-07 |
| 61 | CARDIAC SEPTUM MORPHOGENESIS | 5 | 49 | 1.159e-08 | 8.84e-07 |
| 62 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 7 | 191 | 1.406e-08 | 1.004e-06 |
| 63 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 9 | 437 | 1.376e-08 | 1.004e-06 |
| 64 | ARTERY MORPHOGENESIS | 5 | 51 | 1.425e-08 | 1.004e-06 |
| 65 | MECHANORECEPTOR DIFFERENTIATION | 5 | 51 | 1.425e-08 | 1.004e-06 |
| 66 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 12 | 1004 | 1.396e-08 | 1.004e-06 |
| 67 | ANTERIOR POSTERIOR PATTERN SPECIFICATION | 7 | 194 | 1.565e-08 | 1.087e-06 |
| 68 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 11 | 801 | 1.614e-08 | 1.104e-06 |
| 69 | COLUMNAR CUBOIDAL EPITHELIAL CELL DIFFERENTIATION | 6 | 111 | 1.677e-08 | 1.131e-06 |
| 70 | NEURONAL STEM CELL POPULATION MAINTENANCE | 4 | 19 | 1.8e-08 | 1.184e-06 |
| 71 | REGIONALIZATION | 8 | 311 | 1.806e-08 | 1.184e-06 |
| 72 | REGULATION OF EMBRYONIC DEVELOPMENT | 6 | 114 | 1.969e-08 | 1.272e-06 |
| 73 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 11 | 823 | 2.133e-08 | 1.36e-06 |
| 74 | POSITIVE REGULATION OF CELL DEVELOPMENT | 9 | 472 | 2.675e-08 | 1.682e-06 |
| 75 | AORTA MORPHOGENESIS | 4 | 22 | 3.387e-08 | 2.101e-06 |
| 76 | SOMITOGENESIS | 5 | 62 | 3.878e-08 | 2.374e-06 |
| 77 | EPITHELIAL CELL DIFFERENTIATION | 9 | 495 | 4.025e-08 | 2.432e-06 |
| 78 | NEUROEPITHELIAL CELL DIFFERENTIATION | 5 | 63 | 4.208e-08 | 2.51e-06 |
| 79 | B CELL ACTIVATION | 6 | 132 | 4.741e-08 | 2.757e-06 |
| 80 | MAINTENANCE OF CELL NUMBER | 6 | 132 | 4.741e-08 | 2.757e-06 |
| 81 | IMMUNE SYSTEM PROCESS | 15 | 1984 | 5.412e-08 | 3.109e-06 |
| 82 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 13 | 1395 | 5.502e-08 | 3.122e-06 |
| 83 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 12 | 1142 | 5.813e-08 | 3.259e-06 |
| 84 | CELL DEVELOPMENT | 13 | 1426 | 7.131e-08 | 3.95e-06 |
| 85 | EPIDERMAL CELL DIFFERENTIATION | 6 | 142 | 7.328e-08 | 4.012e-06 |
| 86 | ENDOTHELIAL CELL DIFFERENTIATION | 5 | 72 | 8.294e-08 | 4.488e-06 |
| 87 | EPIDERMIS DEVELOPMENT | 7 | 253 | 9.682e-08 | 5.178e-06 |
| 88 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 10 | 740 | 9.914e-08 | 5.242e-06 |
| 89 | ARTERY DEVELOPMENT | 5 | 75 | 1.02e-07 | 5.332e-06 |
| 90 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 7 | 262 | 1.228e-07 | 6.351e-06 |
| 91 | SOMITE DEVELOPMENT | 5 | 78 | 1.243e-07 | 6.357e-06 |
| 92 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO HYPOXIA | 4 | 32 | 1.647e-07 | 8.331e-06 |
| 93 | CARDIAC SEPTUM DEVELOPMENT | 5 | 85 | 1.917e-07 | 9.592e-06 |
| 94 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 9 | 609 | 2.349e-07 | 1.163e-05 |
| 95 | SEGMENTATION | 5 | 89 | 2.416e-07 | 1.171e-05 |
| 96 | B CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 4 | 35 | 2.391e-07 | 1.171e-05 |
| 97 | NEGATIVE REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 4 | 37 | 3.009e-07 | 1.443e-05 |
| 98 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 7 | 303 | 3.291e-07 | 1.562e-05 |
| 99 | LYMPHOCYTE ACTIVATION INVOLVED IN IMMUNE RESPONSE | 5 | 98 | 3.914e-07 | 1.839e-05 |
| 100 | VASCULATURE DEVELOPMENT | 8 | 469 | 4.23e-07 | 1.968e-05 |
| 101 | AORTA DEVELOPMENT | 4 | 41 | 4.594e-07 | 2.116e-05 |
| 102 | VENTRICULAR TRABECULA MYOCARDIUM MORPHOGENESIS | 3 | 11 | 5.517e-07 | 2.517e-05 |
| 103 | MORPHOGENESIS OF AN EPITHELIAL SHEET | 4 | 43 | 5.586e-07 | 2.523e-05 |
| 104 | CARDIAC VENTRICLE DEVELOPMENT | 5 | 106 | 5.79e-07 | 2.59e-05 |
| 105 | AMYLOID PRECURSOR PROTEIN METABOLIC PROCESS | 3 | 12 | 7.348e-07 | 3.256e-05 |
| 106 | SKIN DEVELOPMENT | 6 | 211 | 7.587e-07 | 3.33e-05 |
| 107 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 8 | 513 | 8.323e-07 | 3.619e-05 |
| 108 | NEGATIVE REGULATION OF EPIDERMAL CELL DIFFERENTIATION | 3 | 13 | 9.542e-07 | 4.111e-05 |
| 109 | BLOOD VESSEL MORPHOGENESIS | 7 | 364 | 1.127e-06 | 4.811e-05 |
| 110 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 5 | 122 | 1.164e-06 | 4.923e-05 |
| 111 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 10 | 983 | 1.355e-06 | 5.68e-05 |
| 112 | POSITIVE REGULATION OF BINDING | 5 | 127 | 1.42e-06 | 5.898e-05 |
| 113 | N TERMINAL PROTEIN AMINO ACID ACETYLATION | 3 | 15 | 1.515e-06 | 6.183e-05 |
| 114 | REGULATION OF CELL PROLIFERATION INVOLVED IN HEART MORPHOGENESIS | 3 | 15 | 1.515e-06 | 6.183e-05 |
| 115 | OUTFLOW TRACT MORPHOGENESIS | 4 | 56 | 1.639e-06 | 6.633e-05 |
| 116 | REGULATION OF ORGAN MORPHOGENESIS | 6 | 242 | 1.686e-06 | 6.762e-05 |
| 117 | MORPHOGENESIS OF AN ENDOTHELIUM | 3 | 16 | 1.863e-06 | 7.345e-05 |
| 118 | NEGATIVE REGULATION OF EPIDERMIS DEVELOPMENT | 3 | 16 | 1.863e-06 | 7.345e-05 |
| 119 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 5 | 139 | 2.216e-06 | 8.666e-05 |
| 120 | EMBRYONIC ORGAN DEVELOPMENT | 7 | 406 | 2.329e-06 | 9.03e-05 |
| 121 | CARDIAC VENTRICLE MORPHOGENESIS | 4 | 62 | 2.474e-06 | 9.434e-05 |
| 122 | EMBRYONIC HEART TUBE MORPHOGENESIS | 4 | 62 | 2.474e-06 | 9.434e-05 |
| 123 | REGULATION OF PROTEIN ACETYLATION | 4 | 64 | 2.811e-06 | 0.0001064 |
| 124 | NEURAL TUBE DEVELOPMENT | 5 | 149 | 3.118e-06 | 0.000117 |
| 125 | REGULATION OF DNA TEMPLATED TRANSCRIPTION IN RESPONSE TO STRESS | 4 | 67 | 3.381e-06 | 0.0001258 |
| 126 | MUSCLE STRUCTURE DEVELOPMENT | 7 | 432 | 3.509e-06 | 0.0001296 |
| 127 | EMBRYONIC ORGAN MORPHOGENESIS | 6 | 279 | 3.835e-06 | 0.0001405 |
| 128 | SKIN EPIDERMIS DEVELOPMENT | 4 | 71 | 4.267e-06 | 0.0001539 |
| 129 | CELL FATE SPECIFICATION | 4 | 71 | 4.267e-06 | 0.0001539 |
| 130 | NEGATIVE REGULATION OF CELL PROLIFERATION | 8 | 643 | 4.478e-06 | 0.0001603 |
| 131 | EMBRYONIC HEART TUBE DEVELOPMENT | 4 | 73 | 4.769e-06 | 0.0001694 |
| 132 | MEMBRANE PROTEIN ECTODOMAIN PROTEOLYSIS | 3 | 22 | 5.09e-06 | 0.0001767 |
| 133 | SOMATIC STEM CELL DIVISION | 3 | 22 | 5.09e-06 | 0.0001767 |
| 134 | ANGIOGENESIS | 6 | 293 | 5.081e-06 | 0.0001767 |
| 135 | REGULATION OF PROTEIN BINDING | 5 | 168 | 5.61e-06 | 0.0001928 |
| 136 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 10 | 1152 | 5.635e-06 | 0.0001928 |
| 137 | RENAL TUBULE DEVELOPMENT | 4 | 78 | 6.217e-06 | 0.0002111 |
| 138 | REGULATION OF CELL PROLIFERATION | 11 | 1496 | 8.246e-06 | 0.000278 |
| 139 | HEART TRABECULA MORPHOGENESIS | 3 | 26 | 8.557e-06 | 0.0002844 |
| 140 | N TERMINAL PROTEIN AMINO ACID MODIFICATION | 3 | 26 | 8.557e-06 | 0.0002844 |
| 141 | TISSUE REMODELING | 4 | 87 | 9.607e-06 | 0.0003151 |
| 142 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER INVOLVED IN CELLULAR RESPONSE TO CHEMICAL STIMULUS | 3 | 27 | 9.617e-06 | 0.0003151 |
| 143 | NEGATIVE REGULATION OF NOTCH SIGNALING PATHWAY | 3 | 28 | 1.076e-05 | 0.0003477 |
| 144 | REGULATION OF SPROUTING ANGIOGENESIS | 3 | 28 | 1.076e-05 | 0.0003477 |
| 145 | STEM CELL DIVISION | 3 | 29 | 1.199e-05 | 0.0003795 |
| 146 | REGULATION OF HEART MORPHOGENESIS | 3 | 29 | 1.199e-05 | 0.0003795 |
| 147 | REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 3 | 29 | 1.199e-05 | 0.0003795 |
| 148 | RESPIRATORY SYSTEM DEVELOPMENT | 5 | 197 | 1.216e-05 | 0.0003823 |
| 149 | NEPHRON EPITHELIUM DEVELOPMENT | 4 | 93 | 1.252e-05 | 0.000391 |
| 150 | COVALENT CHROMATIN MODIFICATION | 6 | 345 | 1.29e-05 | 0.0004003 |
| 151 | CARDIOCYTE DIFFERENTIATION | 4 | 96 | 1.42e-05 | 0.0004375 |
| 152 | CARDIAC ATRIUM DEVELOPMENT | 3 | 31 | 1.472e-05 | 0.0004505 |
| 153 | FOREBRAIN DEVELOPMENT | 6 | 357 | 1.566e-05 | 0.0004763 |
| 154 | ENDOCARDIAL CUSHION DEVELOPMENT | 3 | 32 | 1.622e-05 | 0.0004808 |
| 155 | POSITIVE REGULATION OF GLIAL CELL DIFFERENTIATION | 3 | 32 | 1.622e-05 | 0.0004808 |
| 156 | POSITIVE REGULATION OF BMP SIGNALING PATHWAY | 3 | 32 | 1.622e-05 | 0.0004808 |
| 157 | BLOOD VESSEL REMODELING | 3 | 32 | 1.622e-05 | 0.0004808 |
| 158 | HEART VALVE DEVELOPMENT | 3 | 34 | 1.953e-05 | 0.0005752 |
| 159 | MEMBRANE PROTEIN PROTEOLYSIS | 3 | 35 | 2.134e-05 | 0.0006245 |
| 160 | CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 3 | 36 | 2.325e-05 | 0.0006763 |
| 161 | MYOBLAST DIFFERENTIATION | 3 | 37 | 2.528e-05 | 0.0007306 |
| 162 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 5 | 232 | 2.672e-05 | 0.0007675 |
| 163 | REGULATION OF STEM CELL DIFFERENTIATION | 4 | 113 | 2.702e-05 | 0.0007713 |
| 164 | GLAND DEVELOPMENT | 6 | 395 | 2.769e-05 | 0.0007856 |
| 165 | NEPHRON DEVELOPMENT | 4 | 115 | 2.895e-05 | 0.0008164 |
| 166 | TRABECULA MORPHOGENESIS | 3 | 39 | 2.967e-05 | 0.0008267 |
| 167 | COCHLEA DEVELOPMENT | 3 | 39 | 2.967e-05 | 0.0008267 |
| 168 | SENSORY ORGAN MORPHOGENESIS | 5 | 239 | 3.081e-05 | 0.0008533 |
| 169 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 6 | 408 | 3.32e-05 | 0.0009141 |
| 170 | NEGATIVE REGULATION OF STEM CELL DIFFERENTIATION | 3 | 43 | 3.99e-05 | 0.001092 |
| 171 | KIDNEY EPITHELIUM DEVELOPMENT | 4 | 125 | 4.015e-05 | 0.001092 |
| 172 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 8 | 872 | 4.077e-05 | 0.001103 |
| 173 | LEUKOCYTE CELL CELL ADHESION | 5 | 255 | 4.197e-05 | 0.001129 |
| 174 | NEGATIVE REGULATION OF HEMOPOIESIS | 4 | 128 | 4.405e-05 | 0.001178 |
| 175 | VENTRICULAR CARDIAC MUSCLE TISSUE DEVELOPMENT | 3 | 45 | 4.578e-05 | 0.00121 |
| 176 | REGULATION OF EPIDERMAL CELL DIFFERENTIATION | 3 | 45 | 4.578e-05 | 0.00121 |
| 177 | CELLULAR COMPONENT MORPHOGENESIS | 8 | 900 | 5.103e-05 | 0.001341 |
| 178 | POSITIVE REGULATION OF GLIOGENESIS | 3 | 47 | 5.22e-05 | 0.001365 |
| 179 | MESENCHYMAL CELL DIFFERENTIATION | 4 | 134 | 5.268e-05 | 0.001369 |
| 180 | NEGATIVE REGULATION OF GENE EXPRESSION | 10 | 1493 | 5.398e-05 | 0.001395 |
| 181 | GLIAL CELL DIFFERENTIATION | 4 | 136 | 5.581e-05 | 0.001435 |
| 182 | GLOMERULUS DEVELOPMENT | 3 | 49 | 5.919e-05 | 0.001513 |
| 183 | MUSCLE TISSUE DEVELOPMENT | 5 | 275 | 6.008e-05 | 0.001528 |
| 184 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 10 | 1517 | 6.182e-05 | 0.001563 |
| 185 | MUSCLE ORGAN DEVELOPMENT | 5 | 277 | 6.218e-05 | 0.001564 |
| 186 | CELLULAR RESPONSE TO OXYGEN LEVELS | 4 | 143 | 6.786e-05 | 0.001697 |
| 187 | NEURON DEVELOPMENT | 7 | 687 | 7.003e-05 | 0.001743 |
| 188 | CARDIAC MUSCLE TISSUE MORPHOGENESIS | 3 | 54 | 7.927e-05 | 0.001962 |
| 189 | HEAD DEVELOPMENT | 7 | 709 | 8.537e-05 | 0.002102 |
| 190 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 4 | 152 | 8.6e-05 | 0.002106 |
| 191 | RHYTHMIC PROCESS | 5 | 298 | 8.781e-05 | 0.002128 |
| 192 | IMMUNE EFFECTOR PROCESS | 6 | 486 | 8.768e-05 | 0.002128 |
| 193 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 3 | 56 | 8.841e-05 | 0.002131 |
| 194 | UROGENITAL SYSTEM DEVELOPMENT | 5 | 299 | 8.921e-05 | 0.00214 |
| 195 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 4 | 156 | 9.51e-05 | 0.002258 |
| 196 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 5 | 303 | 9.496e-05 | 0.002258 |
| 197 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 5 | 306 | 9.947e-05 | 0.002349 |
| 198 | REGULATION OF GLIAL CELL DIFFERENTIATION | 3 | 59 | 0.0001034 | 0.002429 |
| 199 | RESPONSE TO OXYGEN LEVELS | 5 | 311 | 0.0001073 | 0.00251 |
| 200 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 4 | 162 | 0.00011 | 0.00256 |
| 201 | REGULATION OF HEMOPOIESIS | 5 | 314 | 0.0001123 | 0.002599 |
| 202 | EMBRYONIC DIGIT MORPHOGENESIS | 3 | 61 | 0.0001142 | 0.00263 |
| 203 | REGULATION OF EPIDERMIS DEVELOPMENT | 3 | 63 | 0.0001257 | 0.002822 |
| 204 | LOOP OF HENLE DEVELOPMENT | 2 | 11 | 0.0001268 | 0.002822 |
| 205 | ENDOCARDIUM DEVELOPMENT | 2 | 11 | 0.0001268 | 0.002822 |
| 206 | NEGATIVE REGULATION OF SPROUTING ANGIOGENESIS | 2 | 11 | 0.0001268 | 0.002822 |
| 207 | NEGATIVE REGULATION OF GLIAL CELL PROLIFERATION | 2 | 11 | 0.0001268 | 0.002822 |
| 208 | EPITHELIAL TO MESENCHYMAL TRANSITION INVOLVED IN ENDOCARDIAL CUSHION FORMATION | 2 | 11 | 0.0001268 | 0.002822 |
| 209 | ENDOTHELIAL TUBE MORPHOGENESIS | 2 | 11 | 0.0001268 | 0.002822 |
| 210 | APPENDAGE DEVELOPMENT | 4 | 169 | 0.0001295 | 0.002857 |
| 211 | LIMB DEVELOPMENT | 4 | 169 | 0.0001295 | 0.002857 |
| 212 | FOREBRAIN GENERATION OF NEURONS | 3 | 66 | 0.0001444 | 0.00317 |
| 213 | GLIOGENESIS | 4 | 175 | 0.0001482 | 0.003237 |
| 214 | CARDIAC LEFT VENTRICLE MORPHOGENESIS | 2 | 12 | 0.000152 | 0.003259 |
| 215 | DISTAL TUBULE DEVELOPMENT | 2 | 12 | 0.000152 | 0.003259 |
| 216 | LATERAL VENTRICLE DEVELOPMENT | 2 | 12 | 0.000152 | 0.003259 |
| 217 | POSITIVE REGULATION OF ASTROCYTE DIFFERENTIATION | 2 | 12 | 0.000152 | 0.003259 |
| 218 | CHROMATIN MODIFICATION | 6 | 539 | 0.0001545 | 0.003298 |
| 219 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 5 | 337 | 0.0001563 | 0.00332 |
| 220 | MUSCLE ORGAN MORPHOGENESIS | 3 | 70 | 0.000172 | 0.003639 |
| 221 | NEURONAL STEM CELL DIVISION | 2 | 13 | 0.0001794 | 0.003694 |
| 222 | NEGATIVE REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 2 | 13 | 0.0001794 | 0.003694 |
| 223 | VASCULAR SMOOTH MUSCLE CELL DIFFERENTIATION | 2 | 13 | 0.0001794 | 0.003694 |
| 224 | LEFT RIGHT AXIS SPECIFICATION | 2 | 13 | 0.0001794 | 0.003694 |
| 225 | CARDIOBLAST DIFFERENTIATION | 2 | 13 | 0.0001794 | 0.003694 |
| 226 | NEUROBLAST DIVISION | 2 | 13 | 0.0001794 | 0.003694 |
| 227 | EPITHELIAL CELL DEVELOPMENT | 4 | 186 | 0.0001872 | 0.003821 |
| 228 | REGULATION OF CELL PROJECTION ORGANIZATION | 6 | 558 | 0.0001865 | 0.003821 |
| 229 | POSITIVE REGULATION OF PROTEIN BINDING | 3 | 73 | 0.0001949 | 0.003925 |
| 230 | POSITIVE REGULATION OF STAT CASCADE | 3 | 73 | 0.0001949 | 0.003925 |
| 231 | POSITIVE REGULATION OF JAK STAT CASCADE | 3 | 73 | 0.0001949 | 0.003925 |
| 232 | MESENCHYME DEVELOPMENT | 4 | 190 | 0.0002031 | 0.004057 |
| 233 | STEM CELL DIFFERENTIATION | 4 | 190 | 0.0002031 | 0.004057 |
| 234 | IMMUNE RESPONSE | 8 | 1100 | 0.0002069 | 0.004115 |
| 235 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY INVOLVED IN NEURAL TUBE CLOSURE | 2 | 14 | 0.0002091 | 0.004124 |
| 236 | BETA AMYLOID METABOLIC PROCESS | 2 | 14 | 0.0002091 | 0.004124 |
| 237 | REGULATION OF BMP SIGNALING PATHWAY | 3 | 77 | 0.0002282 | 0.00448 |
| 238 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 5 | 368 | 0.0002352 | 0.004598 |
| 239 | RESPONSE TO MURAMYL DIPEPTIDE | 2 | 15 | 0.0002411 | 0.004674 |
| 240 | ENDOCARDIAL CUSHION FORMATION | 2 | 15 | 0.0002411 | 0.004674 |
| 241 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 8 | 1135 | 0.0002564 | 0.00495 |
| 242 | CARDIAC RIGHT VENTRICLE MORPHOGENESIS | 2 | 16 | 0.0002753 | 0.005271 |
| 243 | APOPTOTIC PROCESS INVOLVED IN MORPHOGENESIS | 2 | 16 | 0.0002753 | 0.005271 |
| 244 | REGULATION OF CELL DEATH | 9 | 1472 | 0.0002823 | 0.005383 |
| 245 | HAIR CYCLE | 3 | 83 | 0.0002849 | 0.005388 |
| 246 | MOLTING CYCLE | 3 | 83 | 0.0002849 | 0.005388 |
| 247 | CELL CELL ADHESION | 6 | 608 | 0.0002964 | 0.005583 |
| 248 | REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION | 3 | 85 | 0.0003056 | 0.00571 |
| 249 | POSITIVE REGULATION OF CHROMATIN MODIFICATION | 3 | 85 | 0.0003056 | 0.00571 |
| 250 | MEMBRANE PROTEIN INTRACELLULAR DOMAIN PROTEOLYSIS | 2 | 17 | 0.0003117 | 0.005788 |
| 251 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 9 | 1492 | 0.0003122 | 0.005788 |
| 252 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 7 | 876 | 0.0003155 | 0.005825 |
| 253 | LABYRINTHINE LAYER BLOOD VESSEL DEVELOPMENT | 2 | 18 | 0.0003503 | 0.006417 |
| 254 | PERICARDIUM DEVELOPMENT | 2 | 18 | 0.0003503 | 0.006417 |
| 255 | REGULATION OF GLIOGENESIS | 3 | 90 | 0.0003616 | 0.006597 |
| 256 | KIDNEY VASCULATURE DEVELOPMENT | 2 | 19 | 0.0003911 | 0.007026 |
| 257 | REGULATION OF CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 2 | 19 | 0.0003911 | 0.007026 |
| 258 | POSITIVE REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 2 | 19 | 0.0003911 | 0.007026 |
| 259 | RENAL SYSTEM VASCULATURE DEVELOPMENT | 2 | 19 | 0.0003911 | 0.007026 |
| 260 | REGULATION OF DNA BINDING | 3 | 93 | 0.0003981 | 0.007125 |
| 261 | TELENCEPHALON DEVELOPMENT | 4 | 228 | 0.0004064 | 0.007246 |
| 262 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 4 | 229 | 0.0004132 | 0.007339 |
| 263 | CANONICAL WNT SIGNALING PATHWAY | 3 | 95 | 0.0004238 | 0.007497 |
| 264 | MYELOID DENDRITIC CELL DIFFERENTIATION | 2 | 20 | 0.0004342 | 0.007652 |
| 265 | MEMORY | 3 | 98 | 0.0004642 | 0.00812 |
| 266 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 4 | 236 | 0.000463 | 0.00812 |
| 267 | LEFT RIGHT PATTERN FORMATION | 2 | 21 | 0.0004794 | 0.008141 |
| 268 | REGULATION OF GLIAL CELL PROLIFERATION | 2 | 21 | 0.0004794 | 0.008141 |
| 269 | CHROMATIN ORGANIZATION | 6 | 663 | 0.0004704 | 0.008141 |
| 270 | APOPTOTIC PROCESS INVOLVED IN DEVELOPMENT | 2 | 21 | 0.0004794 | 0.008141 |
| 271 | POSITIVE REGULATION OF GROWTH | 4 | 238 | 0.0004779 | 0.008141 |
| 272 | COCHLEA MORPHOGENESIS | 2 | 21 | 0.0004794 | 0.008141 |
| 273 | NEGATIVE REGULATION OF PROTEIN ACETYLATION | 2 | 21 | 0.0004794 | 0.008141 |
| 274 | MUSCLE CELL DIFFERENTIATION | 4 | 237 | 0.0004704 | 0.008141 |
| 275 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 3 | 100 | 0.0004925 | 0.008332 |
| 276 | KERATINOCYTE DIFFERENTIATION | 3 | 101 | 0.000507 | 0.008517 |
| 277 | CELL PROLIFERATION | 6 | 672 | 0.0005052 | 0.008517 |
| 278 | ENDOCARDIAL CUSHION MORPHOGENESIS | 2 | 22 | 0.0005268 | 0.008786 |
| 279 | BETA CATENIN DESTRUCTION COMPLEX DISASSEMBLY | 2 | 22 | 0.0005268 | 0.008786 |
| 280 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 3 | 103 | 0.0005369 | 0.008891 |
| 281 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 3 | 103 | 0.0005369 | 0.008891 |
| 282 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 2 | 23 | 0.0005764 | 0.009444 |
| 283 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO STRESS | 2 | 23 | 0.0005764 | 0.009444 |
| 284 | POSITIVE REGULATION OF COLLAGEN METABOLIC PROCESS | 2 | 23 | 0.0005764 | 0.009444 |
| 285 | SPINAL CORD DEVELOPMENT | 3 | 106 | 0.0005839 | 0.009532 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NOTCH BINDING | 7 | 18 | 3.261e-16 | 3.029e-13 |
| 2 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 10 | 588 | 1.136e-08 | 5.278e-06 |
| 3 | TRANSCRIPTION FACTOR BINDING | 9 | 524 | 6.551e-08 | 2.029e-05 |
| 4 | BETA CATENIN BINDING | 5 | 84 | 1.806e-07 | 4.195e-05 |
| 5 | TRANSCRIPTION COACTIVATOR ACTIVITY | 6 | 296 | 5.387e-06 | 0.001001 |
| 6 | RECEPTOR BINDING | 11 | 1476 | 7.247e-06 | 0.001122 |
| 7 | RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 4 | 104 | 1.949e-05 | 0.002586 |
| 8 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 4 | 133 | 5.116e-05 | 0.005281 |
| 9 | CHROMATIN BINDING | 6 | 435 | 4.747e-05 | 0.005281 |
| 10 | CALCIUM ION BINDING | 7 | 697 | 7.67e-05 | 0.007125 |
| 11 | CORE PROMOTER BINDING | 4 | 152 | 8.6e-05 | 0.007217 |
| 12 | REPRESSING TRANSCRIPTION FACTOR BINDING | 3 | 57 | 9.322e-05 | 0.007217 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
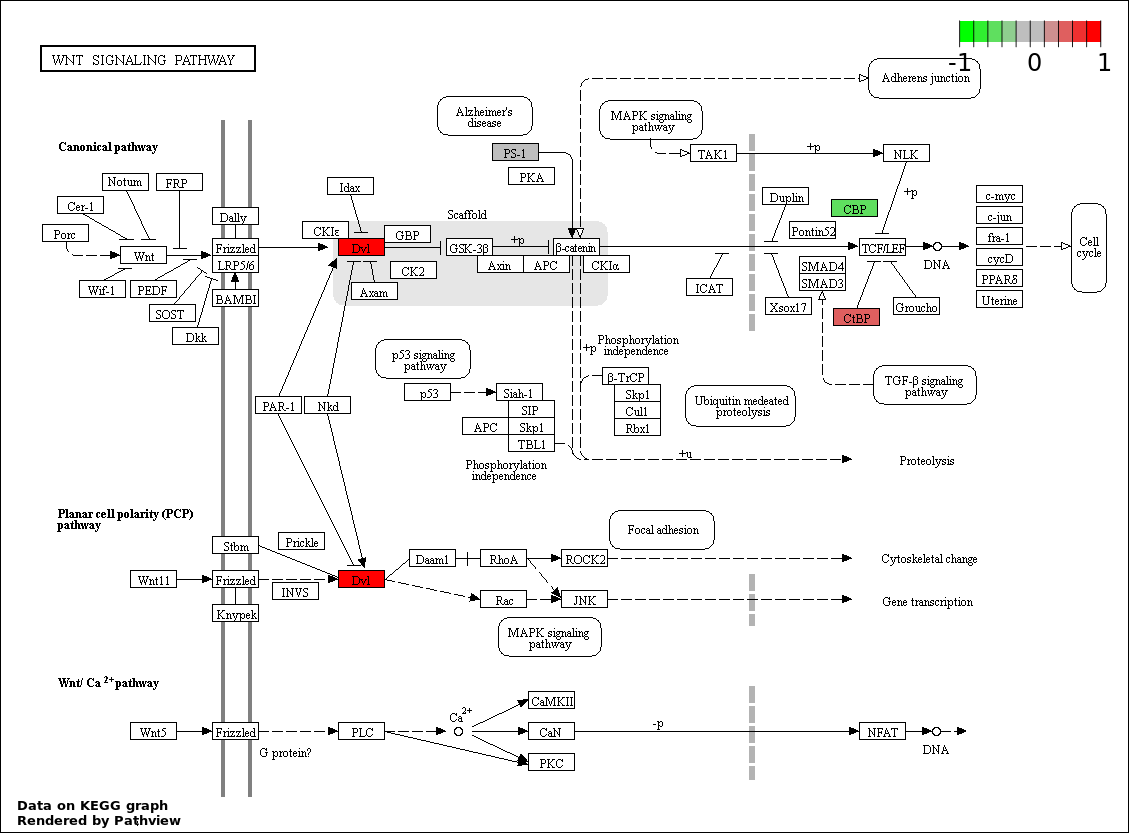
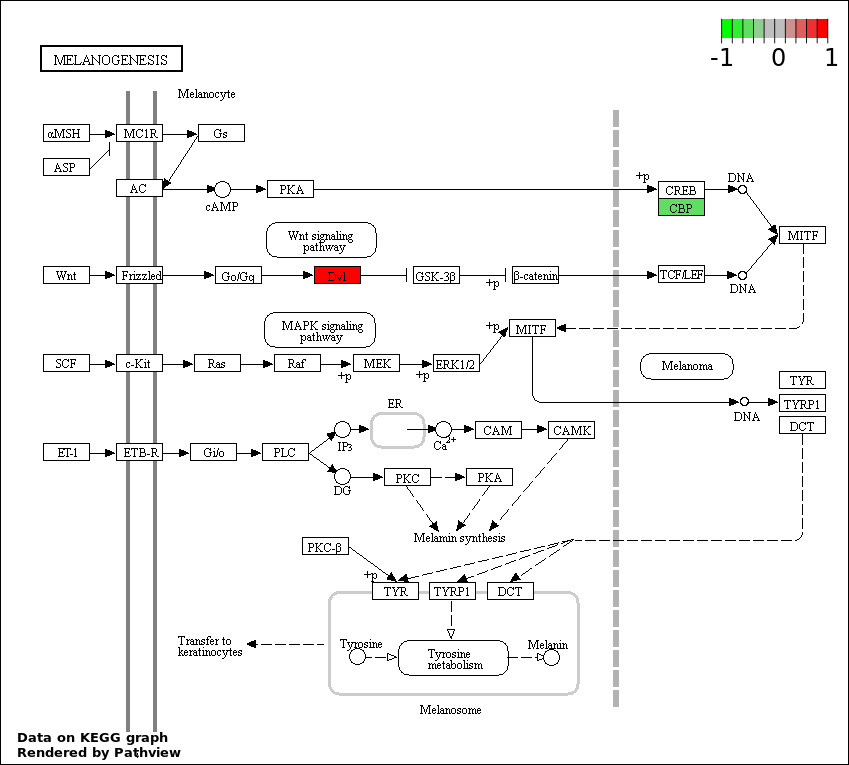
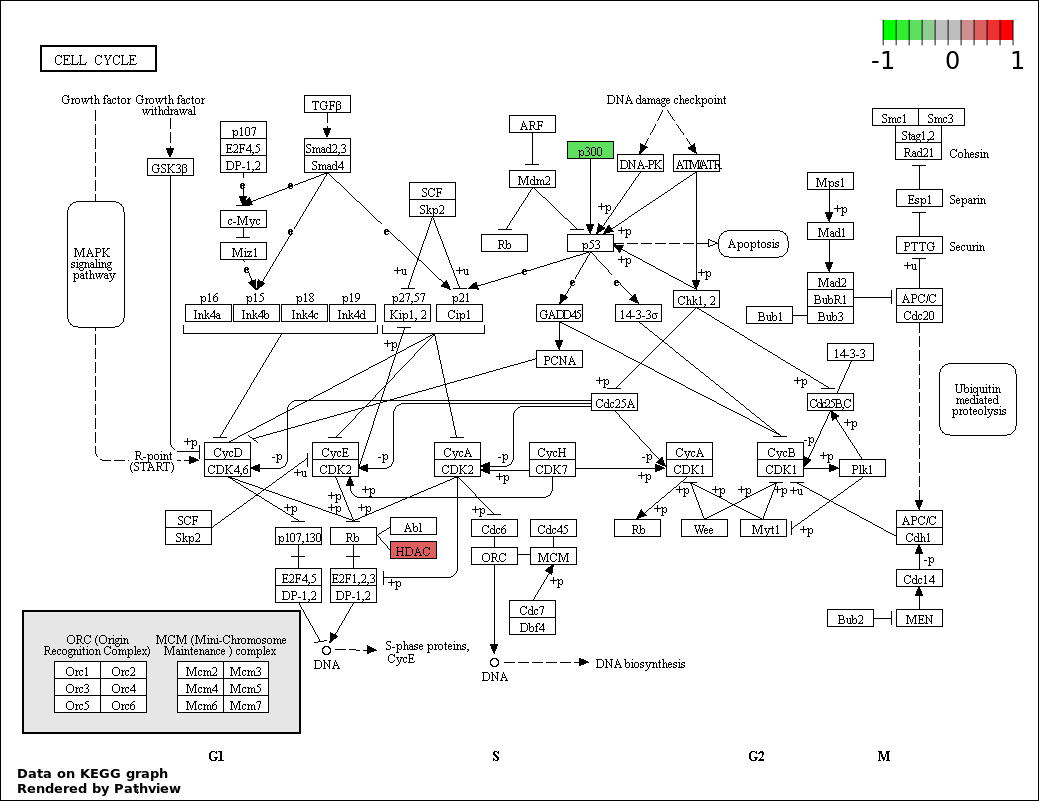
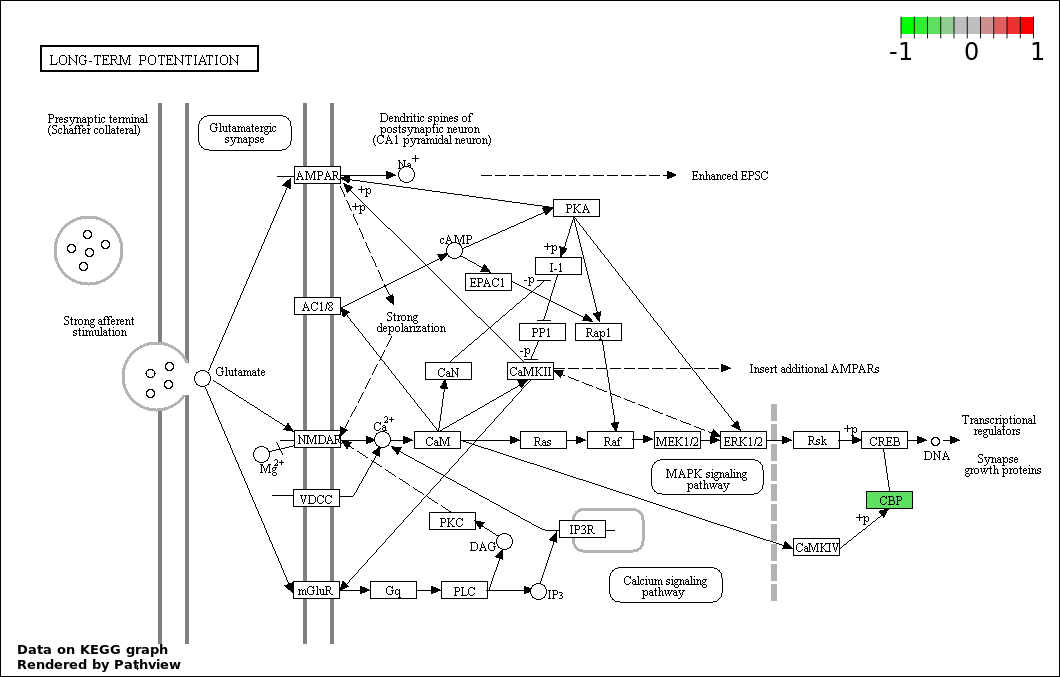
| 1 | hsa04330_Notch_signaling_pathway | 31 | 47 | 5.891e-88 | 1.06e-85 | |
| 2 | hsa04310_Wnt_signaling_pathway | 6 | 151 | 1.056e-07 | 9.506e-06 | |
| 3 | hsa04320_Dorso.ventral_axis_formation | 3 | 25 | 7.578e-06 | 0.0004547 | |
| 4 | hsa04916_Melanogenesis | 4 | 101 | 1.736e-05 | 0.0007811 | |
| 5 | hsa04110_Cell_cycle | 3 | 128 | 0.00101 | 0.03635 | |
| 6 | hsa00514_Other_types_of_O.glycan_biosynthesis | 2 | 46 | 0.002306 | 0.06919 | |
| 7 | hsa04720_Long.term_potentiation | 2 | 70 | 0.005259 | 0.1284 | |
| 8 | hsa04520_Adherens_junction | 2 | 73 | 0.005706 | 0.1284 | |
| 9 | hsa04350_TGF.beta_signaling_pathway | 2 | 85 | 0.007662 | 0.1532 | |
| 10 | hsa04390_Hippo_signaling_pathway | 2 | 154 | 0.02367 | 0.392 | |
| 11 | hsa04630_Jak.STAT_signaling_pathway | 2 | 155 | 0.02396 | 0.392 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | DLL1 | Sponge network | -4.039 | 0 | -0.948 | 0 | 0.598 |
| 2 | MIR143HG |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | DLL1 | Sponge network | -4.451 | 0 | -0.948 | 0 | 0.582 |
| 3 | HOXA-AS2 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-423-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -2.242 | 0 | -0.948 | 0 | 0.557 |
| 4 | RP11-175K6.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -3.312 | 0 | -1.587 | 0 | 0.556 |
| 5 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | DLL1 | Sponge network | -2.333 | 0 | -0.948 | 0 | 0.549 |
| 6 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -1.579 | 0 | -1.587 | 0 | 0.544 |
| 7 | RP11-1024P17.1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -2.541 | 0 | -0.948 | 0 | 0.533 |
| 8 | MEG3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p | 16 | DLL1 | Sponge network | -2.353 | 0 | -0.948 | 0 | 0.531 |
| 9 | ACTA2-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -3.065 | 0 | -0.948 | 0 | 0.527 |
| 10 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | DLL1 | Sponge network | -1.912 | 0 | -0.948 | 0 | 0.525 |
| 11 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 16 | KAT2B | Sponge network | -2.333 | 0 | -1.587 | 0 | 0.52 |
| 12 | RP11-175K6.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p | 11 | DLL1 | Sponge network | -3.312 | 0 | -0.948 | 0 | 0.511 |
| 13 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 15 | KAT2B | Sponge network | -1.618 | 0 | -1.587 | 0 | 0.497 |
| 14 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -0.993 | 0 | -0.948 | 0 | 0.489 |
| 15 | RP11-276H19.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -2.749 | 0 | -0.948 | 0 | 0.487 |
| 16 | RP11-384P7.7 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -4.533 | 0 | -0.948 | 0 | 0.479 |
| 17 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -3.119 | 0 | -0.948 | 0 | 0.471 |
| 18 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 13 | KAT2B | Sponge network | -4.039 | 0 | -1.587 | 0 | 0.47 |
| 19 | MIR22HG |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 15 | KAT2B | Sponge network | -1.551 | 0 | -1.587 | 0 | 0.47 |
| 20 | LINC00968 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -4.245 | 0 | -1.587 | 0 | 0.47 |
| 21 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -1.579 | 0 | -0.948 | 0 | 0.468 |
| 22 | ADAMTS9-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | DLL1 | Sponge network | -4.696 | 0 | -0.948 | 0 | 0.464 |
| 23 | LINC00924 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -3.023 | 0 | -1.587 | 0 | 0.455 |
| 24 | RP11-92A5.2 | hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -9.601 | 0 | -0.948 | 0 | 0.451 |
| 25 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p | 13 | DLL1 | Sponge network | -2.406 | 0 | -0.948 | 0 | 0.449 |
| 26 | ADIPOQ-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -8 | 0 | -1.587 | 0 | 0.448 |
| 27 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p | 16 | KAT2B | Sponge network | -4.451 | 0 | -1.587 | 0 | 0.445 |
| 28 | MIR22HG |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p | 11 | DLL1 | Sponge network | -1.551 | 0 | -0.948 | 0 | 0.44 |
| 29 | RP11-1101K5.1 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -8.619 | 0 | -1.587 | 0 | 0.433 |
| 30 | TRHDE-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p | 14 | KAT2B | Sponge network | -7.569 | 0 | -1.587 | 0 | 0.433 |
| 31 | CTD-2013N24.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -1.842 | 0 | -0.948 | 0 | 0.428 |
| 32 | EMX2OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -2.548 | 0 | -0.948 | 0 | 0.419 |
| 33 | LINC00639 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p | 11 | DLL1 | Sponge network | -2.449 | 0 | -0.948 | 0 | 0.417 |
| 34 | AL035610.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -6.728 | 0 | -1.587 | 0 | 0.416 |
| 35 | ADAMTS9-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 17 | KAT2B | Sponge network | -4.696 | 0 | -1.587 | 0 | 0.41 |
| 36 | RP11-384P7.7 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -4.533 | 0 | -1.587 | 0 | 0.407 |
| 37 | AF131215.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p | 16 | DLL1 | Sponge network | -2.008 | 0 | -0.948 | 0 | 0.403 |
| 38 | MIR497HG |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-589-5p | 13 | DLL1 | Sponge network | -2.527 | 0 | -0.948 | 0 | 0.403 |
| 39 | TRHDE-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | DLL1 | Sponge network | -7.569 | 0 | -0.948 | 0 | 0.402 |
| 40 | AC005682.5 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -3.076 | 0 | -0.948 | 0 | 0.401 |
| 41 | AC003991.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 17 | KAT2B | Sponge network | -2.021 | 0 | -1.587 | 0 | 0.397 |
| 42 | MIR143HG |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-98-5p | 10 | JAG1 | Sponge network | -4.451 | 0 | -0.64 | 0 | 0.396 |
| 43 | TBX5-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-576-5p | 10 | DLL1 | Sponge network | -0.804 | 0 | -0.948 | 0 | 0.393 |
| 44 | ALDH1L1-AS2 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -7.428 | 0 | -0.948 | 0 | 0.391 |
| 45 | RP11-276H19.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | KAT2B | Sponge network | -2.749 | 0 | -1.587 | 0 | 0.39 |
| 46 | LL22NC03-86G7.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | DLL1 | Sponge network | -1.159 | 0 | -0.948 | 0 | 0.388 |
| 47 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -2.33 | 0 | -0.948 | 0 | 0.388 |
| 48 | RP11-182J1.1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-589-5p | 10 | DLL1 | Sponge network | -1.088 | 0 | -0.948 | 0 | 0.385 |
| 49 | RP11-286B14.1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -5.866 | 0 | -0.948 | 0 | 0.384 |
| 50 | LINC01088 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -1.675 | 0 | -0.948 | 0 | 0.383 |
| 51 | LINC00702 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | DLL1 | Sponge network | -1.308 | 0 | -0.948 | 0 | 0.382 |
| 52 | RP11-401P9.4 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-5p | 12 | DLL1 | Sponge network | -0.963 | 0 | -0.948 | 0 | 0.382 |
| 53 | DIO3OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p | 10 | DLL1 | Sponge network | -3.064 | 0 | -0.948 | 0 | 0.379 |
| 54 | RP11-244O19.1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.068 | 0 | -0.948 | 0 | 0.375 |
| 55 | FZD10-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | DLL1 | Sponge network | -1.884 | 0 | -0.948 | 0 | 0.375 |
| 56 | SNHG18 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 15 | DLL1 | Sponge network | -0.741 | 0 | -0.948 | 0 | 0.373 |
| 57 | RP11-251M1.1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p | 10 | DLL1 | Sponge network | -2.455 | 0 | -0.948 | 0 | 0.371 |
| 58 | AC144831.1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-576-5p | 13 | DLL1 | Sponge network | -1.421 | 0 | -0.948 | 0 | 0.37 |
| 59 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | KAT2B | Sponge network | -1.912 | 0 | -1.587 | 0 | 0.369 |
| 60 | HOXB-AS1 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429 | 11 | KAT2B | Sponge network | -2.063 | 0 | -1.587 | 0 | 0.369 |
| 61 | APCDD1L-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p | 10 | DLL1 | Sponge network | -3.451 | 0 | -0.948 | 0 | 0.368 |
| 62 | AF131215.9 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -1.776 | 0 | -0.948 | 0 | 0.368 |
| 63 | ZNF582-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-576-5p | 12 | DLL1 | Sponge network | -1.307 | 0 | -0.948 | 0 | 0.367 |
| 64 | ADAMTS9-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-98-5p | 11 | JAG1 | Sponge network | -4.696 | 0 | -0.64 | 0 | 0.366 |
| 65 | RP11-448G15.3 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.586 | 0 | -0.948 | 0 | 0.364 |
| 66 | LINC00924 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -3.023 | 0 | -0.948 | 0 | 0.363 |
| 67 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 15 | KAT2B | Sponge network | -2.548 | 0 | -1.587 | 0 | 0.362 |
| 68 | AC003991.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | DLL1 | Sponge network | -2.021 | 0 | -0.948 | 0 | 0.361 |
| 69 | LINC00968 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -4.245 | 0 | -0.948 | 0 | 0.361 |
| 70 | RP11-981G7.6 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -2.292 | 0 | -0.948 | 0 | 0.361 |
| 71 | AL035610.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -6.728 | 0 | -0.948 | 0 | 0.359 |
| 72 | PDZRN3-AS1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -2.387 | 0 | -0.948 | 0 | 0.359 |
| 73 | AGAP11 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -2.72 | 0 | -1.587 | 0 | 0.356 |
| 74 | SOCS2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-423-3p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -1.697 | 0 | -0.948 | 0 | 0.352 |
| 75 | MEG3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -2.353 | 0 | -1.587 | 0 | 0.351 |
| 76 | AC074289.1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p | 10 | DLL1 | Sponge network | -2.611 | 0 | -0.948 | 0 | 0.349 |
| 77 | FAM13A-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.451 | 0 | -0.948 | 0 | 0.346 |
| 78 | RP11-276H19.2 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -3.719 | 0 | -0.948 | 0 | 0.343 |
| 79 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | KAT2B | Sponge network | -1.696 | 0 | -1.587 | 0 | 0.343 |
| 80 | RP11-57H14.4 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -1.436 | 0 | -0.948 | 0 | 0.342 |
| 81 | LINC00641 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -1.721 | 0 | -0.948 | 0 | 0.34 |
| 82 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-340-5p | 10 | NOTCH2 | Sponge network | -2.333 | 0 | -0.567 | 0 | 0.339 |
| 83 | AC116366.6 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-92a-3p | 10 | KAT2B | Sponge network | -0.799 | 0 | -1.587 | 0 | 0.337 |
| 84 | LINC00958 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -2.904 | 0 | -0.948 | 0 | 0.335 |
| 85 | ZNF667-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-589-5p | 11 | DLL1 | Sponge network | -1.213 | 0 | -0.948 | 0 | 0.334 |
| 86 | TRHDE-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-98-5p | 11 | JAG1 | Sponge network | -7.569 | 0 | -0.64 | 0 | 0.326 |
| 87 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 17 | KAT2B | Sponge network | -1.308 | 0 | -1.587 | 0 | 0.326 |
| 88 | RBPMS-AS1 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-454-3p | 10 | DLL1 | Sponge network | -2.131 | 0 | -0.948 | 0 | 0.324 |
| 89 | DIO3OS |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p | 10 | KAT2B | Sponge network | -3.064 | 0 | -1.587 | 0 | 0.321 |
| 90 | AC004947.2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 15 | DLL1 | Sponge network | -3.692 | 0 | -0.948 | 0 | 0.32 |
| 91 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -0.935 | 0 | -1.587 | 0 | 0.319 |
| 92 | AC004947.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | KAT2B | Sponge network | -3.692 | 0 | -1.587 | 0 | 0.319 |
| 93 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | KAT2B | Sponge network | -2.527 | 0 | -1.587 | 0 | 0.319 |
| 94 | CTC-429P9.5 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.354 | 0 | -0.948 | 0 | 0.318 |
| 95 | AC141928.1 | hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-423-3p;hsa-miR-454-3p | 11 | DLL1 | Sponge network | -1.583 | 0 | -0.948 | 0 | 0.313 |
| 96 | RP4-799P18.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -4.203 | 0 | -0.948 | 0 | 0.312 |
| 97 | ADIPOQ-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -8 | 0 | -0.948 | 0 | 0.311 |
| 98 | AC009950.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -0.716 | 0.0005 | -1.587 | 0 | 0.31 |
| 99 | CTD-2015G9.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | DLL1 | Sponge network | -5.267 | 0 | -0.948 | 0 | 0.309 |
| 100 | RP11-978I15.10 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-5p | 10 | DLL1 | Sponge network | -3.591 | 0 | -0.948 | 0 | 0.308 |
| 101 | CTD-2619J13.17 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -0.985 | 0 | -1.587 | 0 | 0.308 |
| 102 | AC007246.3 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | KAT2B | Sponge network | -0.496 | 0 | -1.587 | 0 | 0.308 |
| 103 | RP11-631N16.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.389 | 0 | -0.948 | 0 | 0.307 |
| 104 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 17 | KAT2B | Sponge network | -1.168 | 0 | -1.587 | 0 | 0.305 |
| 105 | AF131215.8 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -3.371 | 0 | -0.948 | 0 | 0.304 |
| 106 | TPRG1-AS1 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -1.972 | 0 | -1.587 | 0 | 0.304 |
| 107 | AC108142.1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -2.247 | 0 | -0.948 | 0 | 0.303 |
| 108 | DNM3OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -0.276 | 0.07533 | -0.948 | 0 | 0.301 |
| 109 | HOXD-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-423-3p;hsa-miR-454-3p | 12 | DLL1 | Sponge network | -1.529 | 0 | -0.948 | 0 | 0.3 |
| 110 | CTD-3247F14.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-589-5p | 13 | DLL1 | Sponge network | -2.833 | 0 | -0.948 | 0 | 0.299 |
| 111 | RP11-359E10.1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-576-5p | 11 | DLL1 | Sponge network | -1.868 | 0 | -0.948 | 0 | 0.295 |
| 112 | TP73-AS1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p | 10 | DLL1 | Sponge network | -0.602 | 0 | -0.948 | 0 | 0.294 |
| 113 | CTB-107G13.1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -3.353 | 0 | -0.948 | 0 | 0.29 |
| 114 | RP11-293M10.6 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -0.601 | 0.00056 | -1.587 | 0 | 0.288 |
| 115 | AC007743.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-92b-3p;hsa-miR-93-5p | 14 | KAT2B | Sponge network | -2.327 | 0 | -1.587 | 0 | 0.288 |
| 116 | RP11-54A9.1 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -2.207 | 0 | -1.587 | 0 | 0.287 |
| 117 | LINC00641 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-98-5p | 10 | JAG1 | Sponge network | -1.721 | 0 | -0.64 | 0 | 0.286 |
| 118 | RP11-366L5.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 13 | KAT2B | Sponge network | -0.54 | 0.00309 | -1.587 | 0 | 0.285 |
| 119 | LINC01085 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p | 15 | DLL1 | Sponge network | -3.703 | 0 | -0.948 | 0 | 0.284 |
| 120 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -0.677 | 0 | -1.587 | 0 | 0.283 |
| 121 | CKMT2-AS1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -0.724 | 0 | -0.948 | 0 | 0.283 |
| 122 | AGAP11 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-576-5p | 14 | DLL1 | Sponge network | -2.72 | 0 | -0.948 | 0 | 0.281 |
| 123 | AC108142.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 12 | KAT2B | Sponge network | -2.247 | 0 | -1.587 | 0 | 0.281 |
| 124 | RP1-193H18.2 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -1.975 | 0 | -0.948 | 0 | 0.28 |
| 125 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 14 | KAT2B | Sponge network | -1.884 | 0 | -1.587 | 0 | 0.28 |
| 126 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | DLL1 | Sponge network | -1.167 | 0 | -0.948 | 0 | 0.273 |
| 127 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -1.618 | 0 | -0.948 | 0 | 0.273 |
| 128 | RP11-834C11.4 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | DLL1 | Sponge network | -0.637 | 0 | -0.948 | 0 | 0.27 |
| 129 | MEG3 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-98-5p | 10 | JAG1 | Sponge network | -2.353 | 0 | -0.64 | 0 | 0.27 |
| 130 | GRIK1-AS1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 12 | DLL1 | Sponge network | -3.557 | 0 | -0.948 | 0 | 0.269 |
| 131 | CTB-92J24.3 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429 | 10 | DLL1 | Sponge network | -3.42 | 0 | -0.948 | 0 | 0.268 |
| 132 | HOXD-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-5p | 11 | KAT2B | Sponge network | -1.529 | 0 | -1.587 | 0 | 0.267 |
| 133 | AC016995.3 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -3.825 | 0 | -0.948 | 0 | 0.265 |
| 134 | LINC00654 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -0.902 | 0 | -1.587 | 0 | 0.264 |
| 135 | LINC01024 | hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | KAT2B | Sponge network | -0.493 | 0 | -1.587 | 0 | 0.262 |
| 136 | RP11-116O18.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | KAT2B | Sponge network | -3.1 | 0 | -1.587 | 0 | 0.26 |
| 137 | RP11-545I5.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | KAT2B | Sponge network | -1.007 | 0 | -1.587 | 0 | 0.259 |
| 138 | RP11-403I13.8 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-423-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 13 | DLL1 | Sponge network | -0.541 | 0.0007 | -0.948 | 0 | 0.257 |
| 139 | LINC00648 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 14 | DLL1 | Sponge network | -5.143 | 0 | -0.948 | 0 | 0.255 |
| 140 | CTC-523E23.4 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | DLL1 | Sponge network | -1.207 | 0 | -0.948 | 0 | 0.254 |
| 141 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 18 | KAT2B | Sponge network | -0.884 | 3.0E-5 | -1.587 | 0 | 0.253 |
| 142 | CTD-2619J13.17 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -0.985 | 0 | -0.948 | 0 | 0.252 |
| 143 | RP11-403B2.7 | hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | DLL1 | Sponge network | -1.946 | 0 | -0.948 | 0 | 0.252 |