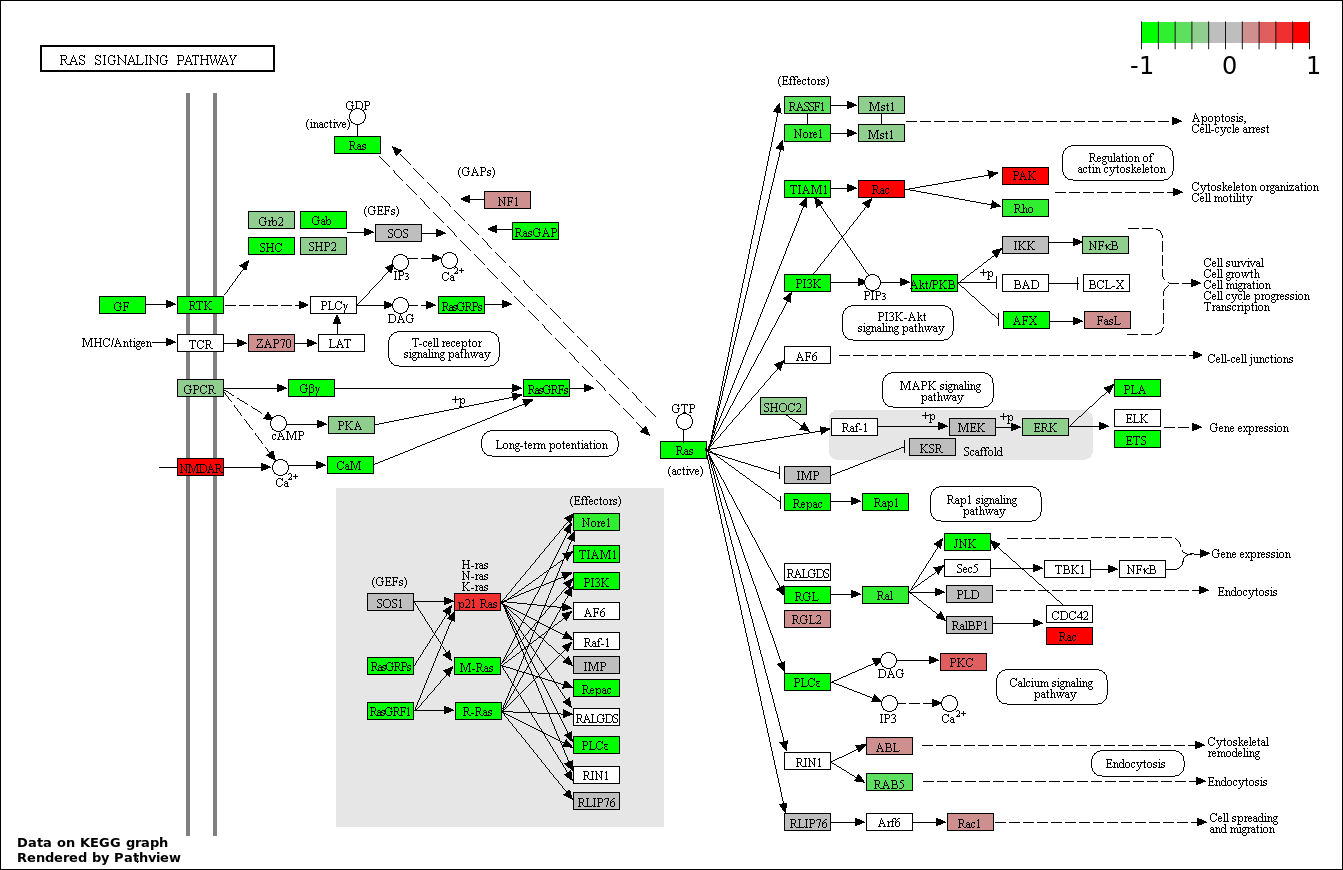
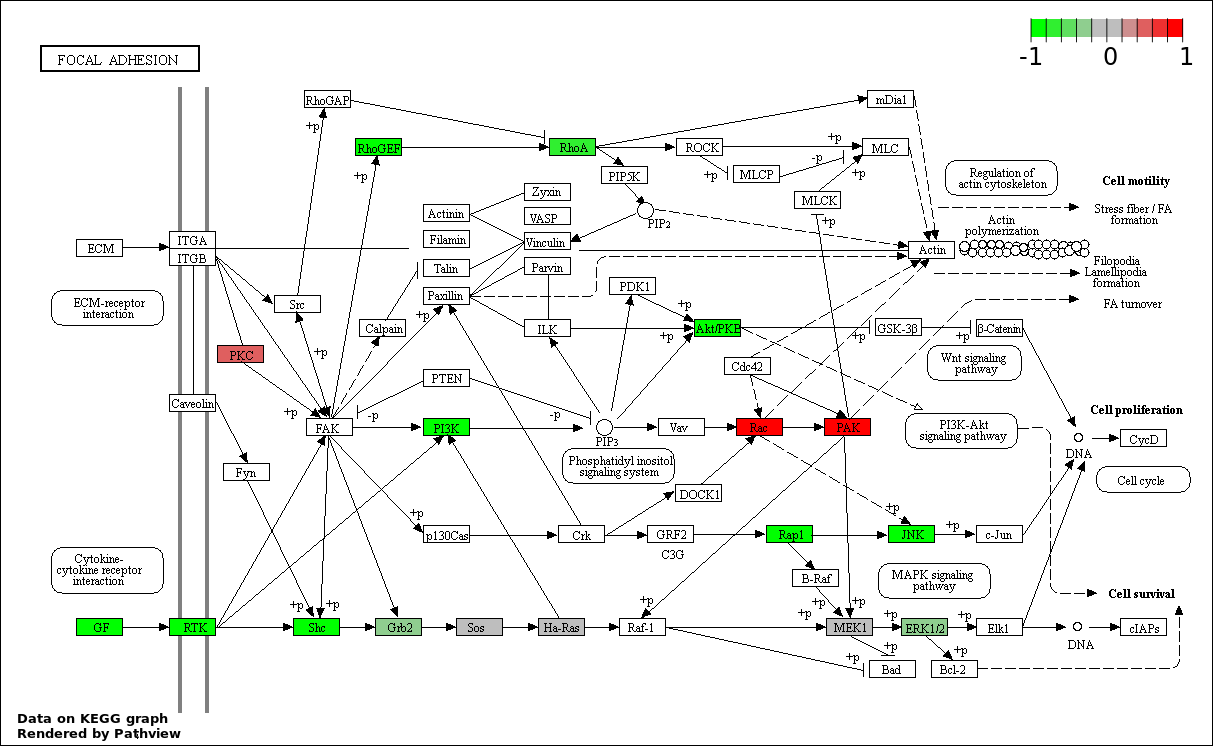
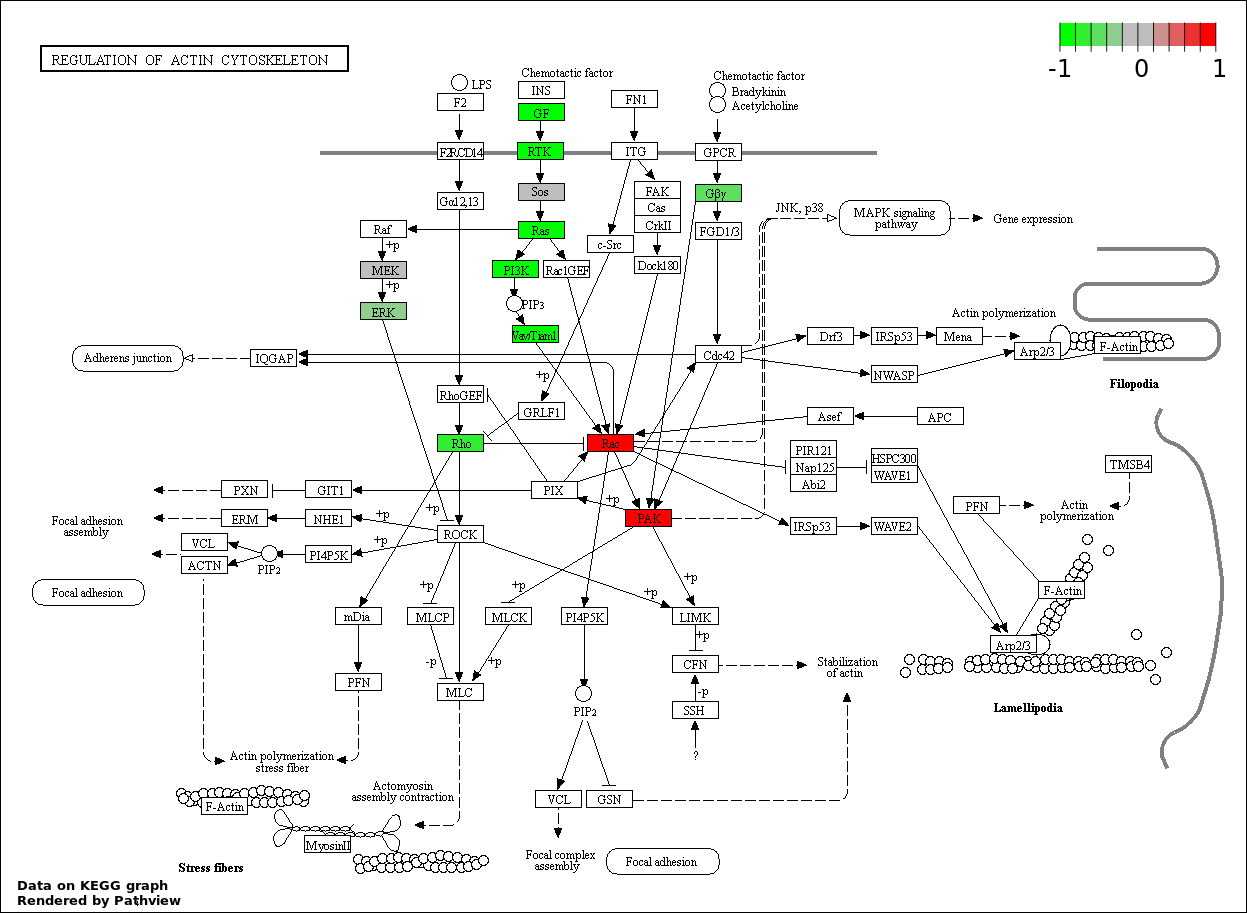
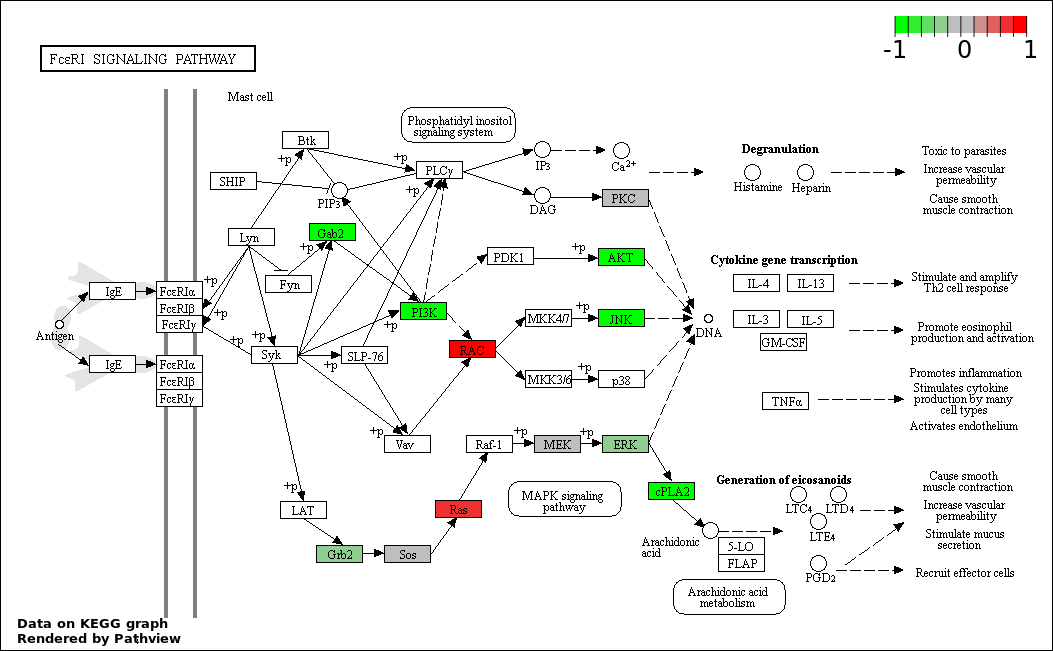
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-30e-5p | ABL1 | 1.6 | 0 | -0.1 | 0.42908 | MirTarget; miRNATAP | -0.11 | 0.00016 | NA | |
| 2 | hsa-let-7b-5p | ABL2 | -1.62 | 0 | 0.31 | 0.01567 | miRNATAP | -0.11 | 0.00013 | NA | |
| 3 | hsa-let-7d-5p | ABL2 | -0.62 | 6.0E-5 | 0.31 | 0.01567 | miRNATAP | -0.13 | 0.00086 | NA | |
| 4 | hsa-miR-125a-5p | ABL2 | -1.05 | 0 | 0.31 | 0.01567 | MirTarget; PITA | -0.13 | 1.0E-5 | 26766902 | MicroRNA 125a 5p modulates human cervical carcinoma proliferation and migration by targeting ABL2; The direct regulation of miR-125a-5p on its target gene ABL proto-oncogene 2 ABL2 in cervical carcinoma was evaluated by quantitative real-time reverse transcription polymerase chain reaction Western blotting and luciferase reporter assays respectively; ABL2 was shown to be directly targeted by miR-125a-5p; In cervical carcinoma ABL2 gene and protein levels were both downregulated by miR-125a-5p |
| 5 | hsa-miR-140-3p | ABL2 | -1.11 | 0 | 0.31 | 0.01567 | miRNAWalker2 validate | -0.1 | 0.00743 | NA | |
| 6 | hsa-miR-200a-5p | ABL2 | 2.27 | 0 | 0.31 | 0.01567 | mirMAP | -0.11 | 0 | NA | |
| 7 | hsa-miR-200c-3p | ABL2 | 0.38 | 0.08422 | 0.31 | 0.01567 | MirTarget | -0.14 | 0 | NA | |
| 8 | hsa-miR-29b-2-5p | ABL2 | 0.35 | 0.19484 | 0.31 | 0.01567 | mirMAP | -0.11 | 0 | NA | |
| 9 | hsa-miR-30a-3p | ABL2 | -2.54 | 0 | 0.31 | 0.01567 | mirMAP | -0.1 | 0 | NA | |
| 10 | hsa-miR-30a-5p | ABL2 | -0.92 | 0.00076 | 0.31 | 0.01567 | mirMAP; miRNATAP | -0.11 | 0 | NA | |
| 11 | hsa-miR-30e-3p | ABL2 | -0.1 | 0.52624 | 0.31 | 0.01567 | mirMAP | -0.17 | 0 | NA | |
| 12 | hsa-let-7a-5p | AKT2 | -1.37 | 0 | 0.16 | 0.1135 | TargetScan | -0.12 | 0 | NA | |
| 13 | hsa-miR-29a-3p | AKT2 | 0.1 | 0.5732 | 0.16 | 0.1135 | MirTarget | -0.1 | 4.0E-5 | 24076586 | Furthermore a feed-back loop comprising of c-Myc miR-29 family and Akt2 were found in myeloid leukemogenesis |
| 14 | hsa-miR-106b-5p | AKT3 | 1.47 | 0 | -1.44 | 0 | miRNATAP | -0.16 | 0.00426 | NA | |
| 15 | hsa-miR-107 | AKT3 | 0.66 | 0 | -1.44 | 0 | PITA; miRanda | -0.26 | 0.0031 | NA | |
| 16 | hsa-miR-146b-5p | AKT3 | 1.09 | 1.0E-5 | -1.44 | 0 | miRNAWalker2 validate | -0.15 | 0.00189 | NA | |
| 17 | hsa-miR-15a-5p | AKT3 | 1.63 | 0 | -1.44 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.41 | 0 | NA | |
| 18 | hsa-miR-16-5p | AKT3 | 0.75 | 0 | -1.44 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.27 | 0.0001 | NA | |
| 19 | hsa-miR-17-5p | AKT3 | 2.07 | 0 | -1.44 | 0 | TargetScan; miRNATAP | -0.22 | 0 | NA | |
| 20 | hsa-miR-181a-5p | AKT3 | -0.38 | 0.05621 | -1.44 | 0 | miRNATAP | -0.23 | 9.0E-5 | NA | |
| 21 | hsa-miR-181b-5p | AKT3 | 0.67 | 0.00024 | -1.44 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 22 | hsa-miR-20a-5p | AKT3 | 2.65 | 0 | -1.44 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 23 | hsa-miR-22-3p | AKT3 | 1.43 | 0 | -1.44 | 0 | miRNATAP | -0.26 | 0.00109 | NA | |
| 24 | hsa-miR-29b-2-5p | AKT3 | 0.35 | 0.19484 | -1.44 | 0 | mirMAP | -0.18 | 2.0E-5 | NA | |
| 25 | hsa-miR-29b-3p | AKT3 | 3.11 | 0 | -1.44 | 0 | miRNATAP | -0.27 | 0 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 26 | hsa-miR-3065-5p | AKT3 | 0.65 | 0.09995 | -1.44 | 0 | mirMAP | -0.18 | 0 | NA | |
| 27 | hsa-miR-362-5p | AKT3 | 0.66 | 0.02433 | -1.44 | 0 | PITA; TargetScan; miRNATAP | -0.23 | 0 | NA | |
| 28 | hsa-miR-542-3p | AKT3 | 1.62 | 0 | -1.44 | 0 | miRanda | -0.19 | 1.0E-5 | NA | |
| 29 | hsa-miR-93-5p | AKT3 | 1.51 | 0 | -1.44 | 0 | miRNATAP | -0.25 | 0 | NA | |
| 30 | hsa-miR-335-3p | ANGPT1 | 1.51 | 0 | -2.86 | 0 | mirMAP | -0.33 | 1.0E-5 | NA | |
| 31 | hsa-miR-429 | ANGPT1 | 2.38 | 0 | -2.86 | 0 | miRanda | -0.25 | 0 | NA | |
| 32 | hsa-miR-590-3p | ANGPT1 | 0.84 | 0.00129 | -2.86 | 0 | PITA; miRanda; mirMAP | -0.4 | 0 | NA | |
| 33 | hsa-miR-125a-5p | ANGPT2 | -1.05 | 0 | -0.36 | 0.23492 | miRanda | -0.23 | 0.0005 | NA | |
| 34 | hsa-miR-135b-5p | ANGPT2 | 3.25 | 0 | -0.36 | 0.23492 | MirTarget | -0.21 | 0 | NA | |
| 35 | hsa-miR-142-5p | ANGPT2 | 1.3 | 0 | -0.36 | 0.23492 | MirTarget | -0.15 | 0.00403 | NA | |
| 36 | hsa-miR-186-5p | ANGPT2 | 0.85 | 0 | -0.36 | 0.23492 | MirTarget; mirMAP | -0.39 | 7.0E-5 | NA | |
| 37 | hsa-miR-34c-5p | ANGPT2 | -1 | 0.07244 | -0.36 | 0.23492 | miRanda | -0.16 | 0 | NA | |
| 38 | hsa-miR-374a-5p | ANGPT2 | -0.2 | 0.29808 | -0.36 | 0.23492 | mirMAP | -0.26 | 0.00171 | NA | |
| 39 | hsa-miR-374b-5p | ANGPT2 | 0.47 | 0.01092 | -0.36 | 0.23492 | mirMAP | -0.22 | 0.00511 | NA | |
| 40 | hsa-miR-429 | ANGPT2 | 2.38 | 0 | -0.36 | 0.23492 | miRanda | -0.22 | 0 | NA | |
| 41 | hsa-miR-664a-3p | ANGPT2 | 0.44 | 0.02142 | -0.36 | 0.23492 | mirMAP | -0.39 | 0 | NA | |
| 42 | hsa-miR-10a-5p | BDNF | 0.79 | 0.00059 | -3.58 | 0 | MirTarget; miRNATAP | -0.28 | 0.00181 | NA | |
| 43 | hsa-miR-146b-5p | BDNF | 1.09 | 1.0E-5 | -3.58 | 0 | miRanda | -0.33 | 0.00012 | NA | |
| 44 | hsa-miR-148a-5p | BDNF | 1.46 | 0 | -3.58 | 0 | mirMAP | -0.24 | 0.00212 | NA | |
| 45 | hsa-miR-155-5p | BDNF | 0.81 | 0.00061 | -3.58 | 0 | miRNATAP | -0.23 | 0.00731 | NA | |
| 46 | hsa-miR-15a-5p | BDNF | 1.63 | 0 | -3.58 | 0 | MirTarget; miRNATAP | -0.3 | 0.00719 | 26581909 | MicroRNA 15a 5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF; BDNF was then overexpressed in HepG2 and SNU-182 cells to evaluate its selective effect on miR-15a-5p in HCC modulation; MiR-15a-5p selectively and negatively regulated BDNF at both gene and protein levels in HCC cells; Forced overexpression of BDNF effectively reversed the tumor suppressive functions of miR-15a-5p on HCC proliferation and cell division in vitro; Our study demonstrated that miR-15a-5p is a tumor suppressor in HCC and its regulation is through BDNF in HCC |
| 47 | hsa-miR-182-5p | BDNF | 3.22 | 0 | -3.58 | 0 | MirTarget; miRNATAP | -0.49 | 0 | NA | |
| 48 | hsa-miR-210-3p | BDNF | 4.89 | 0 | -3.58 | 0 | miRNAWalker2 validate; miRTarBase | -0.22 | 0 | NA | |
| 49 | hsa-miR-29b-1-5p | BDNF | 1.71 | 0 | -3.58 | 0 | mirMAP | -0.25 | 0.00028 | NA | |
| 50 | hsa-miR-30e-5p | BDNF | 1.6 | 0 | -3.58 | 0 | miRNATAP | -0.44 | 9.0E-5 | NA | |
| 51 | hsa-miR-7-1-3p | BDNF | 2.61 | 0 | -3.58 | 0 | MirTarget | -0.3 | 0.00109 | NA | |
| 52 | hsa-miR-140-5p | BRAP | 0.67 | 0.00034 | 0.19 | 0.03293 | miRanda | -0.12 | 0 | NA | |
| 53 | hsa-miR-30a-5p | BRAP | -0.92 | 0.00076 | 0.19 | 0.03293 | miRNATAP | -0.1 | 0 | NA | |
| 54 | hsa-miR-30c-5p | BRAP | -0.33 | 0.1236 | 0.19 | 0.03293 | miRNATAP | -0.12 | 0 | NA | |
| 55 | hsa-miR-15a-5p | CALM1 | 1.63 | 0 | -0.79 | 0 | miRNATAP | -0.11 | 2.0E-5 | NA | |
| 56 | hsa-miR-186-5p | CALM1 | 0.85 | 0 | -0.79 | 0 | mirMAP | -0.18 | 0 | NA | |
| 57 | hsa-miR-576-5p | CALM1 | 1.03 | 0 | -0.79 | 0 | mirMAP | -0.14 | 0 | NA | |
| 58 | hsa-miR-7-1-3p | CALM1 | 2.61 | 0 | -0.79 | 0 | mirMAP | -0.11 | 0 | NA | |
| 59 | hsa-miR-27b-3p | CALM3 | 0.24 | 0.12264 | -0.12 | 0.23429 | miRNAWalker2 validate; miRNATAP | -0.17 | 0 | NA | |
| 60 | hsa-miR-29a-3p | CALM3 | 0.1 | 0.5732 | -0.12 | 0.23429 | miRNATAP | -0.13 | 0 | NA | |
| 61 | hsa-miR-93-5p | CALML4 | 1.51 | 0 | -0.27 | 0.09728 | mirMAP | -0.12 | 0.00073 | NA | |
| 62 | hsa-miR-15a-5p | CHUK | 1.63 | 0 | 0.04 | 0.74424 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.16 | 0 | NA | |
| 63 | hsa-miR-29b-2-5p | CHUK | 0.35 | 0.19484 | 0.04 | 0.74424 | MirTarget | -0.12 | 0 | NA | |
| 64 | hsa-miR-339-5p | CHUK | 0.54 | 0.04881 | 0.04 | 0.74424 | miRanda | -0.1 | 0 | NA | |
| 65 | hsa-miR-342-3p | CHUK | -0.13 | 0.56103 | 0.04 | 0.74424 | miRanda | -0.18 | 0 | NA | |
| 66 | hsa-miR-497-5p | CHUK | -0.05 | 0.78621 | 0.04 | 0.74424 | MirTarget | -0.19 | 0 | NA | |
| 67 | hsa-miR-106a-5p | CSF1 | 1.39 | 6.0E-5 | -0.65 | 0.00787 | miRNATAP | -0.16 | 1.0E-5 | NA | |
| 68 | hsa-miR-106b-5p | CSF1 | 1.47 | 0 | -0.65 | 0.00787 | miRNATAP | -0.31 | 0 | NA | |
| 69 | hsa-miR-107 | CSF1 | 0.66 | 0 | -0.65 | 0.00787 | miRanda | -0.32 | 0.00017 | NA | |
| 70 | hsa-miR-128-3p | CSF1 | 1.04 | 0 | -0.65 | 0.00787 | MirTarget | -0.19 | 0.00593 | 22909061 | By mutations in putative miRNA targets in CSF-1 mRNA 3'UTR we identified a common target for both miR-128 and miR-152; We have also found that both miR-128 and miR-152 down-regulate CSF-1 mRNA and protein expression in ovarian cancer cells leading to decreased cell motility and adhesion in vitro two major aspects of the metastatic potential of cancer cells; Our results provide the evidence for a mechanism by which miR-128 and miR-152 down-regulate CSF-1 an important regulator of ovarian cancer |
| 71 | hsa-miR-130b-3p | CSF1 | 1.83 | 0 | -0.65 | 0.00787 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.14 | 0.00318 | 22005523 | CSF-1 regulated by miR-130b was detected using Dual Luciferase Reporter system; CSF-1 expression was negatively associated with miR-130b level in ovarian tissues and cell lines; Luciferase assay validated CSF-1 is a direct target of miR-130b; Knock-down of CSF-1 sensitized ovarian cancer cells to anticancer drugs and could partially attenuate the resistance inducing effect of miR-130b inhibitors; Downregulation of miR-130b promotes the development of multidrug resistant ovarian cancer partially by targeting the 3'-UTR of CSF-1 and the silencing of miR-130b may be mediated by DNA methylation |
| 72 | hsa-miR-148b-3p | CSF1 | 0.48 | 0.00265 | -0.65 | 0.00787 | miRNAWalker2 validate | -0.42 | 0 | NA | |
| 73 | hsa-miR-17-5p | CSF1 | 2.07 | 0 | -0.65 | 0.00787 | TargetScan; miRNATAP | -0.31 | 0 | NA | |
| 74 | hsa-miR-20a-5p | CSF1 | 2.65 | 0 | -0.65 | 0.00787 | miRNATAP | -0.25 | 0 | NA | |
| 75 | hsa-miR-335-5p | CSF1 | -0.47 | 0.0677 | -0.65 | 0.00787 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 76 | hsa-miR-429 | CSF1 | 2.38 | 0 | -0.65 | 0.00787 | PITA; miRanda; miRNATAP | -0.19 | 0 | NA | |
| 77 | hsa-miR-93-5p | CSF1 | 1.51 | 0 | -0.65 | 0.00787 | miRNATAP | -0.19 | 0.00022 | NA | |
| 78 | hsa-miR-96-5p | CSF1 | 3.04 | 0 | -0.65 | 0.00787 | miRNATAP | -0.15 | 0.00028 | NA | |
| 79 | hsa-miR-107 | CSF1R | 0.66 | 0 | -0.65 | 0.01137 | miRanda | -0.42 | 0 | NA | |
| 80 | hsa-miR-218-5p | EFNA1 | -0.28 | 0.25955 | 0.13 | 0.58969 | miRNAWalker2 validate | -0.13 | 0.00248 | NA | |
| 81 | hsa-miR-145-5p | EFNA3 | -1.35 | 0 | 2.4 | 0 | miRNATAP | -0.17 | 0.00044 | NA | |
| 82 | hsa-miR-2110 | EFNA3 | -1.92 | 0 | 2.4 | 0 | miRNATAP | -0.15 | 0.00104 | NA | |
| 83 | hsa-miR-221-5p | EFNA3 | -2.22 | 0 | 2.4 | 0 | MirTarget | -0.14 | 7.0E-5 | NA | |
| 84 | hsa-miR-30a-5p | EFNA3 | -0.92 | 0.00076 | 2.4 | 0 | MirTarget; mirMAP; miRNATAP | -0.32 | 0 | NA | |
| 85 | hsa-miR-30d-5p | EFNA3 | -0.92 | 4.0E-5 | 2.4 | 0 | MirTarget; mirMAP; miRNATAP | -0.25 | 1.0E-5 | NA | |
| 86 | hsa-miR-361-3p | EFNA3 | 0.1 | 0.54913 | 2.4 | 0 | MirTarget; miRNATAP | -0.21 | 0.00608 | NA | |
| 87 | hsa-miR-423-5p | EFNA3 | -1.8 | 0 | 2.4 | 0 | MirTarget; PITA; miRNATAP | -0.3 | 0 | NA | |
| 88 | hsa-miR-486-5p | EFNA3 | -4.39 | 0 | 2.4 | 0 | miRanda | -0.23 | 0 | NA | |
| 89 | hsa-miR-140-5p | EFNA5 | 0.67 | 0.00034 | 0.77 | 0.00962 | miRanda | -0.2 | 0.00645 | NA | |
| 90 | hsa-miR-150-5p | EFNA5 | -0.7 | 0.02153 | 0.77 | 0.00962 | mirMAP; miRNATAP | -0.12 | 0.00796 | NA | |
| 91 | hsa-miR-15b-3p | EFNA5 | 0.8 | 0.0004 | 0.77 | 0.00962 | mirMAP | -0.17 | 0.007 | NA | |
| 92 | hsa-miR-217 | EFNA5 | 1.88 | 0 | 0.77 | 0.00962 | MirTarget; miRanda | -0.12 | 0.00287 | NA | |
| 93 | hsa-miR-335-3p | EFNA5 | 1.51 | 0 | 0.77 | 0.00962 | mirMAP | -0.19 | 0.00354 | NA | |
| 94 | hsa-miR-335-5p | EFNA5 | -0.47 | 0.0677 | 0.77 | 0.00962 | miRNAWalker2 validate | -0.22 | 0.0002 | NA | |
| 95 | hsa-miR-342-3p | EFNA5 | -0.13 | 0.56103 | 0.77 | 0.00962 | miRNATAP | -0.17 | 0.00565 | NA | |
| 96 | hsa-miR-362-3p | EFNA5 | 0.19 | 0.52808 | 0.77 | 0.00962 | miRanda | -0.15 | 0.00657 | NA | |
| 97 | hsa-miR-378a-3p | EFNA5 | -2.01 | 0 | 0.77 | 0.00962 | miRNATAP | -0.18 | 0.00233 | NA | |
| 98 | hsa-miR-421 | EFNA5 | 0.17 | 0.53528 | 0.77 | 0.00962 | mirMAP | -0.21 | 9.0E-5 | NA | |
| 99 | hsa-miR-501-5p | EFNA5 | 0.41 | 0.10435 | 0.77 | 0.00962 | mirMAP | -0.17 | 0.00174 | NA | |
| 100 | hsa-miR-141-3p | EGFR | 3.37 | 0 | -0.27 | 0.42339 | MirTarget | -0.18 | 0.00032 | 26025929 | Treatment with the EGFR inhibitor AG1478 or overexpression of miR141 blocked the activity of ERK downstream of EGFR and inhibited KLF8-depndent cell invasiveness proliferation and viability in cell culture and invasive growth and lung metastasis in nude mice |
| 101 | hsa-miR-142-3p | EGFR | 3.98 | 0 | -0.27 | 0.42339 | miRanda | -0.14 | 0.00224 | NA | |
| 102 | hsa-miR-148b-5p | EGFR | 1.39 | 0 | -0.27 | 0.42339 | mirMAP | -0.31 | 1.0E-5 | NA | |
| 103 | hsa-miR-17-5p | EGFR | 2.07 | 0 | -0.27 | 0.42339 | TargetScan | -0.23 | 0.00045 | NA | |
| 104 | hsa-miR-192-5p | EGFR | 2.03 | 2.0E-5 | -0.27 | 0.42339 | mirMAP | -0.15 | 0 | NA | |
| 105 | hsa-miR-19b-1-5p | EGFR | 1.71 | 0 | -0.27 | 0.42339 | MirTarget; mirMAP | -0.2 | 0.00608 | NA | |
| 106 | hsa-miR-200a-3p | EGFR | 3.15 | 0 | -0.27 | 0.42339 | MirTarget | -0.14 | 0.0034 | 26184032; 19671845 | MicroRNA 200a Targets EGFR and c Met to Inhibit Migration Invasion and Gefitinib Resistance in Non Small Cell Lung Cancer; In this study we found that miR-200a is downregulated in NSCLC cells where it directly targets the 3'-UTR of both EGFR and c-Met mRNA; Overexpression of miR-200a in NSCLC cells significantly downregulates both EGFR and c-Met levels and severely inhibits cell migration and invasion;Protein expression and signaling pathway modulation as well as intracellular distribution of EGFR and ERRFI-1 were validated through Western blot analysis and confocal microscopy whereas ERRFI-1 direct target of miR-200 members was validated by using the wild-type and mutant 3'-untranslated region/ERRFI-1/luciferse reporters; We identified a tight association between the expression of miRNAs of the miR-200 family epithelial phenotype and sensitivity to EGFR inhibitors-induced growth inhibition in bladder carcinoma cell lines; The changes in EGFR sensitivity by silencing or forced expression of ERRFI-1 or by miR-200 expression have also been validated in additional cell lines UMUC5 and T24; Members of the miR-200 family appear to control the EMT process and sensitivity to EGFR therapy in bladder cancer cells and the expression of miR-200 is sufficient to restore EGFR dependency at least in some of the mesenchymal bladder cancer cells |
| 107 | hsa-miR-320b | EGFR | 0.23 | 0.37882 | -0.27 | 0.42339 | miRanda; mirMAP | -0.17 | 0.00529 | NA | |
| 108 | hsa-miR-335-5p | EGFR | -0.47 | 0.0677 | -0.27 | 0.42339 | miRNAWalker2 validate | -0.18 | 0.00718 | NA | |
| 109 | hsa-miR-375 | EGFR | 0.62 | 0.1492 | -0.27 | 0.42339 | miRanda | -0.18 | 0 | NA | |
| 110 | hsa-miR-940 | EPHA2 | 0.01 | 0.97493 | -0.36 | 0.22595 | miRNAWalker2 validate | -0.12 | 0.00778 | NA | |
| 111 | hsa-miR-1180-3p | ETS1 | -0.44 | 0.06826 | -1.33 | 0 | MirTarget | -0.13 | 0.00054 | NA | |
| 112 | hsa-miR-130b-5p | ETS1 | 1.54 | 0 | -1.33 | 0 | MirTarget | -0.21 | 0 | NA | |
| 113 | hsa-miR-141-3p | ETS1 | 3.37 | 0 | -1.33 | 0 | mirMAP | -0.29 | 0 | NA | |
| 114 | hsa-miR-141-5p | ETS1 | 3.03 | 0 | -1.33 | 0 | mirMAP | -0.26 | 0 | NA | |
| 115 | hsa-miR-148b-5p | ETS1 | 1.39 | 0 | -1.33 | 0 | MirTarget | -0.26 | 0 | NA | |
| 116 | hsa-miR-16-1-3p | ETS1 | 1.5 | 0 | -1.33 | 0 | MirTarget | -0.17 | 0.00023 | NA | |
| 117 | hsa-miR-181b-5p | ETS1 | 0.67 | 0.00024 | -1.33 | 0 | MirTarget; miRNATAP | -0.14 | 0.00674 | NA | |
| 118 | hsa-miR-181d-5p | ETS1 | 1.19 | 0 | -1.33 | 0 | MirTarget | -0.17 | 0.00011 | NA | |
| 119 | hsa-miR-186-5p | ETS1 | 0.85 | 0 | -1.33 | 0 | mirMAP | -0.28 | 3.0E-5 | NA | |
| 120 | hsa-miR-200a-3p | ETS1 | 3.15 | 0 | -1.33 | 0 | mirMAP | -0.22 | 0 | NA | |
| 121 | hsa-miR-200b-3p | ETS1 | 1.55 | 0 | -1.33 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; TargetScan | -0.25 | 0 | NA | |
| 122 | hsa-miR-200c-3p | ETS1 | 0.38 | 0.08422 | -1.33 | 0 | MirTarget; miRNATAP | -0.28 | 0 | NA | |
| 123 | hsa-miR-27b-5p | ETS1 | 1.04 | 0 | -1.33 | 0 | MirTarget; miRNATAP | -0.21 | 0.00019 | NA | |
| 124 | hsa-miR-29a-5p | ETS1 | 1.9 | 0 | -1.33 | 0 | MirTarget; miRNATAP | -0.15 | 9.0E-5 | NA | |
| 125 | hsa-miR-335-3p | ETS1 | 1.51 | 0 | -1.33 | 0 | mirMAP | -0.25 | 0 | NA | |
| 126 | hsa-miR-429 | ETS1 | 2.38 | 0 | -1.33 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.25 | 0 | NA | |
| 127 | hsa-miR-450b-5p | ETS1 | 1.69 | 0 | -1.33 | 0 | miRNATAP | -0.11 | 0.00059 | NA | |
| 128 | hsa-miR-501-5p | ETS1 | 0.41 | 0.10435 | -1.33 | 0 | mirMAP | -0.19 | 0 | NA | |
| 129 | hsa-miR-576-5p | ETS1 | 1.03 | 0 | -1.33 | 0 | mirMAP | -0.13 | 0.00314 | NA | |
| 130 | hsa-miR-590-5p | ETS1 | 2.07 | 0 | -1.33 | 0 | miRanda | -0.24 | 0 | NA | |
| 131 | hsa-miR-660-5p | ETS1 | 2.05 | 0 | -1.33 | 0 | MirTarget | -0.17 | 1.0E-5 | NA | |
| 132 | hsa-miR-664a-3p | ETS1 | 0.44 | 0.02142 | -1.33 | 0 | mirMAP | -0.2 | 4.0E-5 | NA | |
| 133 | hsa-miR-7-1-3p | ETS1 | 2.61 | 0 | -1.33 | 0 | mirMAP | -0.2 | 0 | NA | |
| 134 | hsa-miR-146b-5p | ETS2 | 1.09 | 1.0E-5 | -1.08 | 0 | miRanda | -0.14 | 0.00047 | NA | |
| 135 | hsa-miR-199a-3p | ETS2 | 1.68 | 0 | -1.08 | 0 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 136 | hsa-miR-199a-5p | ETS2 | 1.31 | 0 | -1.08 | 0 | miRNAWalker2 validate; miRanda; miRNATAP | -0.11 | 0.00571 | NA | |
| 137 | hsa-miR-221-3p | ETS2 | -0.1 | 0.65445 | -1.08 | 0 | miRNATAP | -0.15 | 0.00055 | NA | |
| 138 | hsa-miR-222-3p | ETS2 | 0.03 | 0.88194 | -1.08 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 139 | hsa-miR-450b-5p | ETS2 | 1.69 | 0 | -1.08 | 0 | miRNATAP | -0.15 | 1.0E-5 | NA | |
| 140 | hsa-miR-590-3p | ETS2 | 0.84 | 0.00129 | -1.08 | 0 | miRanda | -0.15 | 0.00052 | NA | |
| 141 | hsa-let-7e-5p | FASLG | -0.75 | 7.0E-5 | 0.23 | 0.52524 | miRNATAP | -0.24 | 0.0052 | NA | |
| 142 | hsa-miR-16-1-3p | FGF1 | 1.5 | 0 | -0.58 | 0.0506 | mirMAP | -0.26 | 0.0001 | NA | |
| 143 | hsa-miR-186-5p | FGF1 | 0.85 | 0 | -0.58 | 0.0506 | mirMAP | -0.47 | 0 | NA | |
| 144 | hsa-miR-18a-5p | FGF1 | 1.37 | 1.0E-5 | -0.58 | 0.0506 | MirTarget | -0.31 | 0 | NA | |
| 145 | hsa-miR-320b | FGF1 | 0.23 | 0.37882 | -0.58 | 0.0506 | miRanda | -0.15 | 0.0062 | NA | |
| 146 | hsa-miR-330-5p | FGF1 | 0.17 | 0.33643 | -0.58 | 0.0506 | miRanda; miRNATAP | -0.4 | 0 | NA | |
| 147 | hsa-miR-429 | FGF1 | 2.38 | 0 | -0.58 | 0.0506 | miRNATAP | -0.12 | 0.00906 | NA | |
| 148 | hsa-miR-576-5p | FGF1 | 1.03 | 0 | -0.58 | 0.0506 | PITA; mirMAP | -0.35 | 0 | NA | |
| 149 | hsa-miR-590-5p | FGF1 | 2.07 | 0 | -0.58 | 0.0506 | MirTarget; miRanda; miRNATAP | -0.19 | 0.00157 | NA | |
| 150 | hsa-miR-629-5p | FGF1 | 1.32 | 0 | -0.58 | 0.0506 | MirTarget | -0.26 | 0.00036 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | INTRACELLULAR SIGNAL TRANSDUCTION | 105 | 1572 | 1.287e-78 | 2.994e-75 |
| 2 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 75 | 498 | 8.291e-79 | 2.994e-75 |
| 3 | PROTEIN PHOSPHORYLATION | 84 | 944 | 8.262e-70 | 1.281e-66 |
| 4 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 75 | 689 | 7.758e-68 | 9.024e-65 |
| 5 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 62 | 404 | 1.813e-64 | 1.687e-61 |
| 6 | PHOSPHORYLATION | 86 | 1228 | 7.073e-63 | 5.485e-60 |
| 7 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 100 | 1977 | 5.773e-62 | 3.837e-59 |
| 8 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 90 | 1518 | 4.488e-60 | 2.61e-57 |
| 9 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 95 | 1791 | 7.744e-60 | 4.004e-57 |
| 10 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 88 | 1656 | 2.168e-54 | 1.009e-51 |
| 11 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 91 | 1929 | 3.198e-52 | 1.353e-49 |
| 12 | POSITIVE REGULATION OF KINASE ACTIVITY | 56 | 482 | 8.451e-51 | 3.04e-48 |
| 13 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 68 | 876 | 8.492e-51 | 3.04e-48 |
| 14 | REGULATION OF MAPK CASCADE | 61 | 660 | 1.373e-49 | 4.085e-47 |
| 15 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 71 | 1036 | 1.405e-49 | 4.085e-47 |
| 16 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 71 | 1036 | 1.405e-49 | 4.085e-47 |
| 17 | POSITIVE REGULATION OF CELL COMMUNICATION | 81 | 1532 | 5.651e-49 | 1.547e-46 |
| 18 | POSITIVE REGULATION OF MAPK CASCADE | 54 | 470 | 1.338e-48 | 3.458e-46 |
| 19 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 72 | 1135 | 4.872e-48 | 1.193e-45 |
| 20 | REGULATION OF KINASE ACTIVITY | 61 | 776 | 2.377e-45 | 5.529e-43 |
| 21 | REGULATION OF PROTEIN MODIFICATION PROCESS | 81 | 1710 | 2.572e-45 | 5.699e-43 |
| 22 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 79 | 1618 | 6.088e-45 | 1.288e-42 |
| 23 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 56 | 616 | 8.427e-45 | 1.705e-42 |
| 24 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 35 | 138 | 7.924e-44 | 1.536e-41 |
| 25 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 74 | 1492 | 4.728e-42 | 8.8e-40 |
| 26 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 42 | 289 | 9.084e-42 | 1.626e-39 |
| 27 | REGULATION OF TRANSFERASE ACTIVITY | 61 | 946 | 2.884e-40 | 4.97e-38 |
| 28 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 60 | 905 | 3.113e-40 | 5.173e-38 |
| 29 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 47 | 470 | 3.887e-39 | 6.237e-37 |
| 30 | INOSITOL LIPID MEDIATED SIGNALING | 31 | 124 | 1.368e-38 | 2.121e-36 |
| 31 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 42 | 352 | 4.525e-38 | 6.792e-36 |
| 32 | REGULATION OF GTPASE ACTIVITY | 52 | 673 | 8.734e-38 | 1.27e-35 |
| 33 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 35 | 207 | 4.101e-37 | 5.783e-35 |
| 34 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 34 | 193 | 1.087e-36 | 1.488e-34 |
| 35 | RAS PROTEIN SIGNAL TRANSDUCTION | 31 | 143 | 1.827e-36 | 2.43e-34 |
| 36 | REGULATION OF HYDROLASE ACTIVITY | 65 | 1327 | 6.995e-36 | 9.041e-34 |
| 37 | REGULATION OF MAP KINASE ACTIVITY | 39 | 319 | 1.039e-35 | 1.306e-33 |
| 38 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 37 | 297 | 3.692e-34 | 4.52e-32 |
| 39 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 72 | 1848 | 8.949e-34 | 1.068e-31 |
| 40 | RESPONSE TO ENDOGENOUS STIMULUS | 64 | 1450 | 1.356e-32 | 1.558e-30 |
| 41 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 50 | 771 | 1.373e-32 | 1.558e-30 |
| 42 | GLYCEROLIPID METABOLIC PROCESS | 38 | 356 | 1.615e-32 | 1.789e-30 |
| 43 | POSITIVE REGULATION OF LOCOMOTION | 40 | 420 | 2.734e-32 | 2.958e-30 |
| 44 | PHOSPHOLIPID METABOLIC PROCESS | 38 | 364 | 3.758e-32 | 3.974e-30 |
| 45 | PEPTIDYL TYROSINE MODIFICATION | 30 | 186 | 3.62e-31 | 3.743e-29 |
| 46 | POSITIVE REGULATION OF CELL PROLIFERATION | 48 | 814 | 2.423e-29 | 2.451e-27 |
| 47 | LOCOMOTION | 54 | 1114 | 5.354e-29 | 5.191e-27 |
| 48 | RESPONSE TO GROWTH FACTOR | 39 | 475 | 5.294e-29 | 5.191e-27 |
| 49 | TAXIS | 38 | 464 | 3.315e-28 | 3.148e-26 |
| 50 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 51 | 1008 | 3.887e-28 | 3.617e-26 |
| 51 | REGULATION OF CELL PROLIFERATION | 60 | 1496 | 4.598e-28 | 4.195e-26 |
| 52 | LIPID PHOSPHORYLATION | 23 | 99 | 6.104e-28 | 5.462e-26 |
| 53 | REGULATION OF ERK1 AND ERK2 CASCADE | 29 | 238 | 1.691e-26 | 1.485e-24 |
| 54 | POSITIVE REGULATION OF ERK1 AND ERK2 CASCADE | 26 | 172 | 2.635e-26 | 2.27e-24 |
| 55 | RESPONSE TO NITROGEN COMPOUND | 46 | 859 | 2.863e-26 | 2.422e-24 |
| 56 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 30 | 279 | 9.24e-26 | 7.677e-24 |
| 57 | RESPONSE TO EXTERNAL STIMULUS | 61 | 1821 | 2.188e-24 | 1.786e-22 |
| 58 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 17 | 49 | 2.803e-24 | 2.249e-22 |
| 59 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 26 | 211 | 6.149e-24 | 4.849e-22 |
| 60 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 53 | 1381 | 1.368e-23 | 1.061e-21 |
| 61 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 57 | 1672 | 5.572e-23 | 4.25e-21 |
| 62 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 48 | 1142 | 6.869e-23 | 5.155e-21 |
| 63 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 29 | 323 | 1.137e-22 | 8.399e-21 |
| 64 | PROTEIN AUTOPHOSPHORYLATION | 24 | 192 | 2.669e-22 | 1.941e-20 |
| 65 | FC RECEPTOR SIGNALING PATHWAY | 24 | 206 | 1.47e-21 | 1.053e-19 |
| 66 | REGULATION OF VASCULATURE DEVELOPMENT | 25 | 233 | 1.564e-21 | 1.103e-19 |
| 67 | REGULATION OF CELL DEATH | 52 | 1472 | 1.764e-21 | 1.225e-19 |
| 68 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 25 | 235 | 1.935e-21 | 1.324e-19 |
| 69 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 33 | 505 | 2.173e-21 | 1.466e-19 |
| 70 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 44 | 1021 | 2.546e-21 | 1.693e-19 |
| 71 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 21 | 142 | 3.464e-21 | 2.27e-19 |
| 72 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 50 | 1395 | 7.237e-21 | 4.613e-19 |
| 73 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 48 | 1275 | 7.207e-21 | 4.613e-19 |
| 74 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 26 | 278 | 7.819e-21 | 4.917e-19 |
| 75 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 16 | 62 | 1.545e-20 | 9.583e-19 |
| 76 | LIPID MODIFICATION | 23 | 210 | 4.482e-20 | 2.708e-18 |
| 77 | REGULATION OF RAS PROTEIN SIGNAL TRANSDUCTION | 22 | 184 | 4.435e-20 | 2.708e-18 |
| 78 | ORGANOPHOSPHATE METABOLIC PROCESS | 40 | 885 | 4.552e-20 | 2.715e-18 |
| 79 | PEPTIDYL AMINO ACID MODIFICATION | 39 | 841 | 6.008e-20 | 3.538e-18 |
| 80 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 38 | 799 | 8.353e-20 | 4.858e-18 |
| 81 | VASCULATURE DEVELOPMENT | 30 | 469 | 3.273e-19 | 1.88e-17 |
| 82 | CELL MOTILITY | 38 | 835 | 3.744e-19 | 2.099e-17 |
| 83 | LOCALIZATION OF CELL | 38 | 835 | 3.744e-19 | 2.099e-17 |
| 84 | BLOOD VESSEL MORPHOGENESIS | 27 | 364 | 5.545e-19 | 3.072e-17 |
| 85 | CELL DEVELOPMENT | 48 | 1426 | 7.328e-19 | 4.011e-17 |
| 86 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 18 | 114 | 7.864e-19 | 4.255e-17 |
| 87 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 22 | 213 | 1.108e-18 | 5.924e-17 |
| 88 | REGULATION OF PHOSPHOLIPASE ACTIVITY | 15 | 64 | 1.26e-18 | 6.664e-17 |
| 89 | CELLULAR RESPONSE TO PEPTIDE | 24 | 274 | 1.281e-18 | 6.695e-17 |
| 90 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 13 | 39 | 1.489e-18 | 7.698e-17 |
| 91 | REGULATION OF IMMUNE SYSTEM PROCESS | 47 | 1403 | 2.299e-18 | 1.175e-16 |
| 92 | REGULATION OF LIPASE ACTIVITY | 16 | 83 | 2.545e-18 | 1.287e-16 |
| 93 | PHOSPHATIDYLETHANOLAMINE ACYL CHAIN REMODELING | 11 | 23 | 5.276e-18 | 2.64e-16 |
| 94 | RESPONSE TO WOUNDING | 31 | 563 | 5.702e-18 | 2.822e-16 |
| 95 | CELLULAR LIPID METABOLIC PROCESS | 38 | 913 | 7.548e-18 | 3.697e-16 |
| 96 | NEGATIVE REGULATION OF CELL DEATH | 37 | 872 | 1.194e-17 | 5.787e-16 |
| 97 | CELL PROLIFERATION | 33 | 672 | 1.28e-17 | 6.138e-16 |
| 98 | REGULATION OF CELL ADHESION | 32 | 629 | 1.544e-17 | 7.329e-16 |
| 99 | RESPONSE TO HORMONE | 37 | 893 | 2.587e-17 | 1.193e-15 |
| 100 | REGULATION OF CELL DIFFERENTIATION | 47 | 1492 | 2.616e-17 | 1.193e-15 |
| 101 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 35 | 788 | 2.596e-17 | 1.193e-15 |
| 102 | CIRCULATORY SYSTEM DEVELOPMENT | 35 | 788 | 2.596e-17 | 1.193e-15 |
| 103 | PHOSPHATIDYLCHOLINE ACYL CHAIN REMODELING | 11 | 26 | 2.955e-17 | 1.335e-15 |
| 104 | WOUND HEALING | 28 | 470 | 3.682e-17 | 1.647e-15 |
| 105 | REGULATION OF EPITHELIAL CELL MIGRATION | 19 | 166 | 4.149e-17 | 1.839e-15 |
| 106 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 17 | 120 | 5.143e-17 | 2.257e-15 |
| 107 | REGULATION OF TRANSPORT | 51 | 1804 | 6.878e-17 | 2.991e-15 |
| 108 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 41 | 1152 | 7.32e-17 | 3.154e-15 |
| 109 | ANGIOGENESIS | 23 | 293 | 8.043e-17 | 3.434e-15 |
| 110 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 37 | 926 | 8.349e-17 | 3.532e-15 |
| 111 | POSITIVE REGULATION OF LIPASE ACTIVITY | 14 | 66 | 8.493e-17 | 3.56e-15 |
| 112 | RESPONSE TO PEPTIDE | 26 | 404 | 8.616e-17 | 3.58e-15 |
| 113 | FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 15 | 84 | 1.026e-16 | 4.223e-15 |
| 114 | POSITIVE REGULATION OF PHOSPHOLIPASE ACTIVITY | 13 | 53 | 1.406e-16 | 5.715e-15 |
| 115 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 12 | 40 | 1.412e-16 | 5.715e-15 |
| 116 | NEURON PROJECTION DEVELOPMENT | 29 | 545 | 1.897e-16 | 7.609e-15 |
| 117 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 13 | 55 | 2.393e-16 | 9.516e-15 |
| 118 | POSITIVE REGULATION OF CELL DIVISION | 17 | 132 | 2.69e-16 | 1.061e-14 |
| 119 | TISSUE DEVELOPMENT | 46 | 1518 | 2.789e-16 | 1.09e-14 |
| 120 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 17 | 133 | 3.064e-16 | 1.188e-14 |
| 121 | REGULATION OF IMMUNE RESPONSE | 35 | 858 | 3.556e-16 | 1.367e-14 |
| 122 | NEUROGENESIS | 44 | 1402 | 4.212e-16 | 1.606e-14 |
| 123 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 35 | 867 | 4.886e-16 | 1.848e-14 |
| 124 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 18 | 162 | 4.963e-16 | 1.862e-14 |
| 125 | ACTIVATION OF MAPK ACTIVITY | 17 | 137 | 5.103e-16 | 1.899e-14 |
| 126 | IMMUNE SYSTEM PROCESS | 52 | 1984 | 7.322e-16 | 2.704e-14 |
| 127 | PHOSPHATIDYLINOSITOL ACYL CHAIN REMODELING | 9 | 16 | 8.712e-16 | 3.192e-14 |
| 128 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 13 | 61 | 1.04e-15 | 3.782e-14 |
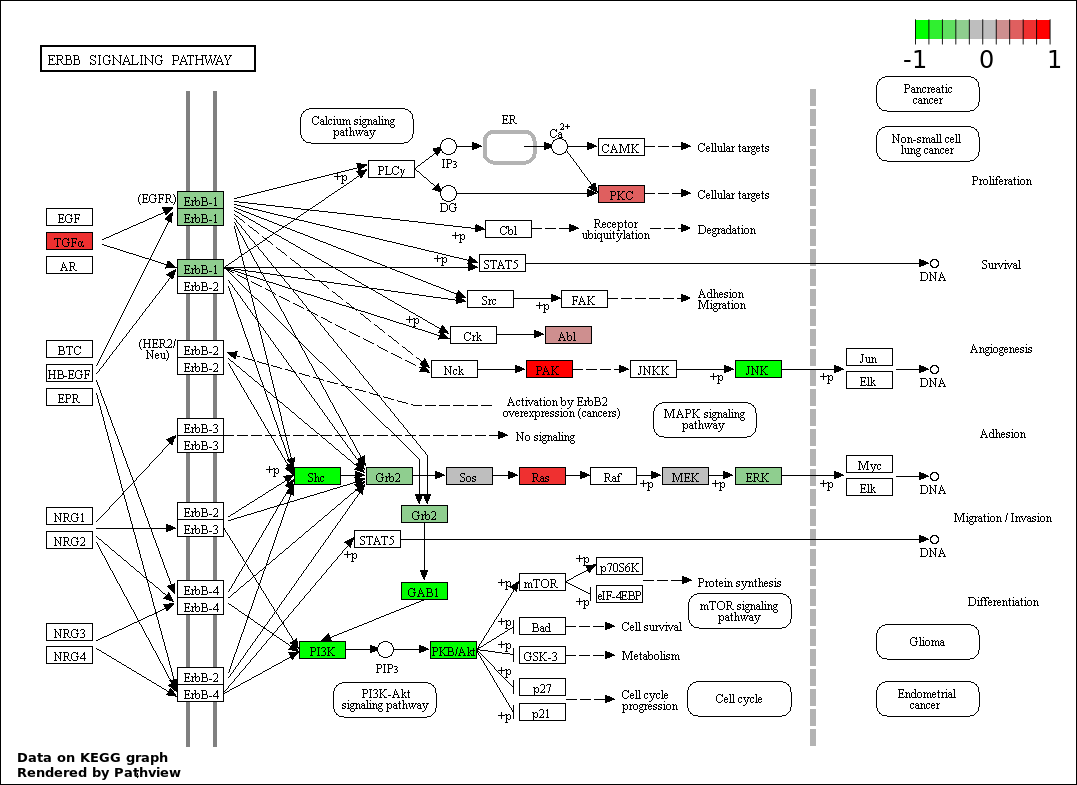
| 129 | ERBB SIGNALING PATHWAY | 14 | 79 | 1.253e-15 | 4.518e-14 |
| 130 | CELL PROJECTION ORGANIZATION | 35 | 902 | 1.622e-15 | 5.804e-14 |
| 131 | REGULATION OF LIPID KINASE ACTIVITY | 12 | 48 | 1.67e-15 | 5.933e-14 |
| 132 | CELLULAR RESPONSE TO HORMONE STIMULUS | 28 | 552 | 2.248e-15 | 7.925e-14 |
| 133 | REGULATION OF CHEMOTAXIS | 18 | 180 | 3.253e-15 | 1.138e-13 |
| 134 | EPHRIN RECEPTOR SIGNALING PATHWAY | 14 | 85 | 3.676e-15 | 1.276e-13 |
| 135 | CELL CELL SIGNALING | 32 | 767 | 4.358e-15 | 1.502e-13 |
| 136 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 33 | 823 | 4.723e-15 | 1.616e-13 |
| 137 | REGULATION OF PHOSPHOLIPASE C ACTIVITY | 11 | 39 | 5.882e-15 | 1.998e-13 |
| 138 | REGULATION OF NEURON DEATH | 20 | 252 | 7.704e-15 | 2.598e-13 |
| 139 | LIPID BIOSYNTHETIC PROCESS | 27 | 539 | 1.023e-14 | 3.423e-13 |
| 140 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 25 | 450 | 1.064e-14 | 3.536e-13 |
| 141 | NEURON DEVELOPMENT | 30 | 687 | 1.077e-14 | 3.555e-13 |
| 142 | REGULATION OF RESPONSE TO STRESS | 43 | 1468 | 1.146e-14 | 3.754e-13 |
| 143 | LEUKOCYTE MIGRATION | 20 | 259 | 1.301e-14 | 4.233e-13 |
| 144 | RESPONSE TO FIBROBLAST GROWTH FACTOR | 15 | 116 | 1.543e-14 | 4.987e-13 |
| 145 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 34 | 917 | 1.667e-14 | 5.35e-13 |
| 146 | LIPID METABOLIC PROCESS | 38 | 1158 | 1.721e-14 | 5.485e-13 |
| 147 | REGULATION OF CELL PROJECTION ORGANIZATION | 27 | 558 | 2.364e-14 | 7.482e-13 |
| 148 | POSITIVE REGULATION OF CHEMOTAXIS | 15 | 120 | 2.583e-14 | 8.12e-13 |
| 149 | POSITIVE REGULATION OF LIPID KINASE ACTIVITY | 10 | 32 | 3.249e-14 | 1.014e-12 |
| 150 | PLATELET DERIVED GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 34 | 6.516e-14 | 2.021e-12 |
| 151 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 16 | 154 | 6.584e-14 | 2.029e-12 |
| 152 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 15 | 128 | 6.845e-14 | 2.095e-12 |
| 153 | PHOSPHATIDYLCHOLINE METABOLIC PROCESS | 12 | 64 | 7.082e-14 | 2.154e-12 |
| 154 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 17 | 185 | 8.108e-14 | 2.45e-12 |
| 155 | POSITIVE REGULATION OF DNA REPLICATION | 13 | 86 | 1.169e-13 | 3.508e-12 |
| 156 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 12 | 67 | 1.268e-13 | 3.781e-12 |
| 157 | TUBE DEVELOPMENT | 26 | 552 | 1.409e-13 | 4.176e-12 |
| 158 | NEURON DIFFERENTIATION | 32 | 874 | 1.59e-13 | 4.684e-12 |
| 159 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 30 | 767 | 1.904e-13 | 5.572e-12 |
| 160 | EPITHELIUM DEVELOPMENT | 33 | 945 | 2.319e-13 | 6.743e-12 |
| 161 | PHOSPHATIDYLSERINE ACYL CHAIN REMODELING | 8 | 17 | 2.486e-13 | 7.139e-12 |
| 162 | PHOSPHATIDYLGLYCEROL ACYL CHAIN REMODELING | 8 | 17 | 2.486e-13 | 7.139e-12 |
| 163 | REGULATION OF CELL DIVISION | 19 | 272 | 3.782e-13 | 1.08e-11 |
| 164 | NEURON PROJECTION MORPHOGENESIS | 22 | 402 | 6.561e-13 | 1.862e-11 |
| 165 | POSITIVE REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 10 | 42 | 6.935e-13 | 1.956e-11 |
| 166 | REGULATION OF LIPID METABOLIC PROCESS | 19 | 282 | 7.202e-13 | 2.019e-11 |
| 167 | REGULATION OF ORGANELLE ORGANIZATION | 36 | 1178 | 8.077e-13 | 2.25e-11 |
| 168 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 13 | 103 | 1.284e-12 | 3.557e-11 |
| 169 | REGULATION OF VESICLE MEDIATED TRANSPORT | 23 | 462 | 1.334e-12 | 3.673e-11 |
| 170 | PHOSPHATIDYLGLYCEROL METABOLIC PROCESS | 9 | 31 | 1.391e-12 | 3.806e-11 |
| 171 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 24 | 513 | 1.559e-12 | 4.242e-11 |
| 172 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 19 | 296 | 1.7e-12 | 4.599e-11 |
| 173 | CELL ACTIVATION | 25 | 568 | 1.949e-12 | 5.242e-11 |
| 174 | CELLULAR COMPONENT MORPHOGENESIS | 31 | 900 | 1.998e-12 | 5.342e-11 |
| 175 | REGULATION OF NEURON APOPTOTIC PROCESS | 16 | 192 | 2.058e-12 | 5.473e-11 |
| 176 | RESPONSE TO ABIOTIC STIMULUS | 33 | 1024 | 2.101e-12 | 5.554e-11 |
| 177 | ETHANOLAMINE CONTAINING COMPOUND METABOLIC PROCESS | 12 | 85 | 2.468e-12 | 6.489e-11 |
| 178 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 19 | 303 | 2.568e-12 | 6.712e-11 |
| 179 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 44 | 1805 | 3.103e-12 | 8.066e-11 |
| 180 | VASCULAR ENDOTHELIAL GROWTH FACTOR SIGNALING PATHWAY | 7 | 14 | 4.805e-12 | 1.242e-10 |
| 181 | NEGATIVE REGULATION OF NEURON DEATH | 15 | 171 | 4.952e-12 | 1.273e-10 |
| 182 | PLATELET ACTIVATION | 14 | 142 | 5.32e-12 | 1.36e-10 |
| 183 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 10 | 51 | 5.674e-12 | 1.443e-10 |
| 184 | CELL DEATH | 32 | 1001 | 5.964e-12 | 1.508e-10 |
| 185 | CELLULAR RESPONSE TO INSULIN STIMULUS | 14 | 146 | 7.788e-12 | 1.959e-10 |
| 186 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 18 | 285 | 8.897e-12 | 2.226e-10 |
| 187 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 20 | 368 | 9.023e-12 | 2.245e-10 |
| 188 | EMBRYO DEVELOPMENT | 30 | 894 | 9.233e-12 | 2.285e-10 |
| 189 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 31 | 957 | 9.679e-12 | 2.383e-10 |
| 190 | POSITIVE REGULATION OF IMMUNE RESPONSE | 24 | 563 | 1.106e-11 | 2.709e-10 |
| 191 | ORGAN MORPHOGENESIS | 29 | 841 | 1.124e-11 | 2.738e-10 |
| 192 | POSITIVE REGULATION OF CELL ADHESION | 20 | 376 | 1.332e-11 | 3.227e-10 |
| 193 | ERBB2 SIGNALING PATHWAY | 9 | 39 | 1.387e-11 | 3.343e-10 |
| 194 | ACTIVATION OF IMMUNE RESPONSE | 21 | 427 | 1.726e-11 | 4.14e-10 |
| 195 | IMMUNE SYSTEM DEVELOPMENT | 24 | 582 | 2.207e-11 | 5.266e-10 |
| 196 | REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 7 | 17 | 2.67e-11 | 6.339e-10 |
| 197 | INSULIN RECEPTOR SIGNALING PATHWAY | 11 | 80 | 2.802e-11 | 6.619e-10 |
| 198 | POSITIVE REGULATION OF TRANSPORT | 30 | 936 | 2.872e-11 | 6.75e-10 |
| 199 | PHOSPHATIDYLSERINE METABOLIC PROCESS | 8 | 28 | 2.957e-11 | 6.914e-10 |
| 200 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 15 | 195 | 3.282e-11 | 7.635e-10 |
| 201 | HEMOSTASIS | 18 | 311 | 3.783e-11 | 8.758e-10 |
| 202 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 13 | 135 | 4.246e-11 | 9.781e-10 |
| 203 | REGULATION OF BODY FLUID LEVELS | 22 | 506 | 5.963e-11 | 1.367e-09 |
| 204 | RESPONSE TO INSULIN | 15 | 205 | 6.686e-11 | 1.518e-09 |
| 205 | NEURON PROJECTION GUIDANCE | 15 | 205 | 6.686e-11 | 1.518e-09 |
| 206 | POSITIVE REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 6 | 11 | 8.797e-11 | 1.987e-09 |
| 207 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 23 | 573 | 1.028e-10 | 2.31e-09 |
| 208 | T CELL RECEPTOR SIGNALING PATHWAY | 13 | 146 | 1.145e-10 | 2.561e-09 |
| 209 | COGNITION | 16 | 251 | 1.184e-10 | 2.636e-09 |
| 210 | REGULATION OF CELL CELL ADHESION | 19 | 380 | 1.29e-10 | 2.858e-09 |
| 211 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 26 | 750 | 1.349e-10 | 2.974e-09 |
| 212 | REGULATION OF DNA METABOLIC PROCESS | 18 | 340 | 1.625e-10 | 3.567e-09 |
| 213 | MODULATION OF SYNAPTIC TRANSMISSION | 17 | 301 | 1.997e-10 | 4.362e-09 |
| 214 | ALDITOL PHOSPHATE METABOLIC PROCESS | 8 | 35 | 2.139e-10 | 4.65e-09 |
| 215 | POSITIVE REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 7 | 22 | 2.267e-10 | 4.906e-09 |
| 216 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 74 | 2.74e-10 | 5.864e-09 |
| 217 | REGULATION OF CELL DEVELOPMENT | 27 | 836 | 2.748e-10 | 5.864e-09 |
| 218 | POSITIVE CHEMOTAXIS | 8 | 36 | 2.732e-10 | 5.864e-09 |
| 219 | MORPHOGENESIS OF AN EPITHELIUM | 19 | 400 | 3.069e-10 | 6.52e-09 |
| 220 | REGULATION OF NEURON DIFFERENTIATION | 22 | 554 | 3.338e-10 | 7.061e-09 |
| 221 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 17 | 312 | 3.476e-10 | 7.318e-09 |
| 222 | REGULATION OF DNA REPLICATION | 13 | 161 | 3.895e-10 | 8.164e-09 |
| 223 | EMBRYONIC ORGAN DEVELOPMENT | 19 | 406 | 3.94e-10 | 8.222e-09 |
| 224 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 19 | 408 | 4.279e-10 | 8.888e-09 |
| 225 | CELLULAR RESPONSE TO GLUCAGON STIMULUS | 8 | 38 | 4.357e-10 | 9.011e-09 |
| 226 | AMINE METABOLIC PROCESS | 12 | 131 | 4.388e-10 | 9.034e-09 |
| 227 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 13 | 167 | 6.135e-10 | 1.258e-08 |
| 228 | REGULATION OF AXONOGENESIS | 13 | 168 | 6.606e-10 | 1.348e-08 |
| 229 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 27 | 872 | 6.933e-10 | 1.409e-08 |
| 230 | REGULATION OF CELLULAR LOCALIZATION | 33 | 1277 | 7.019e-10 | 1.42e-08 |
| 231 | AMMONIUM ION METABOLIC PROCESS | 13 | 169 | 7.109e-10 | 1.432e-08 |
| 232 | CELL PART MORPHOGENESIS | 23 | 633 | 7.182e-10 | 1.44e-08 |
| 233 | POSITIVE REGULATION OF CELL DEVELOPMENT | 20 | 472 | 7.465e-10 | 1.491e-08 |
| 234 | REGULATION OF DEVELOPMENTAL GROWTH | 16 | 289 | 9.384e-10 | 1.866e-08 |
| 235 | REGULATION OF CELL SUBSTRATE ADHESION | 13 | 173 | 9.489e-10 | 1.879e-08 |
| 236 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 17 | 337 | 1.13e-09 | 2.228e-08 |
| 237 | REGULATION OF CELL ACTIVATION | 20 | 484 | 1.152e-09 | 2.261e-08 |
| 238 | HEAD DEVELOPMENT | 24 | 709 | 1.21e-09 | 2.365e-08 |
| 239 | RHYTHMIC PROCESS | 16 | 298 | 1.463e-09 | 2.849e-08 |
| 240 | GLAND DEVELOPMENT | 18 | 395 | 1.795e-09 | 3.48e-08 |
| 241 | ALCOHOL METABOLIC PROCESS | 17 | 348 | 1.838e-09 | 3.549e-08 |
| 242 | REGULATION OF CELL MORPHOGENESIS | 21 | 552 | 1.882e-09 | 3.618e-08 |
| 243 | REGULATION OF CELL MATRIX ADHESION | 10 | 90 | 1.963e-09 | 3.759e-08 |
| 244 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 16 | 307 | 2.247e-09 | 4.284e-08 |
| 245 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 7 | 30 | 2.569e-09 | 4.879e-08 |
| 246 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 18 | 406 | 2.767e-09 | 5.234e-08 |
| 247 | REPRODUCTIVE SYSTEM DEVELOPMENT | 18 | 408 | 2.989e-09 | 5.631e-08 |
| 248 | RESPONSE TO GLUCAGON | 8 | 48 | 3.149e-09 | 5.908e-08 |
| 249 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 33 | 1360 | 3.415e-09 | 6.381e-08 |
| 250 | RESPONSE TO RADIATION | 18 | 413 | 3.618e-09 | 6.734e-08 |
| 251 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 14 | 232 | 3.678e-09 | 6.792e-08 |
| 252 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 14 | 232 | 3.678e-09 | 6.792e-08 |
| 253 | REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 7 | 32 | 4.193e-09 | 7.711e-08 |
| 254 | REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 10 | 98 | 4.559e-09 | 8.351e-08 |
| 255 | TUBE MORPHOGENESIS | 16 | 323 | 4.649e-09 | 8.482e-08 |
| 256 | RESPONSE TO LIPID | 26 | 888 | 4.804e-09 | 8.731e-08 |
| 257 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 12 | 162 | 5.06e-09 | 9.161e-08 |
| 258 | PHOSPHATIDIC ACID METABOLIC PROCESS | 7 | 33 | 5.287e-09 | 9.535e-08 |
| 259 | SECRETION | 21 | 588 | 5.74e-09 | 1.031e-07 |
| 260 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 8 | 53 | 7.158e-09 | 1.281e-07 |
| 261 | SINGLE ORGANISM BEHAVIOR | 17 | 384 | 8.025e-09 | 1.431e-07 |
| 262 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 21 | 602 | 8.658e-09 | 1.538e-07 |
| 263 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 10 | 106 | 9.834e-09 | 1.74e-07 |
| 264 | REGULATION OF SYNAPTIC PLASTICITY | 11 | 140 | 1.219e-08 | 2.148e-07 |
| 265 | REGULATION OF HOMEOSTATIC PROCESS | 18 | 447 | 1.237e-08 | 2.171e-07 |
| 266 | REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 7 | 37 | 1.241e-08 | 2.172e-07 |
| 267 | REGULATION OF CYTOSKELETON ORGANIZATION | 19 | 502 | 1.285e-08 | 2.24e-07 |
| 268 | REGULATION OF FIBROBLAST PROLIFERATION | 9 | 81 | 1.295e-08 | 2.248e-07 |
| 269 | REGULATION OF INTRACELLULAR TRANSPORT | 21 | 621 | 1.484e-08 | 2.567e-07 |
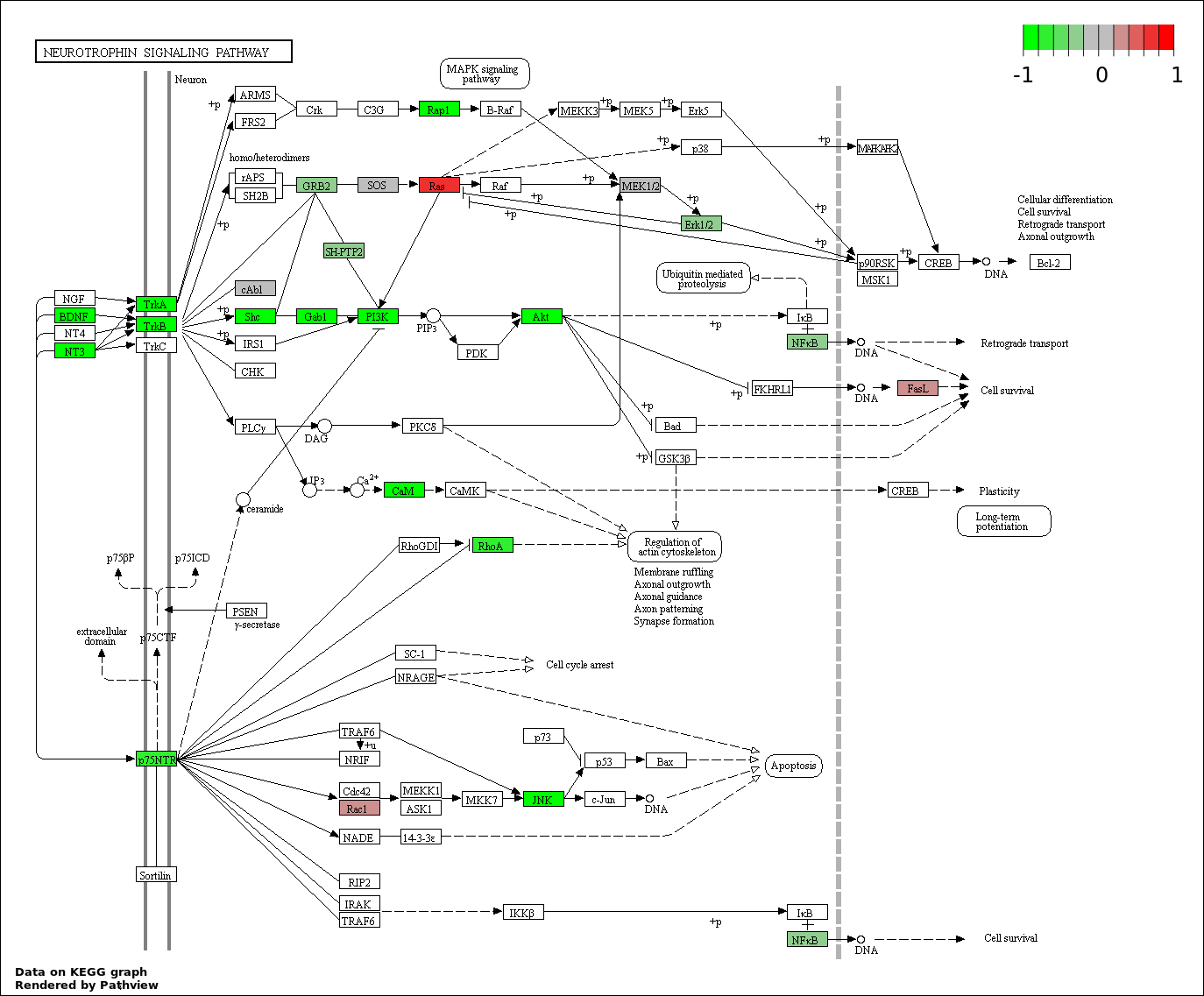
| 270 | NEUROTROPHIN SIGNALING PATHWAY | 6 | 23 | 1.78e-08 | 3.068e-07 |
| 271 | REGULATION OF GLUCOSE IMPORT | 8 | 60 | 1.974e-08 | 3.389e-07 |
| 272 | REGULATION OF GROWTH | 21 | 633 | 2.064e-08 | 3.532e-07 |
| 273 | REGULATION OF LONG TERM NEURONAL SYNAPTIC PLASTICITY | 6 | 24 | 2.358e-08 | 4.019e-07 |
| 274 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 25 | 3.083e-08 | 5.236e-07 |
| 275 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 7 | 42 | 3.149e-08 | 5.329e-07 |
| 276 | TISSUE MORPHOGENESIS | 19 | 533 | 3.34e-08 | 5.63e-07 |
| 277 | REGULATION OF PROTEIN KINASE B SIGNALING | 10 | 121 | 3.537e-08 | 5.942e-07 |
| 278 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 18 | 482 | 3.912e-08 | 6.548e-07 |
| 279 | RESPONSE TO LIGHT STIMULUS | 14 | 280 | 3.991e-08 | 6.657e-07 |
| 280 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 12 | 197 | 4.525e-08 | 7.52e-07 |
| 281 | POSITIVE REGULATION OF CELL CYCLE | 15 | 332 | 4.89e-08 | 8.097e-07 |
| 282 | REGULATION OF ENDOCYTOSIS | 12 | 199 | 5.059e-08 | 8.347e-07 |
| 283 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 9 | 95 | 5.318e-08 | 8.721e-07 |
| 284 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 17 | 437 | 5.323e-08 | 8.721e-07 |
| 285 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 8 | 68 | 5.412e-08 | 8.836e-07 |
| 286 | CELL CHEMOTAXIS | 11 | 162 | 5.549e-08 | 9.027e-07 |
| 287 | POSITIVE REGULATION OF CELL CELL ADHESION | 13 | 243 | 5.659e-08 | 9.175e-07 |
| 288 | POSITIVE REGULATION OF AXONOGENESIS | 8 | 69 | 6.082e-08 | 9.826e-07 |
| 289 | LIPID CATABOLIC PROCESS | 13 | 247 | 6.851e-08 | 1.103e-06 |
| 290 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 10 | 131 | 7.548e-08 | 1.211e-06 |
| 291 | ACTIN FILAMENT BASED PROCESS | 17 | 450 | 8.114e-08 | 1.297e-06 |
| 292 | OSSIFICATION | 13 | 251 | 8.264e-08 | 1.317e-06 |
| 293 | REGULATION OF GLUCOSE TRANSPORT | 9 | 100 | 8.33e-08 | 1.323e-06 |
| 294 | REGULATION OF CELLULAR RESPONSE TO STRESS | 21 | 691 | 9.154e-08 | 1.449e-06 |
| 295 | REGULATION OF NEURONAL SYNAPTIC PLASTICITY | 7 | 49 | 9.583e-08 | 1.512e-06 |
| 296 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 5 | 16 | 1.069e-07 | 1.68e-06 |
| 297 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 9 | 103 | 1.078e-07 | 1.688e-06 |
| 298 | POSITIVE REGULATION OF GENE EXPRESSION | 35 | 1733 | 1.091e-07 | 1.698e-06 |
| 299 | BEHAVIOR | 18 | 516 | 1.089e-07 | 1.698e-06 |
| 300 | REGULATION OF SECRETION | 21 | 699 | 1.11e-07 | 1.722e-06 |
| 301 | HOMEOSTATIC PROCESS | 30 | 1337 | 1.143e-07 | 1.768e-06 |
| 302 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 14 | 306 | 1.198e-07 | 1.844e-06 |
| 303 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 30 | 1340 | 1.201e-07 | 1.844e-06 |
| 304 | GROWTH | 16 | 410 | 1.288e-07 | 1.971e-06 |
| 305 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 17 | 465 | 1.296e-07 | 1.977e-06 |
| 306 | HEART DEVELOPMENT | 17 | 466 | 1.336e-07 | 2.032e-06 |
| 307 | EXOCYTOSIS | 14 | 310 | 1.405e-07 | 2.129e-06 |
| 308 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 5 | 17 | 1.504e-07 | 2.273e-06 |
| 309 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 25 | 983 | 1.516e-07 | 2.283e-06 |
| 310 | LYMPHOCYTE COSTIMULATION | 8 | 78 | 1.608e-07 | 2.414e-06 |
| 311 | REGULATED EXOCYTOSIS | 12 | 224 | 1.843e-07 | 2.757e-06 |
| 312 | REGULATION OF CYTOPLASMIC TRANSPORT | 17 | 481 | 2.093e-07 | 3.121e-06 |
| 313 | SYSTEM PROCESS | 35 | 1785 | 2.234e-07 | 3.321e-06 |
| 314 | PEPTIDYL SERINE MODIFICATION | 10 | 148 | 2.382e-07 | 3.53e-06 |
| 315 | SECRETION BY CELL | 17 | 486 | 2.421e-07 | 3.576e-06 |
| 316 | EMBRYONIC PLACENTA DEVELOPMENT | 8 | 83 | 2.618e-07 | 3.855e-06 |
| 317 | KIDNEY VASCULATURE DEVELOPMENT | 5 | 19 | 2.792e-07 | 4.085e-06 |
| 318 | RENAL SYSTEM VASCULATURE DEVELOPMENT | 5 | 19 | 2.792e-07 | 4.085e-06 |
| 319 | SENSORY ORGAN DEVELOPMENT | 17 | 493 | 2.96e-07 | 4.317e-06 |
| 320 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 10 | 152 | 3.054e-07 | 4.441e-06 |
| 321 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 9 | 118 | 3.478e-07 | 5.042e-06 |
| 322 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 7 | 59 | 3.567e-07 | 5.154e-06 |
| 323 | EMBRYONIC HEMOPOIESIS | 5 | 20 | 3.699e-07 | 5.328e-06 |
| 324 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 10 | 156 | 3.888e-07 | 5.583e-06 |
| 325 | RESPIRATORY SYSTEM DEVELOPMENT | 11 | 197 | 4.03e-07 | 5.769e-06 |
| 326 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 8 | 88 | 4.129e-07 | 5.894e-06 |
| 327 | REGULATION OF ORGAN MORPHOGENESIS | 12 | 242 | 4.23e-07 | 6.019e-06 |
| 328 | LEUKOCYTE DIFFERENTIATION | 13 | 292 | 4.708e-07 | 6.679e-06 |
| 329 | MIDBRAIN DEVELOPMENT | 8 | 90 | 4.915e-07 | 6.951e-06 |
| 330 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 9 | 123 | 4.954e-07 | 6.985e-06 |
| 331 | ASTROCYTE DIFFERENTIATION | 6 | 39 | 5.194e-07 | 7.279e-06 |
| 332 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 6 | 39 | 5.194e-07 | 7.279e-06 |
| 333 | SKELETAL SYSTEM DEVELOPMENT | 16 | 455 | 5.211e-07 | 7.282e-06 |
| 334 | NEGATIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 6 | 40 | 6.071e-07 | 8.458e-06 |
| 335 | EPIDERMIS DEVELOPMENT | 12 | 253 | 6.786e-07 | 9.426e-06 |
| 336 | REPRODUCTION | 28 | 1297 | 7.08e-07 | 9.804e-06 |
| 337 | NUCLEAR TRANSPORT | 14 | 355 | 7.224e-07 | 9.974e-06 |
| 338 | RESPONSE TO MECHANICAL STIMULUS | 11 | 210 | 7.603e-07 | 1.047e-05 |
| 339 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 23 | 7.878e-07 | 1.081e-05 |
| 340 | RESPONSE TO ALCOHOL | 14 | 362 | 9.11e-07 | 1.247e-05 |
| 341 | MEMORY | 8 | 98 | 9.47e-07 | 1.288e-05 |
| 342 | REGULATION OF POLYSACCHARIDE METABOLIC PROCESS | 6 | 43 | 9.46e-07 | 1.288e-05 |
| 343 | REGULATION OF CELL JUNCTION ASSEMBLY | 7 | 68 | 9.563e-07 | 1.292e-05 |
| 344 | RESPONSE TO OXYGEN LEVELS | 13 | 311 | 9.579e-07 | 1.292e-05 |
| 345 | POSITIVE REGULATION OF CELL ACTIVATION | 13 | 311 | 9.579e-07 | 1.292e-05 |
| 346 | VESICLE MEDIATED TRANSPORT | 27 | 1239 | 9.653e-07 | 1.298e-05 |
| 347 | REGULATION OF POSITIVE CHEMOTAXIS | 5 | 24 | 9.89e-07 | 1.326e-05 |
| 348 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 21 | 801 | 1.032e-06 | 1.379e-05 |
| 349 | CHEMICAL HOMEOSTASIS | 22 | 874 | 1.094e-06 | 1.458e-05 |
| 350 | POSITIVE REGULATION OF CELL DEATH | 18 | 605 | 1.109e-06 | 1.475e-05 |
| 351 | HOMEOSTASIS OF NUMBER OF CELLS | 10 | 175 | 1.118e-06 | 1.482e-05 |
| 352 | GLIAL CELL DIFFERENTIATION | 9 | 136 | 1.157e-06 | 1.529e-05 |
| 353 | REGULATION OF PROTEIN LOCALIZATION | 23 | 950 | 1.182e-06 | 1.559e-05 |
| 354 | POSITIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 5 | 25 | 1.228e-06 | 1.615e-05 |
| 355 | SPROUTING ANGIOGENESIS | 6 | 45 | 1.248e-06 | 1.636e-05 |
| 356 | PLACENTA DEVELOPMENT | 9 | 138 | 1.307e-06 | 1.708e-05 |
| 357 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 17 | 554 | 1.481e-06 | 1.931e-05 |
| 358 | CEREBRAL CORTEX DEVELOPMENT | 8 | 105 | 1.603e-06 | 2.083e-05 |
| 359 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 12 | 1.637e-06 | 2.121e-05 |
| 360 | REGULATION OF DEFENSE RESPONSE | 20 | 759 | 1.768e-06 | 2.285e-05 |
| 361 | REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 5 | 27 | 1.843e-06 | 2.37e-05 |
| 362 | POSITIVE REGULATION OF MESENCHYMAL CELL PROLIFERATION | 5 | 27 | 1.843e-06 | 2.37e-05 |
| 363 | PLATELET DEGRANULATION | 8 | 107 | 1.849e-06 | 2.371e-05 |
| 364 | GLOMERULUS DEVELOPMENT | 6 | 49 | 2.088e-06 | 2.669e-05 |
| 365 | CYTOSKELETON ORGANIZATION | 21 | 838 | 2.112e-06 | 2.692e-05 |
| 366 | VISUAL BEHAVIOR | 6 | 50 | 2.358e-06 | 2.998e-05 |
| 367 | POSITIVE REGULATION OF GROWTH | 11 | 238 | 2.584e-06 | 3.276e-05 |
| 368 | POSITIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 6 | 51 | 2.655e-06 | 3.357e-05 |
| 369 | NEGATIVE REGULATION OF VASCULATURE DEVELOPMENT | 7 | 80 | 2.899e-06 | 3.656e-05 |
| 370 | CELLULAR RESPONSE TO STRESS | 30 | 1565 | 3.115e-06 | 3.917e-05 |
| 371 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 7 | 81 | 3.153e-06 | 3.955e-05 |
| 372 | PHOSPHOLIPID CATABOLIC PROCESS | 5 | 30 | 3.194e-06 | 3.984e-05 |
| 373 | NEPHRON DEVELOPMENT | 8 | 115 | 3.185e-06 | 3.984e-05 |
| 374 | ARACHIDONIC ACID SECRETION | 4 | 14 | 3.27e-06 | 4.057e-05 |
| 375 | ARACHIDONATE TRANSPORT | 4 | 14 | 3.27e-06 | 4.057e-05 |
| 376 | REGULATION OF MORPHOGENESIS OF A BRANCHING STRUCTURE | 6 | 53 | 3.342e-06 | 4.125e-05 |
| 377 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 6 | 53 | 3.342e-06 | 4.125e-05 |
| 378 | JNK CASCADE | 7 | 82 | 3.425e-06 | 4.216e-05 |
| 379 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 11 | 246 | 3.553e-06 | 4.362e-05 |
| 380 | BONE DEVELOPMENT | 9 | 156 | 3.612e-06 | 4.423e-05 |
| 381 | HOMEOSTASIS OF NUMBER OF CELLS WITHIN A TISSUE | 5 | 31 | 3.784e-06 | 4.609e-05 |
| 382 | REGULATION OF PLATELET ACTIVATION | 5 | 31 | 3.784e-06 | 4.609e-05 |
| 383 | UROGENITAL SYSTEM DEVELOPMENT | 12 | 299 | 3.871e-06 | 4.702e-05 |
| 384 | TISSUE MIGRATION | 7 | 84 | 4.028e-06 | 4.881e-05 |
| 385 | REGULATION OF CELL CYCLE | 22 | 949 | 4.175e-06 | 5.046e-05 |
| 386 | REGULATION OF INNATE IMMUNE RESPONSE | 13 | 357 | 4.385e-06 | 5.273e-05 |
| 387 | FOREBRAIN DEVELOPMENT | 13 | 357 | 4.385e-06 | 5.273e-05 |
| 388 | NEUROTROPHIN TRK RECEPTOR SIGNALING PATHWAY | 4 | 15 | 4.432e-06 | 5.315e-05 |
| 389 | ACTIVATION OF INNATE IMMUNE RESPONSE | 10 | 204 | 4.444e-06 | 5.316e-05 |
| 390 | EMBRYONIC MORPHOGENESIS | 16 | 539 | 4.684e-06 | 5.588e-05 |
| 391 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 7 | 86 | 4.718e-06 | 5.614e-05 |
| 392 | POSITIVE REGULATION OF DEFENSE RESPONSE | 13 | 364 | 5.412e-06 | 6.424e-05 |
| 393 | REGULATION OF CALCIUM ION TRANSPORT | 10 | 209 | 5.508e-06 | 6.505e-05 |
| 394 | REGULATION OF STEM CELL PROLIFERATION | 7 | 88 | 5.502e-06 | 6.505e-05 |
| 395 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 13 | 365 | 5.574e-06 | 6.566e-05 |
| 396 | EPITHELIAL CELL PROLIFERATION | 7 | 89 | 5.933e-06 | 6.954e-05 |
| 397 | IMMUNE EFFECTOR PROCESS | 15 | 486 | 5.928e-06 | 6.954e-05 |
| 398 | REGULATION OF MESENCHYMAL CELL PROLIFERATION | 5 | 34 | 6.082e-06 | 7.11e-05 |
| 399 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 6 | 59 | 6.313e-06 | 7.343e-05 |
| 400 | VASCULOGENESIS | 6 | 59 | 6.313e-06 | 7.343e-05 |
| 401 | REGULATION OF OXIDOREDUCTASE ACTIVITY | 7 | 90 | 6.392e-06 | 7.417e-05 |
| 402 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 13 | 370 | 6.455e-06 | 7.471e-05 |
| 403 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 6 | 60 | 6.97e-06 | 8.047e-05 |
| 404 | NEGATIVE REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 5 | 35 | 7.052e-06 | 8.121e-05 |
| 405 | RESPONSE TO ACID CHEMICAL | 12 | 319 | 7.478e-06 | 8.592e-05 |
| 406 | TISSUE HOMEOSTASIS | 9 | 171 | 7.637e-06 | 8.731e-05 |
| 407 | POSITIVE REGULATION OF GLYCOGEN METABOLIC PROCESS | 4 | 17 | 7.635e-06 | 8.731e-05 |
| 408 | POSITIVE REGULATION OF STEM CELL PROLIFERATION | 6 | 61 | 7.681e-06 | 8.76e-05 |
| 409 | RESPONSE TO STEROID HORMONE | 15 | 497 | 7.746e-06 | 8.813e-05 |
| 410 | RESPONSE TO ESTROGEN | 10 | 218 | 7.985e-06 | 9.062e-05 |
| 411 | LEARNING | 8 | 131 | 8.392e-06 | 9.501e-05 |
| 412 | POSITIVE REGULATION OF NUCLEAR DIVISION | 6 | 62 | 8.45e-06 | 9.543e-05 |
| 413 | B CELL ACTIVATION | 8 | 132 | 8.876e-06 | 1e-04 |
| 414 | GLIOGENESIS | 9 | 175 | 9.206e-06 | 0.0001033 |
| 415 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 11 | 272 | 9.234e-06 | 0.0001033 |
| 416 | CELLULAR MACROMOLECULE LOCALIZATION | 25 | 1234 | 9.24e-06 | 0.0001033 |
| 417 | MAST CELL MEDIATED IMMUNITY | 4 | 18 | 9.757e-06 | 0.0001089 |
| 418 | MYELOID LEUKOCYTE DIFFERENTIATION | 7 | 96 | 9.81e-06 | 0.0001092 |
| 419 | GLAND MORPHOGENESIS | 7 | 97 | 1.05e-05 | 0.0001167 |
| 420 | DEVELOPMENTAL GROWTH | 12 | 333 | 1.151e-05 | 0.0001276 |
| 421 | MYELOID LEUKOCYTE MIGRATION | 7 | 99 | 1.202e-05 | 0.0001328 |
| 422 | ACID SECRETION | 6 | 66 | 1.217e-05 | 0.0001342 |
| 423 | REGULATION OF CELL ADHESION MEDIATED BY INTEGRIN | 5 | 39 | 1.22e-05 | 0.0001342 |
| 424 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 10 | 229 | 1.227e-05 | 0.0001345 |
| 425 | POSITIVE REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 4 | 19 | 1.228e-05 | 0.0001345 |
| 426 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 5 | 40 | 1.385e-05 | 0.0001513 |
| 427 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT PROTEIN | 6 | 68 | 1.448e-05 | 0.0001578 |
| 428 | REGULATION OF SMOOTH MUSCLE CELL DIFFERENTIATION | 4 | 20 | 1.526e-05 | 0.0001652 |
| 429 | ICOSANOID TRANSPORT | 4 | 20 | 1.526e-05 | 0.0001652 |
| 430 | FATTY ACID DERIVATIVE TRANSPORT | 4 | 20 | 1.526e-05 | 0.0001652 |
| 431 | POSITIVE REGULATION OF KIDNEY DEVELOPMENT | 5 | 41 | 1.568e-05 | 0.0001689 |
| 432 | MYELOID CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 5 | 41 | 1.568e-05 | 0.0001689 |
| 433 | NEGATIVE REGULATION OF CELL COMMUNICATION | 24 | 1192 | 1.581e-05 | 0.0001699 |
| 434 | POSITIVE REGULATION OF ION TRANSPORT | 10 | 236 | 1.593e-05 | 0.0001708 |
| 435 | APOPTOTIC SIGNALING PATHWAY | 11 | 289 | 1.627e-05 | 0.0001741 |
| 436 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 7 | 104 | 1.66e-05 | 0.0001772 |
| 437 | MYELOID CELL DIFFERENTIATION | 9 | 189 | 1.705e-05 | 0.0001816 |
| 438 | REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 5 | 42 | 1.769e-05 | 0.0001877 |
| 439 | SENSORY ORGAN MORPHOGENESIS | 10 | 239 | 1.777e-05 | 0.0001877 |
| 440 | STEM CELL DIFFERENTIATION | 9 | 190 | 1.779e-05 | 0.0001877 |
| 441 | PHAGOCYTOSIS | 9 | 190 | 1.779e-05 | 0.0001877 |
| 442 | RESPONSE TO ESTRADIOL | 8 | 146 | 1.853e-05 | 0.0001951 |
| 443 | MAST CELL ACTIVATION | 4 | 21 | 1.874e-05 | 0.0001964 |
| 444 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 4 | 21 | 1.874e-05 | 0.0001964 |
| 445 | POSITIVE REGULATION OF CALCIUM ION TRANSPORT | 7 | 106 | 1.88e-05 | 0.0001966 |
| 446 | PROTEIN LOCALIZATION | 31 | 1805 | 1.904e-05 | 0.0001986 |
| 447 | MYELOID LEUKOCYTE MEDIATED IMMUNITY | 5 | 43 | 1.989e-05 | 0.0002071 |
| 448 | REGULATION OF RESPONSE TO WOUNDING | 13 | 413 | 2.072e-05 | 0.0002152 |
| 449 | CONNECTIVE TISSUE DEVELOPMENT | 9 | 194 | 2.098e-05 | 0.0002174 |
| 450 | ASSOCIATIVE LEARNING | 6 | 73 | 2.182e-05 | 0.0002246 |
| 451 | POSITIVE REGULATION OF STAT CASCADE | 6 | 73 | 2.182e-05 | 0.0002246 |
| 452 | POSITIVE REGULATION OF JAK STAT CASCADE | 6 | 73 | 2.182e-05 | 0.0002246 |
| 453 | LABYRINTHINE LAYER DEVELOPMENT | 5 | 44 | 2.23e-05 | 0.0002291 |
| 454 | RESPONSE TO INORGANIC SUBSTANCE | 14 | 479 | 2.251e-05 | 0.0002307 |
| 455 | SOMATIC STEM CELL DIVISION | 4 | 22 | 2.277e-05 | 0.0002328 |
| 456 | SUBSTANTIA NIGRA DEVELOPMENT | 5 | 45 | 2.493e-05 | 0.0002539 |
| 457 | EXOCRINE SYSTEM DEVELOPMENT | 5 | 45 | 2.493e-05 | 0.0002539 |
| 458 | ACTIVATION OF GTPASE ACTIVITY | 6 | 75 | 2.548e-05 | 0.0002583 |
| 459 | POSITIVE REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 6 | 75 | 2.548e-05 | 0.0002583 |
| 460 | PALLIUM DEVELOPMENT | 8 | 153 | 2.599e-05 | 0.0002629 |
| 461 | AMEBOIDAL TYPE CELL MIGRATION | 8 | 154 | 2.724e-05 | 0.0002749 |
| 462 | LYSOSOME LOCALIZATION | 4 | 23 | 2.74e-05 | 0.0002759 |
| 463 | REGULATION OF ION HOMEOSTASIS | 9 | 201 | 2.776e-05 | 0.0002789 |
| 464 | OVULATION CYCLE | 7 | 113 | 2.848e-05 | 0.0002855 |
| 465 | REGULATION OF CELL PROJECTION ASSEMBLY | 8 | 155 | 2.853e-05 | 0.0002855 |
| 466 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 8 | 156 | 2.988e-05 | 0.0002983 |
| 467 | POSITIVE REGULATION OF OXIDOREDUCTASE ACTIVITY | 5 | 47 | 3.092e-05 | 0.000308 |
| 468 | IN UTERO EMBRYONIC DEVELOPMENT | 11 | 311 | 3.198e-05 | 0.0003179 |
| 469 | EPITHELIAL CELL DIFFERENTIATION | 14 | 495 | 3.227e-05 | 0.0003201 |
| 470 | POSITIVE REGULATION OF CELL JUNCTION ASSEMBLY | 4 | 24 | 3.268e-05 | 0.0003235 |
| 471 | RESPONSE TO AMINE | 5 | 48 | 3.43e-05 | 0.0003381 |
| 472 | REGULATION OF JNK CASCADE | 8 | 159 | 3.424e-05 | 0.0003381 |
| 473 | LEUKOCYTE CHEMOTAXIS | 7 | 117 | 3.564e-05 | 0.0003499 |
| 474 | MAMMARY GLAND DEVELOPMENT | 7 | 117 | 3.564e-05 | 0.0003499 |
| 475 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 5 | 49 | 3.796e-05 | 0.0003718 |
| 476 | CELLULAR CHEMICAL HOMEOSTASIS | 15 | 570 | 3.832e-05 | 0.0003745 |
| 477 | REGULATION OF VOLTAGE GATED CALCIUM CHANNEL ACTIVITY | 4 | 25 | 3.867e-05 | 0.0003772 |
| 478 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 5 | 50 | 4.191e-05 | 0.000408 |
| 479 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 11 | 321 | 4.264e-05 | 0.0004142 |
| 480 | SEX DIFFERENTIATION | 10 | 266 | 4.424e-05 | 0.0004288 |
| 481 | DEVELOPMENTAL PROGRAMMED CELL DEATH | 4 | 26 | 4.542e-05 | 0.0004394 |
| 482 | REGULATION OF METAL ION TRANSPORT | 11 | 325 | 4.769e-05 | 0.0004603 |
| 483 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 9 | 216 | 4.87e-05 | 0.0004691 |
| 484 | EYE DEVELOPMENT | 11 | 326 | 4.903e-05 | 0.0004713 |
| 485 | POSITIVE REGULATION OF HEART GROWTH | 4 | 27 | 5.3e-05 | 0.0005064 |
| 486 | REGULATION OF PROTEIN SECRETION | 12 | 389 | 5.283e-05 | 0.0005064 |
| 487 | NEGATIVE REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 4 | 27 | 5.3e-05 | 0.0005064 |
| 488 | INFLAMMATORY RESPONSE | 13 | 454 | 5.499e-05 | 0.0005244 |
| 489 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 5 | 53 | 5.571e-05 | 0.0005301 |
| 490 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 10 | 274 | 5.673e-05 | 0.0005387 |
| 491 | REGULATION OF WOUND HEALING | 7 | 126 | 5.726e-05 | 0.0005426 |
| 492 | REGULATION OF ION TRANSPORT | 15 | 592 | 5.884e-05 | 0.0005564 |
| 493 | REGULATION OF CARBOHYDRATE BIOSYNTHETIC PROCESS | 6 | 87 | 5.92e-05 | 0.0005587 |
| 494 | POSITIVE REGULATION OF BINDING | 7 | 127 | 6.021e-05 | 0.000566 |
| 495 | NEGATIVE REGULATION OF TRANSPORT | 13 | 458 | 6.011e-05 | 0.000566 |
| 496 | POSITIVE REGULATION OF CALCIUM ION IMPORT | 5 | 54 | 6.102e-05 | 0.0005713 |
| 497 | B CELL RECEPTOR SIGNALING PATHWAY | 5 | 54 | 6.102e-05 | 0.0005713 |
| 498 | REGULATION OF SPROUTING ANGIOGENESIS | 4 | 28 | 6.146e-05 | 0.0005731 |
| 499 | MAMMARY GLAND DUCT MORPHOGENESIS | 4 | 28 | 6.146e-05 | 0.0005731 |
| 500 | NEGATIVE REGULATION OF CELL ADHESION | 9 | 223 | 6.231e-05 | 0.0005799 |
| 501 | OVULATION CYCLE PROCESS | 6 | 88 | 6.313e-05 | 0.0005851 |
| 502 | REGULATION OF CATION CHANNEL ACTIVITY | 6 | 88 | 6.313e-05 | 0.0005851 |
| 503 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 7 | 128 | 6.328e-05 | 0.0005854 |
| 504 | REGULATION OF KIDNEY DEVELOPMENT | 5 | 55 | 6.67e-05 | 0.0006146 |
| 505 | ACTIN CYTOSKELETON REORGANIZATION | 5 | 55 | 6.67e-05 | 0.0006146 |
| 506 | CELLULAR RESPONSE TO ACID CHEMICAL | 8 | 175 | 6.756e-05 | 0.0006212 |
| 507 | REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 4 | 29 | 7.087e-05 | 0.0006478 |
| 508 | STEM CELL DIVISION | 4 | 29 | 7.087e-05 | 0.0006478 |
| 509 | POSITIVE REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 4 | 29 | 7.087e-05 | 0.0006478 |
| 510 | CALCIUM MEDIATED SIGNALING | 6 | 90 | 7.161e-05 | 0.0006521 |
| 511 | ENDOTHELIUM DEVELOPMENT | 6 | 90 | 7.161e-05 | 0.0006521 |
| 512 | CHEMICAL HOMEOSTASIS WITHIN A TISSUE | 3 | 11 | 7.197e-05 | 0.0006541 |
| 513 | NEURON NEURON SYNAPTIC TRANSMISSION | 5 | 56 | 7.279e-05 | 0.0006602 |
| 514 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 9 | 228 | 7.389e-05 | 0.0006676 |
| 515 | TELENCEPHALON DEVELOPMENT | 9 | 228 | 7.389e-05 | 0.0006676 |
| 516 | REGULATION OF BINDING | 10 | 283 | 7.426e-05 | 0.0006696 |
| 517 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 7 | 132 | 7.689e-05 | 0.000692 |
| 518 | DIVALENT INORGANIC CATION HOMEOSTASIS | 11 | 343 | 7.73e-05 | 0.0006944 |
| 519 | ENDOTHELIAL CELL MIGRATION | 5 | 57 | 7.929e-05 | 0.0007109 |
| 520 | LEUKOCYTE DEGRANULATION | 4 | 30 | 8.128e-05 | 0.0007231 |
| 521 | RESPONSE TO AMPHETAMINE | 4 | 30 | 8.128e-05 | 0.0007231 |
| 522 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 4 | 30 | 8.128e-05 | 0.0007231 |
| 523 | OSTEOCLAST DIFFERENTIATION | 4 | 30 | 8.128e-05 | 0.0007231 |
| 524 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 9 | 232 | 8.441e-05 | 0.0007496 |
| 525 | NEUROLOGICAL SYSTEM PROCESS | 23 | 1242 | 8.771e-05 | 0.0007774 |
| 526 | POSITIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 7 | 135 | 8.859e-05 | 0.0007822 |
| 527 | CELL GROWTH | 7 | 135 | 8.859e-05 | 0.0007822 |
| 528 | REGULATION OF DNA BIOSYNTHETIC PROCESS | 6 | 94 | 9.13e-05 | 0.0008046 |
| 529 | POSITIVE REGULATION OF DNA BIOSYNTHETIC PROCESS | 5 | 59 | 9.363e-05 | 0.0008236 |
| 530 | REGULATION OF RAC PROTEIN SIGNAL TRANSDUCTION | 3 | 12 | 9.542e-05 | 0.0008361 |
| 531 | ACTIVATION OF TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 3 | 12 | 9.542e-05 | 0.0008361 |
| 532 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 25 | 1423 | 9.724e-05 | 0.0008489 |
| 533 | RESPONSE TO ALKALOID | 7 | 137 | 9.717e-05 | 0.0008489 |
| 534 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 1004 | 0.0001002 | 0.0008727 |
| 535 | REGULATION OF MONOOXYGENASE ACTIVITY | 5 | 60 | 0.0001015 | 0.0008829 |
| 536 | ICOSANOID METABOLIC PROCESS | 6 | 96 | 0.0001026 | 0.0008893 |
| 537 | FATTY ACID DERIVATIVE METABOLIC PROCESS | 6 | 96 | 0.0001026 | 0.0008893 |
| 538 | EPITHELIAL CELL DEVELOPMENT | 8 | 186 | 0.0001034 | 0.0008928 |
| 539 | REGULATION OF EXOCYTOSIS | 8 | 186 | 0.0001034 | 0.0008928 |
| 540 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 14 | 552 | 0.0001036 | 0.0008929 |
| 541 | SALIVARY GLAND DEVELOPMENT | 4 | 32 | 0.0001054 | 0.0009063 |
| 542 | POSITIVE REGULATION OF CYCLASE ACTIVITY | 5 | 61 | 0.0001099 | 0.0009434 |
| 543 | NON CANONICAL WNT SIGNALING PATHWAY | 7 | 140 | 0.0001113 | 0.0009536 |
| 544 | MYELOID LEUKOCYTE ACTIVATION | 6 | 98 | 0.0001151 | 0.0009842 |
| 545 | MESENCHYME DEVELOPMENT | 8 | 190 | 0.0001199 | 0.001023 |
| 546 | INTRACELLULAR PROTEIN TRANSPORT | 17 | 781 | 0.0001212 | 0.001033 |
| 547 | POSITIVE REGULATION OF CELL SUBSTRATE ADHESION | 6 | 99 | 0.0001217 | 0.001035 |
| 548 | NEURONAL STEM CELL DIVISION | 3 | 13 | 0.0001233 | 0.001043 |
| 549 | REGULATION OF CELL PROLIFERATION INVOLVED IN KIDNEY DEVELOPMENT | 3 | 13 | 0.0001233 | 0.001043 |
| 550 | NEUROBLAST DIVISION | 3 | 13 | 0.0001233 | 0.001043 |
| 551 | REGULATION OF TRANSMEMBRANE TRANSPORT | 12 | 426 | 0.0001249 | 0.001055 |
| 552 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 6 | 100 | 0.0001287 | 0.001085 |
| 553 | FOREBRAIN NEURON DEVELOPMENT | 4 | 34 | 0.0001343 | 0.001126 |
| 554 | NEGATIVE REGULATION OF CATION CHANNEL ACTIVITY | 4 | 34 | 0.0001343 | 0.001126 |
| 555 | PROTEIN KINASE B SIGNALING | 4 | 34 | 0.0001343 | 0.001126 |
| 556 | NEGATIVE REGULATION OF TRANSPORTER ACTIVITY | 5 | 64 | 0.0001382 | 0.001157 |
| 557 | RESPONSE TO CYTOKINE | 16 | 714 | 0.0001393 | 0.001163 |
| 558 | MUSCLE STRUCTURE DEVELOPMENT | 12 | 432 | 0.0001423 | 0.001186 |
| 559 | EAR DEVELOPMENT | 8 | 195 | 0.0001434 | 0.001193 |
| 560 | REGULATION OF AUTOPHAGY | 9 | 249 | 0.0001443 | 0.001199 |
| 561 | REGULATION OF GLYCOGEN METABOLIC PROCESS | 4 | 35 | 0.0001507 | 0.001248 |
| 562 | POSITIVE REGULATION OF SECRETION | 11 | 370 | 0.0001506 | 0.001248 |
| 563 | REGULATION OF RESPIRATORY BURST | 3 | 14 | 0.0001561 | 0.001279 |
| 564 | POSITIVE REGULATION OF METANEPHROS DEVELOPMENT | 3 | 14 | 0.0001561 | 0.001279 |
| 565 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 3 | 14 | 0.0001561 | 0.001279 |
| 566 | REGULATION OF GLOMERULUS DEVELOPMENT | 3 | 14 | 0.0001561 | 0.001279 |
| 567 | REGULATION OF PROTEIN KINASE C SIGNALING | 3 | 14 | 0.0001561 | 0.001279 |
| 568 | BONE MATURATION | 3 | 14 | 0.0001561 | 0.001279 |
| 569 | DIGESTIVE SYSTEM DEVELOPMENT | 7 | 148 | 0.0001573 | 0.001286 |
| 570 | FOREBRAIN GENERATION OF NEURONS | 5 | 66 | 0.00016 | 0.001299 |
| 571 | REGULATION OF MULTICELLULAR ORGANISM GROWTH | 5 | 66 | 0.00016 | 0.001299 |
| 572 | REGULATION OF TRANSPORTER ACTIVITY | 8 | 198 | 0.0001592 | 0.001299 |
| 573 | NEURAL NUCLEUS DEVELOPMENT | 5 | 66 | 0.00016 | 0.001299 |
| 574 | REGULATION OF SYSTEM PROCESS | 13 | 507 | 0.0001656 | 0.001342 |
| 575 | POSITIVE REGULATION OF GLUCOSE METABOLIC PROCESS | 4 | 36 | 0.0001685 | 0.001364 |
| 576 | POSITIVE REGULATION OF NEURON DEATH | 5 | 67 | 0.0001719 | 0.001388 |
| 577 | REGULATION OF HEMOPOIESIS | 10 | 314 | 0.000174 | 0.001403 |
| 578 | RESPONSE TO NERVE GROWTH FACTOR | 4 | 37 | 0.0001878 | 0.001507 |
| 579 | POSITIVE REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 4 | 37 | 0.0001878 | 0.001507 |
| 580 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 4 | 37 | 0.0001878 | 0.001507 |
| 581 | REGULATION OF CYTOSOLIC CALCIUM ION CONCENTRATION | 8 | 203 | 0.0001889 | 0.001512 |
| 582 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 13 | 514 | 0.0001893 | 0.001514 |
| 583 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 3 | 15 | 0.000194 | 0.001538 |
| 584 | POSITIVE REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 3 | 15 | 0.000194 | 0.001538 |
| 585 | CHEMOSENSORY BEHAVIOR | 3 | 15 | 0.000194 | 0.001538 |
| 586 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 153 | 0.0001931 | 0.001538 |
| 587 | REGULATION OF HYDROGEN PEROXIDE METABOLIC PROCESS | 3 | 15 | 0.000194 | 0.001538 |
| 588 | REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 6 | 108 | 0.0001963 | 0.001554 |
| 589 | RESPONSE TO ACTIVITY | 5 | 69 | 0.0001975 | 0.00156 |
| 590 | POSITIVE REGULATION OF LEUKOCYTE MIGRATION | 6 | 109 | 0.0002064 | 0.001625 |
| 591 | REGULATION OF RHO PROTEIN SIGNAL TRANSDUCTION | 6 | 109 | 0.0002064 | 0.001625 |
| 592 | REGULATION OF LEUKOCYTE PROLIFERATION | 8 | 206 | 0.0002087 | 0.001629 |
| 593 | ESTABLISHMENT OF LOCALIZATION IN CELL | 27 | 1676 | 0.0002084 | 0.001629 |
| 594 | POSITIVE REGULATION OF CALCIUM MEDIATED SIGNALING | 4 | 38 | 0.0002086 | 0.001629 |
| 595 | SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 4 | 38 | 0.0002086 | 0.001629 |
| 596 | POSITIVE REGULATION OF ORGAN GROWTH | 4 | 38 | 0.0002086 | 0.001629 |
| 597 | CENTRAL NERVOUS SYSTEM NEURON DEVELOPMENT | 5 | 70 | 0.0002114 | 0.001645 |
| 598 | REGULATION OF CALCIUM ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 5 | 70 | 0.0002114 | 0.001645 |
| 599 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION | 6 | 110 | 0.000217 | 0.001684 |
| 600 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 9 | 263 | 0.0002171 | 0.001684 |
| 601 | CELL PROJECTION ASSEMBLY | 9 | 264 | 0.0002233 | 0.001729 |
| 602 | SKIN EPIDERMIS DEVELOPMENT | 5 | 71 | 0.000226 | 0.001747 |
| 603 | WNT SIGNALING PATHWAY CALCIUM MODULATING PATHWAY | 4 | 39 | 0.0002311 | 0.00178 |
| 604 | ANATOMICAL STRUCTURE MATURATION | 4 | 39 | 0.0002311 | 0.00178 |
| 605 | POSITIVE REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 3 | 16 | 0.0002374 | 0.00182 |
| 606 | POSITIVE REGULATION OF PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 3 | 16 | 0.0002374 | 0.00182 |
| 607 | BRANCHING INVOLVED IN SALIVARY GLAND MORPHOGENESIS | 3 | 16 | 0.0002374 | 0.00182 |
| 608 | RESPONSE TO BACTERIUM | 13 | 528 | 0.0002457 | 0.001877 |
| 609 | SKIN DEVELOPMENT | 8 | 211 | 0.0002456 | 0.001877 |
| 610 | CELLULAR HOMEOSTASIS | 15 | 676 | 0.000252 | 0.001922 |
| 611 | SECOND MESSENGER MEDIATED SIGNALING | 7 | 160 | 0.000254 | 0.001935 |
| 612 | MAMMARY GLAND MORPHOGENESIS | 4 | 40 | 0.0002552 | 0.00194 |
| 613 | REGULATION OF ORGAN GROWTH | 5 | 73 | 0.0002575 | 0.001955 |
| 614 | POSITIVE REGULATION OF HEMOPOIESIS | 7 | 163 | 0.0002845 | 0.002156 |
| 615 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 14 | 609 | 0.0002854 | 0.002159 |
| 616 | NEGATIVE REGULATION OF PLATELET ACTIVATION | 3 | 17 | 0.0002866 | 0.002165 |
| 617 | POSITIVE REGULATION OF HOMEOSTATIC PROCESS | 8 | 216 | 0.0002876 | 0.002169 |
| 618 | REGULATION OF CALCIUM ION TRANSMEMBRANE TRANSPORT | 6 | 116 | 0.0002894 | 0.002175 |
| 619 | FEMALE SEX DIFFERENTIATION | 6 | 116 | 0.0002894 | 0.002175 |
| 620 | SENSORY PERCEPTION OF PAIN | 5 | 75 | 0.0002922 | 0.002193 |
| 621 | REGULATION OF MITOTIC CELL CYCLE | 12 | 468 | 0.0002965 | 0.002222 |
| 622 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 6 | 117 | 0.0003031 | 0.002267 |
| 623 | REGULATION OF CELLULAR COMPONENT SIZE | 10 | 337 | 0.0003064 | 0.002288 |
| 624 | MITOTIC CELL CYCLE | 16 | 766 | 0.0003071 | 0.00229 |
| 625 | REGULATION OF HEART GROWTH | 4 | 42 | 0.0003088 | 0.002296 |
| 626 | LONG CHAIN FATTY ACID TRANSPORT | 4 | 42 | 0.0003088 | 0.002296 |
| 627 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 13 | 541 | 0.0003104 | 0.0023 |
| 628 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 13 | 541 | 0.0003104 | 0.0023 |
| 629 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 12 | 472 | 0.0003202 | 0.002369 |
| 630 | CELLULAR MODIFIED AMINO ACID METABOLIC PROCESS | 8 | 220 | 0.0003253 | 0.002403 |
| 631 | IMMUNE RESPONSE | 20 | 1100 | 0.0003346 | 0.002467 |
| 632 | CEREBRAL CORTEX CELL MIGRATION | 4 | 43 | 0.0003385 | 0.002492 |
| 633 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 3 | 18 | 0.000342 | 0.002502 |
| 634 | REGULATION OF COLLATERAL SPROUTING | 3 | 18 | 0.000342 | 0.002502 |
| 635 | REGULATION OF PROTEIN BINDING | 7 | 168 | 0.0003417 | 0.002502 |
| 636 | ORGAN MATURATION | 3 | 18 | 0.000342 | 0.002502 |
| 637 | REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 5 | 78 | 0.0003509 | 0.002563 |
| 638 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 9 | 282 | 0.0003626 | 0.00264 |
| 639 | MUSCLE SYSTEM PROCESS | 9 | 282 | 0.0003626 | 0.00264 |
| 640 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 4 | 44 | 0.0003701 | 0.002691 |
| 641 | DENDRITE DEVELOPMENT | 5 | 79 | 0.0003723 | 0.002698 |
| 642 | REGULATION OF SYNAPSE ASSEMBLY | 5 | 79 | 0.0003723 | 0.002698 |
| 643 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 7 | 171 | 0.0003803 | 0.002752 |
| 644 | ANATOMICAL STRUCTURE HOMEOSTASIS | 9 | 285 | 0.0003916 | 0.00283 |
| 645 | LEUKOCYTE ACTIVATION | 11 | 414 | 0.0003941 | 0.002843 |
| 646 | DENDRITIC SPINE DEVELOPMENT | 3 | 19 | 0.0004038 | 0.002895 |
| 647 | LUNG MORPHOGENESIS | 4 | 45 | 0.0004038 | 0.002895 |
| 648 | POSITIVE REGULATION OF RECEPTOR ACTIVITY | 4 | 45 | 0.0004038 | 0.002895 |
| 649 | MACROPHAGE DIFFERENTIATION | 3 | 19 | 0.0004038 | 0.002895 |
| 650 | REGULATION OF CELL CYCLE PROCESS | 13 | 558 | 0.0004164 | 0.002981 |
| 651 | POSITIVE REGULATION OF MYELOID CELL DIFFERENTIATION | 5 | 81 | 0.000418 | 0.002988 |
| 652 | RESPONSE TO OXIDATIVE STRESS | 10 | 352 | 0.0004316 | 0.00308 |
| 653 | POSITIVE REGULATION OF CYTOSKELETON ORGANIZATION | 7 | 175 | 0.000437 | 0.003114 |
| 654 | ICOSANOID BIOSYNTHETIC PROCESS | 4 | 46 | 0.0004396 | 0.003118 |
| 655 | PEPTIDYL THREONINE MODIFICATION | 4 | 46 | 0.0004396 | 0.003118 |
| 656 | FATTY ACID DERIVATIVE BIOSYNTHETIC PROCESS | 4 | 46 | 0.0004396 | 0.003118 |
| 657 | RESPONSE TO CORTICOSTEROID | 7 | 176 | 0.0004522 | 0.003203 |
| 658 | SINGLE ORGANISM CATABOLIC PROCESS | 18 | 957 | 0.000454 | 0.00321 |
| 659 | ORGAN REGENERATION | 5 | 83 | 0.0004678 | 0.003288 |
| 660 | REGULATION OF ERBB SIGNALING PATHWAY | 5 | 83 | 0.0004678 | 0.003288 |
| 661 | HAIR CYCLE | 5 | 83 | 0.0004678 | 0.003288 |
| 662 | MOLTING CYCLE | 5 | 83 | 0.0004678 | 0.003288 |
| 663 | REGULATION OF G PROTEIN COUPLED RECEPTOR PROTEIN SIGNALING PATHWAY | 6 | 127 | 0.0004702 | 0.0033 |
| 664 | MICROVILLUS ORGANIZATION | 3 | 20 | 0.0004724 | 0.003311 |
| 665 | SYNAPTIC SIGNALING | 11 | 424 | 0.0004813 | 0.003368 |
| 666 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 4 | 48 | 0.0005179 | 0.003607 |
| 667 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 4 | 48 | 0.0005179 | 0.003607 |
| 668 | POSITIVE REGULATION OF ADENYLATE CYCLASE ACTIVITY | 4 | 48 | 0.0005179 | 0.003607 |
| 669 | RESPONSE TO BIOTIC STIMULUS | 17 | 886 | 0.000526 | 0.003658 |
| 670 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 10 | 363 | 0.0005484 | 0.003809 |
| 671 | POSITIVE REGULATION OF TRANSMEMBRANE TRANSPORT | 6 | 131 | 0.0005543 | 0.003838 |
| 672 | POSITIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 6 | 131 | 0.0005543 | 0.003838 |
| 673 | REGULATION OF NITRIC OXIDE SYNTHASE ACTIVITY | 4 | 49 | 0.0005606 | 0.003876 |
| 674 | REGULATION OF MYELOID CELL DIFFERENTIATION | 7 | 183 | 0.0005707 | 0.00394 |
| 675 | REGULATION OF CATABOLIC PROCESS | 15 | 731 | 0.0005722 | 0.003944 |
| 676 | NEGATIVE REGULATION OF TRANSMEMBRANE TRANSPORT | 5 | 87 | 0.0005808 | 0.003998 |
| 677 | RESPONSE TO TOXIC SUBSTANCE | 8 | 241 | 0.0005955 | 0.004093 |
| 678 | POSITIVE REGULATION OF PURINE NUCLEOTIDE METABOLIC PROCESS | 6 | 133 | 0.0006004 | 0.004115 |
| 679 | POSITIVE REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 6 | 133 | 0.0006004 | 0.004115 |
| 680 | REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 4 | 50 | 0.0006057 | 0.004132 |
| 681 | FACE DEVELOPMENT | 4 | 50 | 0.0006057 | 0.004132 |
| 682 | POSITIVE REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 4 | 50 | 0.0006057 | 0.004132 |
| 683 | REGULATION OF COAGULATION | 5 | 88 | 0.000612 | 0.004163 |
| 684 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 5 | 88 | 0.000612 | 0.004163 |
| 685 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 11 | 437 | 0.0006183 | 0.0042 |
| 686 | MESENCHYMAL CELL DIFFERENTIATION | 6 | 134 | 0.0006246 | 0.004237 |
| 687 | ENDOCYTOSIS | 12 | 509 | 0.0006266 | 0.004244 |
| 688 | ACTIVATION OF PROTEIN KINASE B ACTIVITY | 3 | 22 | 0.000631 | 0.004249 |
| 689 | SYNAPTIC TRANSMISSION GLUTAMATERGIC | 3 | 22 | 0.000631 | 0.004249 |
| 690 | POSITIVE REGULATION OF NITRIC OXIDE SYNTHASE ACTIVITY | 3 | 22 | 0.000631 | 0.004249 |
| 691 | ERK1 AND ERK2 CASCADE | 3 | 22 | 0.000631 | 0.004249 |
| 692 | B CELL DIFFERENTIATION | 5 | 89 | 0.0006444 | 0.004327 |
| 693 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 5 | 89 | 0.0006444 | 0.004327 |
| 694 | REGULATION OF NEUROTRANSMITTER SECRETION | 4 | 51 | 0.0006533 | 0.004367 |
| 695 | RESPONSE TO NICOTINE | 4 | 51 | 0.0006533 | 0.004367 |
| 696 | MECHANORECEPTOR DIFFERENTIATION | 4 | 51 | 0.0006533 | 0.004367 |
| 697 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 8 | 247 | 0.0006994 | 0.004669 |
| 698 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 19 | 1079 | 0.0007006 | 0.00467 |
| 699 | POSITIVE REGULATION OF CALCIUM ION TRANSPORT INTO CYTOSOL | 4 | 52 | 0.0007035 | 0.004676 |
| 700 | ACTIVATION OF MAPKK ACTIVITY | 4 | 52 | 0.0007035 | 0.004676 |
| 701 | REGULATION OF OSTEOBLAST PROLIFERATION | 3 | 23 | 0.0007215 | 0.004782 |
| 702 | REGULATION OF METANEPHROS DEVELOPMENT | 3 | 23 | 0.0007215 | 0.004782 |
| 703 | REGULATION OF NITRIC OXIDE BIOSYNTHETIC PROCESS | 4 | 53 | 0.0007564 | 0.004997 |
| 704 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 6 | 139 | 0.0007571 | 0.004997 |
| 705 | CELLULAR RESPONSE TO AMINO ACID STIMULUS | 4 | 53 | 0.0007564 | 0.004997 |
| 706 | NEGATIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 3 | 24 | 0.0008199 | 0.005389 |
| 707 | BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 3 | 24 | 0.0008199 | 0.005389 |
| 708 | POSITIVE REGULATION OF RECEPTOR INTERNALIZATION | 3 | 24 | 0.0008199 | 0.005389 |
| 709 | REGULATION OF LIPID TRANSPORT | 5 | 95 | 0.0008667 | 0.005688 |
| 710 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 4 | 55 | 0.0008704 | 0.005705 |
| 711 | CELLULAR RESPONSE TO LIPID | 11 | 457 | 0.0008916 | 0.005835 |
| 712 | REGULATION OF JAK STAT CASCADE | 6 | 144 | 0.0009103 | 0.00594 |
| 713 | REGULATION OF STAT CASCADE | 6 | 144 | 0.0009103 | 0.00594 |
| 714 | SINGLE ORGANISM CELL ADHESION | 11 | 459 | 0.0009238 | 0.00602 |
| 715 | DETECTION OF MECHANICAL STIMULUS INVOLVED IN SENSORY PERCEPTION | 3 | 25 | 0.0009264 | 0.006021 |
| 716 | POSITIVE REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 25 | 0.0009264 | 0.006021 |
| 717 | INOSITOL PHOSPHATE METABOLIC PROCESS | 4 | 56 | 0.0009318 | 0.00603 |
| 718 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 4 | 56 | 0.0009318 | 0.00603 |
| 719 | FATTY ACID TRANSPORT | 4 | 56 | 0.0009318 | 0.00603 |
| 720 | NEUROMUSCULAR PROCESS | 5 | 97 | 0.0009522 | 0.006153 |
| 721 | REGULATION OF CELL GROWTH | 10 | 391 | 0.0009681 | 0.006248 |
| 722 | REGULATION OF T CELL PROLIFERATION | 6 | 147 | 0.001013 | 0.006529 |
| 723 | REPLACEMENT OSSIFICATION | 3 | 26 | 0.001041 | 0.006665 |
| 724 | NEGATIVE REGULATION OF LIPID TRANSPORT | 3 | 26 | 0.001041 | 0.006665 |
| 725 | ENDOCHONDRAL OSSIFICATION | 3 | 26 | 0.001041 | 0.006665 |
| 726 | REGULATION OF P38MAPK CASCADE | 3 | 26 | 0.001041 | 0.006665 |
| 727 | REGULATION OF CELL FATE COMMITMENT | 3 | 26 | 0.001041 | 0.006665 |
| 728 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 99 | 0.001044 | 0.006671 |
| 729 | NEGATIVE REGULATION OF WOUND HEALING | 4 | 58 | 0.001064 | 0.006788 |
| 730 | REGULATION OF LEUKOCYTE MIGRATION | 6 | 149 | 0.001086 | 0.006924 |
| 731 | LIMBIC SYSTEM DEVELOPMENT | 5 | 100 | 0.001092 | 0.00695 |
| 732 | POSITIVE REGULATION OF CALCIUM ION TRANSMEMBRANE TRANSPORT | 4 | 59 | 0.001134 | 0.007209 |
| 733 | REGULATION OF EXTENT OF CELL GROWTH | 5 | 101 | 0.001142 | 0.007247 |
| 734 | REGULATION OF LAMELLIPODIUM ASSEMBLY | 3 | 27 | 0.001165 | 0.007375 |
| 735 | REGULATION OF FATTY ACID TRANSPORT | 3 | 27 | 0.001165 | 0.007375 |
| 736 | STEM CELL PROLIFERATION | 4 | 60 | 0.001208 | 0.007619 |
| 737 | CHONDROCYTE DIFFERENTIATION | 4 | 60 | 0.001208 | 0.007619 |
| 738 | REGULATION OF CATION TRANSMEMBRANE TRANSPORT | 7 | 208 | 0.001208 | 0.007619 |
| 739 | LYMPHOCYTE DIFFERENTIATION | 7 | 209 | 0.001242 | 0.007823 |
| 740 | REGULATION OF CALCIUM ION IMPORT | 5 | 103 | 0.001246 | 0.007826 |
| 741 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 26 | 1784 | 0.001245 | 0.007826 |
| 742 | REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 4 | 61 | 0.001285 | 0.008003 |
| 743 | POSITIVE REGULATION OF SYNAPSE ASSEMBLY | 4 | 61 | 0.001285 | 0.008003 |
| 744 | POSITIVE REGULATION OF LYASE ACTIVITY | 4 | 61 | 0.001285 | 0.008003 |
| 745 | NEGATIVE REGULATION OF CATION TRANSMEMBRANE TRANSPORT | 4 | 61 | 0.001285 | 0.008003 |
| 746 | OVARIAN FOLLICLE DEVELOPMENT | 4 | 61 | 0.001285 | 0.008003 |
| 747 | CELL CYCLE | 21 | 1316 | 0.001279 | 0.008003 |
| 748 | REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 3 | 28 | 0.001297 | 0.008049 |
| 749 | LONG TERM MEMORY | 3 | 28 | 0.001297 | 0.008049 |
| 750 | POSITIVE REGULATION OF PHOSPHATASE ACTIVITY | 3 | 28 | 0.001297 | 0.008049 |
| 751 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 7 | 211 | 0.001312 | 0.008131 |
| 752 | DEFENSE RESPONSE | 20 | 1231 | 0.001355 | 0.008382 |
| 753 | FOREBRAIN CELL MIGRATION | 4 | 62 | 0.001365 | 0.008437 |
| 754 | MULTI MULTICELLULAR ORGANISM PROCESS | 7 | 213 | 0.001386 | 0.008539 |
| 755 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 7 | 213 | 0.001386 | 0.008539 |
| 756 | POSITIVE REGULATION OF MONOOXYGENASE ACTIVITY | 3 | 29 | 0.001439 | 0.008832 |
| 757 | NEGATIVE REGULATION OF CALCIUM ION TRANSMEMBRANE TRANSPORT | 3 | 29 | 0.001439 | 0.008832 |
| 758 | NEUROBLAST PROLIFERATION | 3 | 29 | 0.001439 | 0.008832 |
| 759 | REGULATION OF OSTEOCLAST DIFFERENTIATION | 4 | 63 | 0.001449 | 0.008884 |
| 760 | REGULATION OF SEQUESTERING OF CALCIUM ION | 5 | 107 | 0.001477 | 0.00904 |
| 761 | EMBRYONIC ORGAN MORPHOGENESIS | 8 | 279 | 0.001527 | 0.009335 |
| 762 | REGULATION OF NEUROTRANSMITTER TRANSPORT | 4 | 64 | 0.001537 | 0.009383 |
| 763 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 7 | 218 | 0.001582 | 0.009649 |
| 764 | NEGATIVE REGULATION OF CELL MATRIX ADHESION | 3 | 30 | 0.00159 | 0.009656 |
| 765 | REGULATION OF DENDRITIC SPINE MORPHOGENESIS | 3 | 30 | 0.00159 | 0.009656 |
| 766 | REGULATION OF VASCULAR PERMEABILITY | 3 | 30 | 0.00159 | 0.009656 |
| 767 | UNSATURATED FATTY ACID METABOLIC PROCESS | 5 | 109 | 0.001603 | 0.00971 |
| 768 | RESPONSE TO HYDROGEN PEROXIDE | 5 | 109 | 0.001603 | 0.00971 |
| 769 | REGENERATION | 6 | 161 | 0.001615 | 0.009774 |
| 770 | REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 4 | 65 | 0.001628 | 0.009822 |
| 771 | NEGATIVE REGULATION OF AXONOGENESIS | 4 | 65 | 0.001628 | 0.009822 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | KINASE ACTIVITY | 62 | 842 | 1.893e-44 | 1.758e-41 |
| 2 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 62 | 992 | 3.414e-40 | 1.586e-37 |
| 3 | PROTEIN KINASE ACTIVITY | 50 | 640 | 1.776e-36 | 5.499e-34 |
| 4 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 35 | 228 | 1.434e-35 | 3.331e-33 |
| 5 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 37 | 303 | 7.842e-34 | 1.388e-31 |
| 6 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 24 | 70 | 8.962e-34 | 1.388e-31 |
| 7 | PROTEIN TYROSINE KINASE ACTIVITY | 30 | 176 | 6.315e-32 | 8.381e-30 |
| 8 | MOLECULAR FUNCTION REGULATOR | 61 | 1353 | 2.162e-31 | 2.511e-29 |
| 9 | RECEPTOR BINDING | 59 | 1476 | 1.82e-27 | 1.879e-25 |
| 10 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 20 | 64 | 2.53e-27 | 2.351e-25 |
| 11 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 17 | 43 | 1.889e-25 | 1.596e-23 |
| 12 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 20 | 81 | 5.424e-25 | 4.199e-23 |
| 13 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 17 | 51 | 6.288e-24 | 4.432e-22 |
| 14 | RIBONUCLEOTIDE BINDING | 61 | 1860 | 6.679e-24 | 4.432e-22 |
| 15 | GROWTH FACTOR ACTIVITY | 23 | 160 | 7.861e-23 | 4.869e-21 |
| 16 | SIGNAL TRANSDUCER ACTIVITY | 56 | 1731 | 1.915e-21 | 1.112e-19 |
| 17 | ENZYME BINDING | 52 | 1737 | 2.602e-18 | 1.422e-16 |
| 18 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 19 | 172 | 8.143e-17 | 3.981e-15 |
| 19 | GROWTH FACTOR BINDING | 17 | 123 | 7.91e-17 | 3.981e-15 |
| 20 | GROWTH FACTOR RECEPTOR BINDING | 17 | 129 | 1.808e-16 | 8.398e-15 |
| 21 | PHOSPHOLIPASE A2 ACTIVITY | 11 | 31 | 3.133e-16 | 1.386e-14 |
| 22 | ADENYL NUCLEOTIDE BINDING | 45 | 1514 | 1.329e-15 | 5.613e-14 |
| 23 | KINASE BINDING | 28 | 606 | 2.325e-14 | 9.392e-13 |
| 24 | GTPASE ACTIVITY | 19 | 246 | 6.197e-14 | 2.399e-12 |
| 25 | PHOSPHOLIPASE ACTIVITY | 13 | 94 | 3.836e-13 | 1.425e-11 |
| 26 | INSULIN RECEPTOR SUBSTRATE BINDING | 7 | 11 | 4.711e-13 | 1.683e-11 |
| 27 | NEUROTROPHIN RECEPTOR BINDING | 7 | 14 | 4.805e-12 | 1.653e-10 |
| 28 | RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY | 12 | 92 | 6.499e-12 | 2.156e-10 |
| 29 | LIPASE ACTIVITY | 13 | 117 | 6.769e-12 | 2.168e-10 |
| 30 | PLATELET DERIVED GROWTH FACTOR RECEPTOR BINDING | 7 | 15 | 8.951e-12 | 2.772e-10 |
| 31 | FIBROBLAST GROWTH FACTOR RECEPTOR BINDING | 8 | 28 | 2.957e-11 | 8.861e-10 |
| 32 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 21 | 445 | 3.729e-11 | 1.083e-09 |
| 33 | ENZYME ACTIVATOR ACTIVITY | 21 | 471 | 1.066e-10 | 3e-09 |
| 34 | PROTEIN TYROSINE KINASE BINDING | 9 | 54 | 3.153e-10 | 8.614e-09 |
| 35 | KINASE ACTIVATOR ACTIVITY | 9 | 62 | 1.141e-09 | 3.028e-08 |
| 36 | ENZYME REGULATOR ACTIVITY | 27 | 959 | 5.368e-09 | 1.385e-07 |
| 37 | CARBOXYLIC ESTER HYDROLASE ACTIVITY | 11 | 135 | 8.315e-09 | 2.088e-07 |
| 38 | GUANYL NUCLEOTIDE BINDING | 17 | 390 | 1.01e-08 | 2.468e-07 |
| 39 | PHOSPHATIDYLCHOLINE 1 ACYLHYDROLASE ACTIVITY | 5 | 11 | 1.166e-08 | 2.778e-07 |
| 40 | KINASE REGULATOR ACTIVITY | 12 | 186 | 2.393e-08 | 5.557e-07 |
| 41 | CHEMOATTRACTANT ACTIVITY | 6 | 27 | 5.088e-08 | 1.153e-06 |
| 42 | PHOSPHATIDYLINOSITOL 3 KINASE BINDING | 6 | 30 | 1.001e-07 | 2.214e-06 |
| 43 | GDP BINDING | 7 | 51 | 1.275e-07 | 2.754e-06 |
| 44 | GTP DEPENDENT PROTEIN BINDING | 5 | 17 | 1.504e-07 | 3.176e-06 |
| 45 | IDENTICAL PROTEIN BINDING | 28 | 1209 | 1.711e-07 | 3.532e-06 |
| 46 | PROTEIN COMPLEX BINDING | 24 | 935 | 2.347e-07 | 4.74e-06 |
| 47 | PROTEIN SERINE THREONINE KINASE ACTIVATOR ACTIVITY | 5 | 19 | 2.792e-07 | 5.518e-06 |
| 48 | LYSOPHOSPHOLIPASE ACTIVITY | 5 | 20 | 3.699e-07 | 7.159e-06 |
| 49 | HEPARIN BINDING | 10 | 157 | 4.125e-07 | 7.821e-06 |
| 50 | DIACYLGLYCEROL BINDING | 4 | 11 | 1.098e-06 | 2e-05 |
| 51 | PLATELET DERIVED GROWTH FACTOR BINDING | 4 | 11 | 1.098e-06 | 2e-05 |
| 52 | SULFUR COMPOUND BINDING | 11 | 234 | 2.193e-06 | 3.918e-05 |
| 53 | MAP KINASE ACTIVITY | 4 | 14 | 3.27e-06 | 5.732e-05 |
| 54 | PROTEIN PHOSPHATASE BINDING | 8 | 120 | 4.38e-06 | 7.394e-05 |
| 55 | INSULIN RECEPTOR BINDING | 5 | 32 | 4.457e-06 | 7.394e-05 |
| 56 | INSULIN LIKE GROWTH FACTOR RECEPTOR BINDING | 4 | 15 | 4.432e-06 | 7.394e-05 |
| 57 | GLYCOSAMINOGLYCAN BINDING | 10 | 205 | 4.641e-06 | 7.564e-05 |
| 58 | PHOSPHATASE BINDING | 9 | 162 | 4.921e-06 | 7.881e-05 |
| 59 | HYDROLASE ACTIVITY ACTING ON ACID ANHYDRIDES | 20 | 820 | 5.654e-06 | 8.902e-05 |
| 60 | CYTOKINE RECEPTOR BINDING | 11 | 271 | 8.92e-06 | 0.0001381 |
| 61 | PROTEIN HOMODIMERIZATION ACTIVITY | 18 | 722 | 1.285e-05 | 0.0001957 |
| 62 | GTPASE BINDING | 11 | 295 | 1.969e-05 | 0.000295 |
| 63 | SIGNALING ADAPTOR ACTIVITY | 6 | 74 | 2.359e-05 | 0.0003479 |
| 64 | PROTEIN DIMERIZATION ACTIVITY | 23 | 1149 | 2.66e-05 | 0.0003848 |
| 65 | MACROMOLECULAR COMPLEX BINDING | 26 | 1399 | 2.692e-05 | 0.0003848 |
| 66 | EPHRIN RECEPTOR BINDING | 4 | 24 | 3.268e-05 | 0.00046 |
| 67 | SIGNALING RECEPTOR ACTIVITY | 25 | 1393 | 6.923e-05 | 0.0009599 |
| 68 | CALCIUM DEPENDENT PROTEIN KINASE ACTIVITY | 3 | 12 | 9.542e-05 | 0.001285 |
| 69 | MAP KINASE KINASE ACTIVITY | 3 | 12 | 9.542e-05 | 0.001285 |
| 70 | THIOESTERASE BINDING | 3 | 14 | 0.0001561 | 0.002071 |
| 71 | RECEPTOR ACTIVITY | 27 | 1649 | 0.00016 | 0.002094 |
| 72 | GLUTAMATE RECEPTOR BINDING | 4 | 37 | 0.0001878 | 0.002423 |
| 73 | PROTEIN SERINE THREONINE TYROSINE KINASE ACTIVITY | 4 | 39 | 0.0002311 | 0.002941 |
| 74 | PROTEIN KINASE C ACTIVITY | 3 | 16 | 0.0002374 | 0.002941 |
| 75 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 3 | 16 | 0.0002374 | 0.002941 |
| 76 | NUCLEOSIDE TRIPHOSPHATASE REGULATOR ACTIVITY | 10 | 329 | 0.0002531 | 0.003094 |
| 77 | PHOSPHOLIPASE BINDING | 3 | 19 | 0.0004038 | 0.00481 |
| 78 | EPHRIN RECEPTOR ACTIVITY | 3 | 19 | 0.0004038 | 0.00481 |
| 79 | HYDROLASE ACTIVITY ACTING ON ESTER BONDS | 15 | 739 | 0.0006398 | 0.007524 |
| 80 | FIBROBLAST GROWTH FACTOR BINDING | 3 | 23 | 0.0007215 | 0.008379 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYTOPLASMIC SIDE OF MEMBRANE | 21 | 170 | 1.654e-19 | 9.662e-17 |
| 2 | SIDE OF MEMBRANE | 24 | 428 | 3.117e-14 | 9.101e-12 |
| 3 | EXTRINSIC COMPONENT OF MEMBRANE | 18 | 252 | 1.12e-12 | 2.181e-10 |
| 4 | EXTRINSIC COMPONENT OF CYTOPLASMIC SIDE OF PLASMA MEMBRANE | 12 | 98 | 1.398e-11 | 2.041e-09 |
| 5 | EXTRINSIC COMPONENT OF PLASMA MEMBRANE | 13 | 136 | 4.664e-11 | 5.448e-09 |
| 6 | MEMBRANE REGION | 31 | 1134 | 6.61e-10 | 6.434e-08 |
| 7 | CELL JUNCTION | 31 | 1151 | 9.475e-10 | 6.916e-08 |
| 8 | INTRINSIC COMPONENT OF THE CYTOPLASMIC SIDE OF THE PLASMA MEMBRANE | 6 | 15 | 9.289e-10 | 6.916e-08 |
| 9 | INTRACELLULAR VESICLE | 31 | 1259 | 7.982e-09 | 5.18e-07 |
| 10 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 36 | 1649 | 9.303e-09 | 5.433e-07 |
| 11 | CELL SUBSTRATE JUNCTION | 17 | 398 | 1.362e-08 | 7.232e-07 |
| 12 | CELL SURFACE | 23 | 757 | 2.099e-08 | 1.022e-06 |
| 13 | VACUOLE | 29 | 1180 | 2.758e-08 | 1.239e-06 |
| 14 | PLASMA MEMBRANE REGION | 25 | 929 | 5.149e-08 | 2.148e-06 |
| 15 | CELL PROJECTION | 36 | 1786 | 7.187e-08 | 2.798e-06 |
| 16 | CELL LEADING EDGE | 15 | 350 | 9.744e-08 | 3.557e-06 |
| 17 | HETEROTRIMERIC G PROTEIN COMPLEX | 6 | 32 | 1.509e-07 | 5.182e-06 |
| 18 | ANCHORING JUNCTION | 17 | 489 | 2.64e-07 | 8.566e-06 |
| 19 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 5 | 20 | 3.699e-07 | 1.137e-05 |
| 20 | MEMBRANE MICRODOMAIN | 13 | 288 | 4.026e-07 | 1.176e-05 |
| 21 | PERINUCLEAR REGION OF CYTOPLASM | 19 | 642 | 5.905e-07 | 1.642e-05 |
| 22 | VESICLE LUMEN | 8 | 106 | 1.722e-06 | 4.572e-05 |
| 23 | RUFFLE MEMBRANE | 7 | 80 | 2.899e-06 | 7.362e-05 |
| 24 | NEURON PROJECTION | 22 | 942 | 3.708e-06 | 8.662e-05 |
| 25 | RUFFLE | 9 | 156 | 3.612e-06 | 8.662e-05 |
| 26 | CELL PROJECTION PART | 22 | 946 | 3.969e-06 | 8.914e-05 |
| 27 | PLATELET ALPHA GRANULE LUMEN | 6 | 55 | 4.167e-06 | 9.012e-05 |
| 28 | EXTRACELLULAR SPACE | 27 | 1376 | 6.914e-06 | 0.0001442 |
| 29 | LEADING EDGE MEMBRANE | 8 | 134 | 9.913e-06 | 0.000193 |
| 30 | RECEPTOR COMPLEX | 12 | 327 | 9.597e-06 | 0.000193 |
| 31 | PLASMA MEMBRANE PROTEIN COMPLEX | 15 | 510 | 1.052e-05 | 0.0001983 |
| 32 | SOMATODENDRITIC COMPARTMENT | 17 | 650 | 1.231e-05 | 0.0002247 |
| 33 | NEURON PART | 25 | 1265 | 1.411e-05 | 0.0002496 |
| 34 | ACTIN FILAMENT | 6 | 70 | 1.713e-05 | 0.0002942 |
| 35 | MAST CELL GRANULE | 4 | 21 | 1.874e-05 | 0.0003127 |
| 36 | PLATELET ALPHA GRANULE | 6 | 75 | 2.548e-05 | 0.0004133 |
| 37 | ENDOSOME | 18 | 793 | 4.426e-05 | 0.0006985 |
| 38 | GOLGI APPARATUS | 26 | 1445 | 4.652e-05 | 0.0007149 |
| 39 | SECRETORY GRANULE LUMEN | 6 | 85 | 5.193e-05 | 0.0007581 |
| 40 | DENDRITE | 13 | 451 | 5.141e-05 | 0.0007581 |
| 41 | NEURONAL POSTSYNAPTIC DENSITY | 5 | 53 | 5.571e-05 | 0.0007936 |
| 42 | SECRETORY VESICLE | 13 | 461 | 6.422e-05 | 0.000893 |
| 43 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 9 | 237 | 9.931e-05 | 0.001349 |
| 44 | MEMBRANE PROTEIN COMPLEX | 20 | 1020 | 0.000124 | 0.001609 |
| 45 | EARLY ENDOSOME | 10 | 301 | 0.0001234 | 0.001609 |
| 46 | SITE OF POLARIZED GROWTH | 7 | 149 | 0.000164 | 0.002082 |
| 47 | CELL CELL JUNCTION | 11 | 383 | 0.0002032 | 0.002525 |
| 48 | ENDOPLASMIC RETICULUM | 26 | 1631 | 0.0003269 | 0.003977 |
| 49 | SECRETORY GRANULE | 10 | 352 | 0.0004316 | 0.005144 |
| 50 | CELL BODY | 12 | 494 | 0.0004813 | 0.005621 |
| 51 | CELL PROJECTION MEMBRANE | 9 | 298 | 0.0005405 | 0.006189 |
| 52 | EXCITATORY SYNAPSE | 7 | 197 | 0.0008814 | 0.009898 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04014_Ras_signaling_pathway | 155 | 236 | 0 | 0 | |
| 2 | hsa04010_MAPK_signaling_pathway | 72 | 268 | 3.829e-95 | 3.446e-93 | |
| 3 | hsa04151_PI3K_AKT_signaling_pathway | 77 | 351 | 9.056e-95 | 5.434e-93 | |
| 4 | hsa04510_Focal_adhesion | 47 | 200 | 1.823e-57 | 8.202e-56 | |
| 5 | hsa04810_Regulation_of_actin_cytoskeleton | 45 | 214 | 1.52e-52 | 5.473e-51 | |
| 6 | hsa04664_Fc_epsilon_RI_signaling_pathway | 32 | 79 | 9.333e-48 | 2.8e-46 | |
| 7 | hsa04012_ErbB_signaling_pathway | 32 | 87 | 4.269e-46 | 1.098e-44 | |
| 8 | hsa04722_Neurotrophin_signaling_pathway | 35 | 127 | 2.931e-45 | 6.595e-44 | |
| 9 | hsa04062_Chemokine_signaling_pathway | 38 | 189 | 2.155e-43 | 4.31e-42 | |
| 10 | hsa04370_VEGF_signaling_pathway | 29 | 76 | 2.373e-42 | 4.271e-41 | |
| 11 | hsa04912_GnRH_signaling_pathway | 26 | 101 | 8.598e-33 | 1.407e-31 | |
| 12 | hsa04660_T_cell_receptor_signaling_pathway | 26 | 108 | 6.035e-32 | 9.053e-31 | |
| 13 | hsa04662_B_cell_receptor_signaling_pathway | 22 | 75 | 2.857e-29 | 3.956e-28 | |
| 14 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 22 | 95 | 1.001e-26 | 1.287e-25 | |
| 15 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 24 | 136 | 5.023e-26 | 6.027e-25 | |
| 16 | hsa04730_Long.term_depression | 19 | 70 | 1.148e-24 | 1.291e-23 | |
| 17 | hsa04910_Insulin_signaling_pathway | 23 | 138 | 2.275e-24 | 2.409e-23 | |
| 18 | hsa04540_Gap_junction | 18 | 90 | 8.701e-21 | 8.701e-20 | |
| 19 | hsa04380_Osteoclast_differentiation | 20 | 128 | 1.02e-20 | 9.662e-20 | |
| 20 | hsa04360_Axon_guidance | 20 | 130 | 1.408e-20 | 1.267e-19 | |
| 21 | hsa04270_Vascular_smooth_muscle_contraction | 18 | 116 | 1.089e-18 | 9.336e-18 | |
| 22 | hsa04720_Long.term_potentiation | 15 | 70 | 5.479e-18 | 4.444e-17 | |
| 23 | hsa04914_Progesterone.mediated_oocyte_maturation | 16 | 87 | 5.679e-18 | 4.444e-17 | |
| 24 | hsa04972_Pancreatic_secretion | 16 | 101 | 6.992e-17 | 5.244e-16 | |
| 25 | hsa04620_Toll.like_receptor_signaling_pathway | 16 | 102 | 8.239e-17 | 5.932e-16 | |
| 26 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 12 | 42 | 2.759e-16 | 1.91e-15 | |
| 27 | hsa00592_alpha.Linolenic_acid_metabolism | 9 | 20 | 1.246e-14 | 8.305e-14 | |
| 28 | hsa04670_Leukocyte_transendothelial_migration | 15 | 117 | 1.759e-14 | 1.13e-13 | |
| 29 | hsa00565_Ether_lipid_metabolism | 10 | 36 | 1.247e-13 | 7.737e-13 | |
| 30 | hsa04210_Apoptosis | 13 | 89 | 1.851e-13 | 1.11e-12 | |
| 31 | hsa04916_Melanogenesis | 13 | 101 | 9.922e-13 | 5.588e-12 | |
| 32 | hsa00591_Linoleic_acid_metabolism | 9 | 30 | 9.935e-13 | 5.588e-12 | |
| 33 | hsa04144_Endocytosis | 16 | 203 | 4.838e-12 | 2.639e-11 | |
| 34 | hsa04150_mTOR_signaling_pathway | 10 | 52 | 6.979e-12 | 3.695e-11 | |
| 35 | hsa04320_Dorso.ventral_axis_formation | 8 | 25 | 1.049e-11 | 5.397e-11 | |
| 36 | hsa04070_Phosphatidylinositol_signaling_system | 11 | 78 | 2.108e-11 | 1.054e-10 | |
| 37 | hsa04975_Fat_digestion_and_absorption | 9 | 46 | 6.885e-11 | 3.349e-10 | |
| 38 | hsa04520_Adherens_junction | 10 | 73 | 2.386e-10 | 1.13e-09 | |
| 39 | hsa04530_Tight_junction | 12 | 133 | 5.235e-10 | 2.416e-09 | |
| 40 | hsa00564_Glycerophospholipid_metabolism | 10 | 80 | 6.036e-10 | 2.716e-09 | |
| 41 | hsa00590_Arachidonic_acid_metabolism | 9 | 59 | 7.208e-10 | 3.164e-09 | |
| 42 | hsa04020_Calcium_signaling_pathway | 13 | 177 | 1.257e-09 | 5.388e-09 | |
| 43 | hsa04973_Carbohydrate_digestion_and_absorption | 8 | 44 | 1.518e-09 | 6.356e-09 | |
| 44 | hsa04920_Adipocytokine_signaling_pathway | 9 | 68 | 2.664e-09 | 1.09e-08 | |
| 45 | hsa04310_Wnt_signaling_pathway | 11 | 151 | 2.683e-08 | 1.073e-07 | |
| 46 | hsa04621_NOD.like_receptor_signaling_pathway | 7 | 59 | 3.567e-07 | 1.396e-06 | |
| 47 | hsa04630_Jak.STAT_signaling_pathway | 10 | 155 | 3.663e-07 | 1.403e-06 | |
| 48 | hsa04622_RIG.I.like_receptor_signaling_pathway | 6 | 71 | 1.859e-05 | 6.971e-05 | |
| 49 | hsa04971_Gastric_acid_secretion | 6 | 74 | 2.359e-05 | 8.666e-05 | |
| 50 | hsa04114_Oocyte_meiosis | 7 | 114 | 3.015e-05 | 0.0001085 | |
| 51 | hsa04970_Salivary_secretion | 6 | 89 | 6.726e-05 | 0.0002374 | |
| 52 | hsa04640_Hematopoietic_cell_lineage | 5 | 88 | 0.000612 | 0.002118 | |
| 53 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 0.0009961 | 0.003383 | |
| 54 | hsa04744_Phototransduction | 3 | 29 | 0.001439 | 0.004796 | |
| 55 | hsa04962_Vasopressin.regulated_water_reabsorption | 3 | 44 | 0.004791 | 0.01568 | |
| 56 | hsa04742_Taste_transduction | 3 | 52 | 0.007643 | 0.02457 | |
| 57 | hsa04623_Cytosolic_DNA.sensing_pathway | 3 | 56 | 0.009374 | 0.0296 | |
| 58 | hsa04145_Phagosome | 3 | 156 | 0.1211 | 0.3759 | |
| 59 | hsa04350_TGF.beta_signaling_pathway | 2 | 85 | 0.1409 | 0.4261 | |
| 60 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 3 | 168 | 0.142 | 0.4261 | |
| 61 | hsa04390_Hippo_signaling_pathway | 2 | 154 | 0.3356 | 0.9902 | |
| 62 | hsa04740_Olfactory_transduction | 3 | 388 | 0.5814 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TBX5-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | PDGFRA | Sponge network | -2.108 | 0 | -0.92 | 0.00039 | 0.659 |
| 2 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | PDGFRA | Sponge network | 0.053 | 0.85755 | -0.92 | 0.00039 | 0.612 |
| 3 | LINC00702 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | PDGFRA | Sponge network | -2.856 | 0 | -0.92 | 0.00039 | 0.589 |
| 4 | TBX5-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | FGF7 | Sponge network | -2.108 | 0 | -1.351 | 0 | 0.587 |
| 5 | LINC00702 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 18 | ETS1 | Sponge network | -2.856 | 0 | -1.335 | 0 | 0.581 |
| 6 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | ETS1 | Sponge network | -2.039 | 0 | -1.335 | 0 | 0.573 |
| 7 | AC109642.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PDGFRA | Sponge network | -2.791 | 0 | -0.92 | 0.00039 | 0.557 |
| 8 | GAS6-AS2 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p | 12 | FGF2 | Sponge network | -1.761 | 0 | -2.545 | 0 | 0.556 |
| 9 | RP11-354E11.2 |
hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-9-5p;hsa-miR-940;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -2.138 | 0 | -1.412 | 0 | 0.549 |
| 10 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-629-3p | 20 | PIK3R1 | Sponge network | -2.108 | 0 | -1.285 | 0 | 0.544 |
| 11 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PDGFRA | Sponge network | -1.892 | 0 | -0.92 | 0.00039 | 0.534 |
| 12 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | -2.791 | 0 | -1.285 | 0 | 0.529 |
| 13 | LINC00702 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p | 18 | FGF2 | Sponge network | -2.856 | 0 | -2.545 | 0 | 0.527 |
| 14 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 16 | PIK3R1 | Sponge network | -2.039 | 0 | -1.285 | 0 | 0.52 |
| 15 | MIR497HG |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-3127-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-9-5p;hsa-miR-940;hsa-miR-96-5p | 11 | GNG7 | Sponge network | -2.142 | 0 | -1.412 | 0 | 0.519 |
| 16 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PDGFRA | Sponge network | -1.583 | 0 | -0.92 | 0.00039 | 0.516 |
| 17 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FGF7 | Sponge network | -1.892 | 0 | -1.351 | 0 | 0.514 |
| 18 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -4.19 | 0 | -1.285 | 0 | 0.51 |
| 19 | LINC00702 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | FGF7 | Sponge network | -2.856 | 0 | -1.351 | 0 | 0.505 |
| 20 | TBX5-AS1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-590-5p;hsa-miR-93-3p | 15 | FGF2 | Sponge network | -2.108 | 0 | -2.545 | 0 | 0.504 |
| 21 | LINC00968 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-590-5p;hsa-miR-93-3p | 12 | FGF2 | Sponge network | -4.19 | 0 | -2.545 | 0 | 0.504 |
| 22 | MAGI2-AS3 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-93-3p | 15 | FGF2 | Sponge network | -1.892 | 0 | -2.545 | 0 | 0.499 |
| 23 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | GAB1 | Sponge network | -2.952 | 0 | -1.505 | 0 | 0.498 |
| 24 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -2.305 | 0 | -1.285 | 0 | 0.496 |
| 25 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | PIK3R1 | Sponge network | -2.856 | 0 | -1.285 | 0 | 0.494 |
| 26 | LINC00968 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 10 | ETS1 | Sponge network | -4.19 | 0 | -1.335 | 0 | 0.491 |
| 27 | FENDRR |
hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-93-3p | 10 | FGF2 | Sponge network | -4.222 | 0 | -2.545 | 0 | 0.489 |
| 28 | TBX5-AS1 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ETS1 | Sponge network | -2.108 | 0 | -1.335 | 0 | 0.489 |
| 29 | MAGI2-AS3 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 17 | ETS1 | Sponge network | -1.892 | 0 | -1.335 | 0 | 0.487 |
| 30 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | -1.892 | 0 | -1.285 | 0 | 0.486 |
| 31 | AC109642.1 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -2.791 | 0 | -1.412 | 0 | 0.473 |
| 32 | LINC00968 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-9-5p;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -4.19 | 0 | -1.412 | 0 | 0.472 |
| 33 | AC109642.1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-503-5p;hsa-miR-93-3p | 12 | FGF2 | Sponge network | -2.791 | 0 | -2.545 | 0 | 0.467 |
| 34 | RP11-462G12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | RGL1 | Sponge network | -1.071 | 0.01175 | -0.968 | 0 | 0.462 |
| 35 | TBX5-AS1 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-3127-5p;hsa-miR-766-3p;hsa-miR-9-5p;hsa-miR-92a-3p | 10 | GNG7 | Sponge network | -2.108 | 0 | -1.412 | 0 | 0.46 |
| 36 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-93-5p | 10 | RGL1 | Sponge network | -2.305 | 0 | -0.968 | 0 | 0.46 |
| 37 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PDGFRA | Sponge network | -0.582 | 0.05253 | -0.92 | 0.00039 | 0.459 |
| 38 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | -2.952 | 0 | -1.285 | 0 | 0.458 |
| 39 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PDGFRA | Sponge network | -1.761 | 0 | -0.92 | 0.00039 | 0.451 |
| 40 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-32-5p;hsa-miR-582-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | RGL1 | Sponge network | -2.791 | 0 | -0.968 | 0 | 0.45 |
| 41 | RP11-166D19.1 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ETS1 | Sponge network | -0.582 | 0.05253 | -1.335 | 0 | 0.449 |
| 42 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 11 | CSF1 | Sponge network | -0.672 | 0.02084 | -0.65 | 0.00787 | 0.444 |
| 43 | LINC00968 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | PDGFRA | Sponge network | -4.19 | 0 | -0.92 | 0.00039 | 0.443 |
| 44 | CTD-2013N24.2 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429 | 12 | FGF2 | Sponge network | -1.745 | 0 | -2.545 | 0 | 0.441 |
| 45 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -4.222 | 0 | -1.285 | 0 | 0.438 |
| 46 | AC007743.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -2.595 | 0 | -1.285 | 0 | 0.436 |
| 47 | AC011899.9 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-181d-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 11 | ETS1 | Sponge network | -2.611 | 0 | -1.335 | 0 | 0.436 |
| 48 | RP11-1024P17.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-576-5p | 12 | ETS1 | Sponge network | -2.062 | 0 | -1.335 | 0 | 0.435 |
| 49 | SH3RF3-AS1 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ETS1 | Sponge network | -1.583 | 0 | -1.335 | 0 | 0.433 |
| 50 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RAPGEF5 | Sponge network | -2.039 | 0 | -1.092 | 0 | 0.431 |
| 51 | CTD-2013N24.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 16 | ETS1 | Sponge network | -1.745 | 0 | -1.335 | 0 | 0.431 |
| 52 | TBX5-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-629-5p | 14 | IGF1 | Sponge network | -2.108 | 0 | -0.879 | 0.00545 | 0.429 |
| 53 | RP11-456K23.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | IGF1 | Sponge network | -1.488 | 0 | -0.879 | 0.00545 | 0.429 |
| 54 | RP11-389C8.2 |
hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p | 11 | FGF2 | Sponge network | -2.039 | 0 | -2.545 | 0 | 0.428 |
| 55 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 10 | NTRK2 | Sponge network | -2.952 | 0 | -2.564 | 0 | 0.427 |
| 56 | MIR497HG |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | PDGFRA | Sponge network | -2.142 | 0 | -0.92 | 0.00039 | 0.426 |
| 57 | GAS6-AS2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ETS1 | Sponge network | -1.761 | 0 | -1.335 | 0 | 0.424 |
| 58 | BZRAP1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-582-3p | 10 | RGL1 | Sponge network | -0.785 | 0.00723 | -0.968 | 0 | 0.424 |
| 59 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-708-5p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | RGL1 | Sponge network | -4.19 | 0 | -0.968 | 0 | 0.422 |
| 60 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | IGF1 | Sponge network | -2.856 | 0 | -0.879 | 0.00545 | 0.42 |
| 61 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | RGL1 | Sponge network | -0.761 | 0.05061 | -0.968 | 0 | 0.42 |
| 62 | FENDRR |
hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RAPGEF5 | Sponge network | -4.222 | 0 | -1.092 | 0 | 0.418 |
| 63 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | -2.062 | 0 | -1.285 | 0 | 0.418 |
| 64 | SH3RF3-AS1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-503-5p;hsa-miR-590-5p | 13 | FGF2 | Sponge network | -1.583 | 0 | -2.545 | 0 | 0.417 |
| 65 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 23 | PIK3R1 | Sponge network | -1.488 | 0 | -1.285 | 0 | 0.416 |
| 66 | RP11-354E11.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -2.138 | 0 | -1.285 | 0 | 0.41 |
| 67 | AC109642.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | IGF1 | Sponge network | -2.791 | 0 | -0.879 | 0.00545 | 0.41 |
| 68 | CTD-2013N24.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PDGFRA | Sponge network | -1.745 | 0 | -0.92 | 0.00039 | 0.41 |
| 69 | RP11-1024P17.1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RAPGEF5 | Sponge network | -2.062 | 0 | -1.092 | 0 | 0.409 |
| 70 | RP11-456K23.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 22 | PDGFRA | Sponge network | -1.488 | 0 | -0.92 | 0.00039 | 0.409 |
| 71 | TBX5-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RAPGEF5 | Sponge network | -2.108 | 0 | -1.092 | 0 | 0.409 |
| 72 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | -3.04 | 0 | -1.285 | 0 | 0.408 |
| 73 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | -1.745 | 0 | -1.285 | 0 | 0.407 |
| 74 | RP11-536K7.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-93-5p | 10 | PDGFRA | Sponge network | -1.239 | 5.0E-5 | -0.92 | 0.00039 | 0.407 |
| 75 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | GAB1 | Sponge network | -2.791 | 0 | -1.505 | 0 | 0.407 |
| 76 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | GAB1 | Sponge network | -4.19 | 0 | -1.505 | 0 | 0.403 |
| 77 | RP11-401P9.4 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-590-5p;hsa-miR-93-3p | 10 | FGF2 | Sponge network | -3.04 | 0 | -2.545 | 0 | 0.402 |
| 78 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RGL1 | Sponge network | -2.108 | 0 | -0.968 | 0 | 0.402 |
| 79 | LINC00968 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 14 | NTRK2 | Sponge network | -4.19 | 0 | -2.564 | 0 | 0.4 |
| 80 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | RGL1 | Sponge network | -2.039 | 0 | -0.968 | 0 | 0.396 |
| 81 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -2.028 | 0 | -1.285 | 0 | 0.395 |
| 82 | RP11-456K23.1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p | 14 | FGF2 | Sponge network | -1.488 | 0 | -2.545 | 0 | 0.393 |
| 83 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | -2.142 | 0 | -1.285 | 0 | 0.391 |
| 84 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | GAB1 | Sponge network | -2.856 | 0 | -1.505 | 0 | 0.391 |
| 85 | RP11-456K23.1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-93-5p | 10 | AKT3 | Sponge network | -1.488 | 0 | -1.44 | 0 | 0.389 |
| 86 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | GAB1 | Sponge network | -1.297 | 0 | -1.505 | 0 | 0.389 |
| 87 | LINC00702 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-3127-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -2.856 | 0 | -1.412 | 0 | 0.389 |
| 88 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-93-5p | 10 | PDGFRA | Sponge network | -2.305 | 0 | -0.92 | 0.00039 | 0.388 |
| 89 | FENDRR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 14 | NTRK2 | Sponge network | -4.222 | 0 | -2.564 | 0 | 0.387 |
| 90 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 14 | GAB1 | Sponge network | -2.062 | 0 | -1.505 | 0 | 0.387 |
| 91 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | IGF1 | Sponge network | -1.892 | 0 | -0.879 | 0.00545 | 0.384 |
| 92 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | GAB1 | Sponge network | -2.807 | 0 | -1.505 | 0 | 0.383 |
| 93 | RP5-1042I8.7 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | GAB1 | Sponge network | -0.733 | 0.00018 | -1.505 | 0 | 0.381 |
| 94 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -0.873 | 0.00072 | -1.285 | 0 | 0.38 |
| 95 | FGF14-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -2.159 | 0 | -1.285 | 0 | 0.378 |
| 96 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-93-5p | 10 | GAB1 | Sponge network | -1.193 | 0.00359 | -1.505 | 0 | 0.378 |
| 97 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RAPGEF5 | Sponge network | -1.892 | 0 | -1.092 | 0 | 0.377 |
| 98 | AC011899.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-708-5p | 12 | RGL1 | Sponge network | -2.611 | 0 | -0.968 | 0 | 0.377 |
| 99 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p | 13 | NTRK2 | Sponge network | -2.039 | 0 | -2.564 | 0 | 0.377 |
| 100 | RP11-456K23.1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RAPGEF5 | Sponge network | -1.488 | 0 | -1.092 | 0 | 0.376 |
| 101 | RP11-166D19.1 |
hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p | 11 | FGF2 | Sponge network | -0.582 | 0.05253 | -2.545 | 0 | 0.375 |
| 102 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | PIK3R1 | Sponge network | -2.807 | 0 | -1.285 | 0 | 0.374 |
| 103 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 11 | IGF1 | Sponge network | -1.583 | 0 | -0.879 | 0.00545 | 0.374 |
| 104 | RP11-365O16.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-708-5p;hsa-miR-93-5p | 12 | RGL1 | Sponge network | -2.765 | 0.00017 | -0.968 | 0 | 0.374 |
| 105 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 10 | RASGRF2 | Sponge network | -0.582 | 0.05253 | -0.287 | 0.25734 | 0.373 |
| 106 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 15 | GAB1 | Sponge network | -2.108 | 0 | -1.505 | 0 | 0.371 |
| 107 | LINC00092 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | -2.383 | 0 | -1.285 | 0 | 0.369 |
| 108 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | RGL1 | Sponge network | -1.892 | 0 | -0.968 | 0 | 0.369 |
| 109 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | NTRK2 | Sponge network | -2.856 | 0 | -2.564 | 0 | 0.369 |
| 110 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | GAB1 | Sponge network | -4.222 | 0 | -1.505 | 0 | 0.369 |
| 111 | LINC00702 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29b-3p;hsa-miR-92a-3p | 10 | RASAL2 | Sponge network | -2.856 | 0 | -0.753 | 0 | 0.368 |
| 112 | RP11-354E11.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-96-5p | 12 | RGL1 | Sponge network | -2.138 | 0 | -0.968 | 0 | 0.368 |
| 113 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | GAB1 | Sponge network | -1.892 | 0 | -1.505 | 0 | 0.368 |
| 114 | RP11-378A13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -1.713 | 0 | -1.285 | 0 | 0.367 |
| 115 | SH3RF3-AS1 |
hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-590-5p | 12 | PIK3R1 | Sponge network | -1.583 | 0 | -1.285 | 0 | 0.364 |
| 116 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | GAB1 | Sponge network | -2.039 | 0 | -1.505 | 0 | 0.364 |
| 117 | AC007743.1 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-212-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | GAB1 | Sponge network | -2.595 | 0 | -1.505 | 0 | 0.364 |
| 118 | AC011899.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | PIK3R1 | Sponge network | -2.611 | 0 | -1.285 | 0 | 0.364 |
| 119 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-576-5p | 11 | PDGFRA | Sponge network | -2.028 | 0 | -0.92 | 0.00039 | 0.363 |
| 120 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | CSF1 | Sponge network | -0.761 | 0.05061 | -0.65 | 0.00787 | 0.36 |
| 121 | AC079630.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -3.758 | 0 | -1.285 | 0 | 0.359 |
| 122 | RP5-1042I8.7 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -0.733 | 0.00018 | -1.285 | 0 | 0.358 |
| 123 | AC004947.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | NTRK2 | Sponge network | -3.94 | 0 | -2.564 | 0 | 0.358 |
| 124 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | NTRK2 | Sponge network | -2.09 | 0 | -2.564 | 0 | 0.358 |
| 125 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -3.16 | 2.0E-5 | -1.285 | 0 | 0.358 |
| 126 | CTD-2269F5.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | PDGFRA | Sponge network | -1.576 | 0.00334 | -0.92 | 0.00039 | 0.358 |
| 127 | RP11-352D13.6 |
hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-9-5p;hsa-miR-940;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -4.634 | 0 | -1.412 | 0 | 0.357 |
| 128 | RP11-354E11.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NTRK2 | Sponge network | -2.138 | 0 | -2.564 | 0 | 0.355 |
| 129 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 10 | FLT1 | Sponge network | -2.039 | 0 | -0.854 | 4.0E-5 | 0.354 |
| 130 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-590-5p | 15 | PIK3R1 | Sponge network | -1.761 | 0 | -1.285 | 0 | 0.354 |
| 131 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-96-5p | 11 | TIAM1 | Sponge network | -2.856 | 0 | -1.152 | 2.0E-5 | 0.351 |
| 132 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 13 | PDGFRA | Sponge network | -0.672 | 0.02084 | -0.92 | 0.00039 | 0.348 |
| 133 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PDGFRA | Sponge network | -2.039 | 0 | -0.92 | 0.00039 | 0.347 |
| 134 | LINC00961 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -2.724 | 0 | -1.285 | 0 | 0.346 |
| 135 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | IGF1 | Sponge network | -2.039 | 0 | -0.879 | 0.00545 | 0.345 |
| 136 | AC109642.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -2.791 | 0 | -2.564 | 0 | 0.344 |
| 137 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | RGL1 | Sponge network | -0.873 | 0.00072 | -0.968 | 0 | 0.342 |
| 138 | LINC00968 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RAPGEF5 | Sponge network | -4.19 | 0 | -1.092 | 0 | 0.341 |
| 139 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | GAB1 | Sponge network | -1.745 | 0 | -1.505 | 0 | 0.338 |
| 140 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -2.783 | 0 | -1.285 | 0 | 0.338 |
| 141 | AC079630.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -3.758 | 0 | -2.564 | 0 | 0.338 |
| 142 | RP11-1024P17.1 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-3p | 13 | FGF2 | Sponge network | -2.062 | 0 | -2.545 | 0 | 0.338 |
| 143 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p | 11 | ETS1 | Sponge network | -1.84 | 0 | -1.335 | 0 | 0.337 |
| 144 | LINC00702 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RAPGEF5 | Sponge network | -2.856 | 0 | -1.092 | 0 | 0.334 |
| 145 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | NTRK2 | Sponge network | -1.892 | 0 | -2.564 | 0 | 0.333 |
| 146 | AC004947.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -3.94 | 0 | -1.285 | 0 | 0.332 |
| 147 | RP11-354E11.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | GAB1 | Sponge network | -2.138 | 0 | -1.505 | 0 | 0.33 |
| 148 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -1.297 | 0 | -1.285 | 0 | 0.329 |
| 149 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-93-5p | 10 | CSF1 | Sponge network | -1.892 | 0 | -0.65 | 0.00787 | 0.326 |
| 150 | CASC2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | NTRK2 | Sponge network | -1.086 | 0 | -2.564 | 0 | 0.326 |
| 151 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-5p | 10 | NTRK2 | Sponge network | -2.142 | 0 | -2.564 | 0 | 0.325 |
| 152 | AC079630.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | RGL1 | Sponge network | -3.758 | 0 | -0.968 | 0 | 0.324 |
| 153 | CTD-2013N24.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | IGF1 | Sponge network | -1.745 | 0 | -0.879 | 0.00545 | 0.323 |
| 154 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | RGL1 | Sponge network | -2.952 | 0 | -0.968 | 0 | 0.323 |
| 155 | RP11-354E11.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | PDGFRA | Sponge network | -2.138 | 0 | -0.92 | 0.00039 | 0.323 |
| 156 | RP11-1008C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 11 | RGL1 | Sponge network | -1.249 | 0 | -0.968 | 0 | 0.322 |
| 157 | RP11-456K23.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | ETS1 | Sponge network | -1.488 | 0 | -1.335 | 0 | 0.322 |
| 158 | FAM225B | hsa-miR-1180-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-501-5p;hsa-miR-664a-3p | 10 | ETS1 | Sponge network | 0.212 | 0.51382 | -1.335 | 0 | 0.321 |
| 159 | RP11-1008C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-3p;hsa-miR-93-5p | 13 | GAB1 | Sponge network | -1.249 | 0 | -1.505 | 0 | 0.321 |
| 160 | RP11-1008C21.1 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-181d-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 10 | ETS1 | Sponge network | -1.826 | 3.0E-5 | -1.335 | 0 | 0.32 |
| 161 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-93-5p | 16 | PDGFRA | Sponge network | -3.04 | 0 | -0.92 | 0.00039 | 0.32 |
| 162 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-629-5p | 10 | IGF1 | Sponge network | -0.582 | 0.05253 | -0.879 | 0.00545 | 0.319 |
| 163 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | PIK3R1 | Sponge network | -0.427 | 0.1559 | -1.285 | 0 | 0.318 |
| 164 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | RGL1 | Sponge network | -0.427 | 0.1559 | -0.968 | 0 | 0.317 |
| 165 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | RGL1 | Sponge network | -0.672 | 0.02084 | -0.968 | 0 | 0.317 |
| 166 | LINC00607 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -2.277 | 0 | -1.285 | 0 | 0.316 |
| 167 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | RGL1 | Sponge network | -2.142 | 0 | -0.968 | 0 | 0.315 |
| 168 | AC011899.9 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PDGFRA | Sponge network | -2.611 | 0 | -0.92 | 0.00039 | 0.315 |
| 169 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | GAB1 | Sponge network | -0.873 | 0.00072 | -1.505 | 0 | 0.315 |
| 170 | RP11-354E11.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | IGF1 | Sponge network | -2.138 | 0 | -0.879 | 0.00545 | 0.313 |
| 171 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | RGL1 | Sponge network | -2.856 | 0 | -0.968 | 0 | 0.311 |
| 172 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | PDGFRA | Sponge network | -1.84 | 0 | -0.92 | 0.00039 | 0.311 |
| 173 | RP11-399O19.9 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PDGFRA | Sponge network | -0.873 | 0.00072 | -0.92 | 0.00039 | 0.31 |
| 174 | AC011899.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -2.611 | 0 | -2.564 | 0 | 0.31 |
| 175 | MIR497HG |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ETS1 | Sponge network | -2.142 | 0 | -1.335 | 0 | 0.31 |
| 176 | RP11-352D13.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | NTRK2 | Sponge network | -4.634 | 0 | -2.564 | 0 | 0.31 |
| 177 | AF131215.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -1.808 | 0 | -2.564 | 0 | 0.307 |
| 178 | DNM3OS |
hsa-miR-1180-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ETS1 | Sponge network | 0.053 | 0.85755 | -1.335 | 0 | 0.303 |
| 179 | RP11-462G12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.071 | 0.01175 | -1.285 | 0 | 0.302 |
| 180 | RP11-378A13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | GAB1 | Sponge network | -1.713 | 0 | -1.505 | 0 | 0.301 |
| 181 | AC079630.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | GAB1 | Sponge network | -3.758 | 0 | -1.505 | 0 | 0.3 |
| 182 | TBX5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-5p | 14 | NTRK2 | Sponge network | -2.108 | 0 | -2.564 | 0 | 0.299 |
| 183 | RP11-378A13.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 14 | NTRK2 | Sponge network | -1.713 | 0 | -2.564 | 0 | 0.298 |
| 184 | AC016735.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -2.711 | 0.00282 | -1.285 | 0 | 0.298 |
| 185 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-5p | 13 | NTRK2 | Sponge network | -3.04 | 0 | -2.564 | 0 | 0.298 |
| 186 | RP11-88I21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -8.789 | 0 | -1.285 | 0 | 0.297 |
| 187 | RP11-1008C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -1.249 | 0 | -1.285 | 0 | 0.297 |
| 188 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 11 | PDGFRA | Sponge network | -1.193 | 0.00359 | -0.92 | 0.00039 | 0.297 |
| 189 | HLA-F-AS1 |
hsa-miR-1180-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-181d-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | ETS1 | Sponge network | -0.495 | 0.12126 | -1.335 | 0 | 0.296 |
| 190 | CTD-2013N24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29b-3p;hsa-miR-429 | 10 | RASAL2 | Sponge network | -1.745 | 0 | -0.753 | 0 | 0.295 |
| 191 | CASC2 |
hsa-miR-106a-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | GAB1 | Sponge network | -1.086 | 0 | -1.505 | 0 | 0.294 |
| 192 | RP11-365O16.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -2.765 | 0.00017 | -1.285 | 0 | 0.293 |
| 193 | CASC2 |
hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -1.086 | 0 | -1.285 | 0 | 0.29 |
| 194 | CTD-2013N24.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-335-3p | 10 | TIAM1 | Sponge network | -1.745 | 0 | -1.152 | 2.0E-5 | 0.29 |
| 195 | RP1-78O14.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 10 | GAB1 | Sponge network | -4.409 | 0 | -1.505 | 0 | 0.287 |
| 196 | LINC00968 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | IGF1 | Sponge network | -4.19 | 0 | -0.879 | 0.00545 | 0.287 |
| 197 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-92a-3p | 11 | RASAL2 | Sponge network | -0.582 | 0.05253 | -0.753 | 0 | 0.287 |
| 198 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-5p | 13 | NTRK2 | Sponge network | -1.761 | 0 | -2.564 | 0 | 0.287 |
| 199 | AC004947.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-93-5p | 11 | RGL1 | Sponge network | -3.94 | 0 | -0.968 | 0 | 0.285 |
| 200 | LINC00619 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | PDGFRA | Sponge network | -2.307 | 0.02217 | -0.92 | 0.00039 | 0.285 |
| 201 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-3p | 11 | PDGFRA | Sponge network | -2.09 | 0 | -0.92 | 0.00039 | 0.285 |
| 202 | RP11-476D10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181d-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | GAB1 | Sponge network | -4.519 | 0 | -1.505 | 0 | 0.285 |
| 203 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -0.761 | 0.05061 | -1.285 | 0 | 0.285 |
| 204 | HLA-F-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | CSF1 | Sponge network | -0.495 | 0.12126 | -0.65 | 0.00787 | 0.284 |
| 205 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p | 10 | RGL1 | Sponge network | -2.062 | 0 | -0.968 | 0 | 0.283 |
| 206 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | PIK3R1 | Sponge network | -1.808 | 0 | -1.285 | 0 | 0.282 |
| 207 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-93-5p | 12 | RGL1 | Sponge network | -1.488 | 0 | -0.968 | 0 | 0.282 |
| 208 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 11 | NTRK2 | Sponge network | -2.028 | 0 | -2.564 | 0 | 0.281 |
| 209 | RP11-352D13.6 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -4.634 | 0 | -1.285 | 0 | 0.28 |
| 210 | LINC00473 | hsa-let-7i-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-500a-3p;hsa-miR-532-5p | 10 | IGF1R | Sponge network | -0.765 | 0.56027 | -0.179 | 0.43653 | 0.28 |
| 211 | LIPE-AS1 |
hsa-miR-106a-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -0.734 | 0.00039 | -1.285 | 0 | 0.28 |
| 212 | CASC2 |
hsa-miR-106a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-93-5p | 10 | RGL1 | Sponge network | -1.086 | 0 | -0.968 | 0 | 0.28 |
| 213 | CTC-297N7.9 | hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-5p | 10 | NTRK2 | Sponge network | -1.562 | 6.0E-5 | -2.564 | 0 | 0.28 |
| 214 | RP11-476D10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -4.519 | 0 | -1.285 | 0 | 0.279 |
| 215 | RP11-284N8.3 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PDGFRA | Sponge network | -0.761 | 0.05061 | -0.92 | 0.00039 | 0.278 |
| 216 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -0.568 | 0.02011 | -1.285 | 0 | 0.277 |
| 217 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RAPGEF5 | Sponge network | -0.427 | 0.1559 | -1.092 | 0 | 0.277 |
| 218 | LINC00443 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p | 10 | PIK3R1 | Sponge network | -3.704 | 0.0003 | -1.285 | 0 | 0.276 |
| 219 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FLT1 | Sponge network | -2.856 | 0 | -0.854 | 4.0E-5 | 0.276 |
| 220 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | RGL1 | Sponge network | -4.222 | 0 | -0.968 | 0 | 0.275 |
| 221 | RP11-1008C21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | PIK3R1 | Sponge network | -1.826 | 3.0E-5 | -1.285 | 0 | 0.272 |
| 222 | RP5-839B4.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | RGL1 | Sponge network | -5.037 | 0 | -0.968 | 0 | 0.271 |
| 223 | MIR497HG |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p | 13 | FGF2 | Sponge network | -2.142 | 0 | -2.545 | 0 | 0.271 |
| 224 | SNHG18 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.073 | 0.00533 | -1.285 | 0 | 0.27 |
| 225 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PDGFRA | Sponge network | -0.427 | 0.1559 | -0.92 | 0.00039 | 0.27 |
| 226 | CTD-2013N24.2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RAPGEF5 | Sponge network | -1.745 | 0 | -1.092 | 0 | 0.27 |
| 227 | C1orf132 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-182-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -0.86 | 0.02429 | -1.285 | 0 | 0.269 |
| 228 | TBX5-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 12 | GRIN2A | Sponge network | -2.108 | 0 | -1.285 | 0.03401 | 0.264 |
| 229 | RP1-78O14.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -4.409 | 0 | -1.285 | 0 | 0.264 |
| 230 | RP5-839B4.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -5.037 | 0 | -1.285 | 0 | 0.262 |
| 231 | DIO3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -1.936 | 0.00085 | -1.285 | 0 | 0.262 |
| 232 | CYP1B1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | GAB1 | Sponge network | -1.073 | 0.00045 | -1.505 | 0 | 0.261 |
| 233 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | GAB1 | Sponge network | -0.568 | 0.02011 | -1.505 | 0 | 0.261 |
| 234 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-96-5p | 14 | RGL1 | Sponge network | -1.297 | 0 | -0.968 | 0 | 0.26 |
| 235 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-93-5p | 10 | CSF1 | Sponge network | -2.039 | 0 | -0.65 | 0.00787 | 0.26 |
| 236 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p | 14 | GAB1 | Sponge network | -1.761 | 0 | -1.505 | 0 | 0.259 |
| 237 | LINC00261 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 16 | PIK3R1 | Sponge network | -2.566 | 0.00025 | -1.285 | 0 | 0.259 |
| 238 | LINC00961 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-708-5p;hsa-miR-96-5p | 11 | RGL1 | Sponge network | -2.724 | 0 | -0.968 | 0 | 0.258 |
| 239 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | RGL1 | Sponge network | -1.745 | 0 | -0.968 | 0 | 0.258 |
| 240 | LL22NC03-86G7.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PDGFRA | Sponge network | -1.177 | 3.0E-5 | -0.92 | 0.00039 | 0.257 |
| 241 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-96-5p | 10 | CSF1 | Sponge network | -0.582 | 0.05253 | -0.65 | 0.00787 | 0.257 |
| 242 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 12 | GRIN2A | Sponge network | -1.892 | 0 | -1.285 | 0.03401 | 0.256 |
| 243 | LINC00702 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 13 | GRIN2A | Sponge network | -2.856 | 0 | -1.285 | 0.03401 | 0.255 |
| 244 | LINC00619 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p | 10 | PIK3R1 | Sponge network | -2.307 | 0.02217 | -1.285 | 0 | 0.255 |
| 245 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | 0.053 | 0.85755 | -1.285 | 0 | 0.255 |
| 246 | AC011526.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | PDGFRA | Sponge network | -2.783 | 0 | -0.92 | 0.00039 | 0.254 |
| 247 | CTA-221G9.11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.645 | 0.06404 | -1.285 | 0 | 0.254 |
| 248 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -0.672 | 0.02084 | -1.285 | 0 | 0.254 |
| 249 | AC011899.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181d-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GAB1 | Sponge network | -2.611 | 0 | -1.505 | 0 | 0.254 |
| 250 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | CSF1 | Sponge network | -4.19 | 0 | -0.65 | 0.00787 | 0.253 |
| 251 | CTD-2013N24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | NTRK2 | Sponge network | -1.745 | 0 | -2.564 | 0 | 0.252 |
| 252 | RP11-378A13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-708-5p;hsa-miR-93-5p | 12 | RGL1 | Sponge network | -1.713 | 0 | -0.968 | 0 | 0.251 |
| 253 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-92a-3p | 11 | RASAL2 | Sponge network | -1.892 | 0 | -0.753 | 0 | 0.251 |