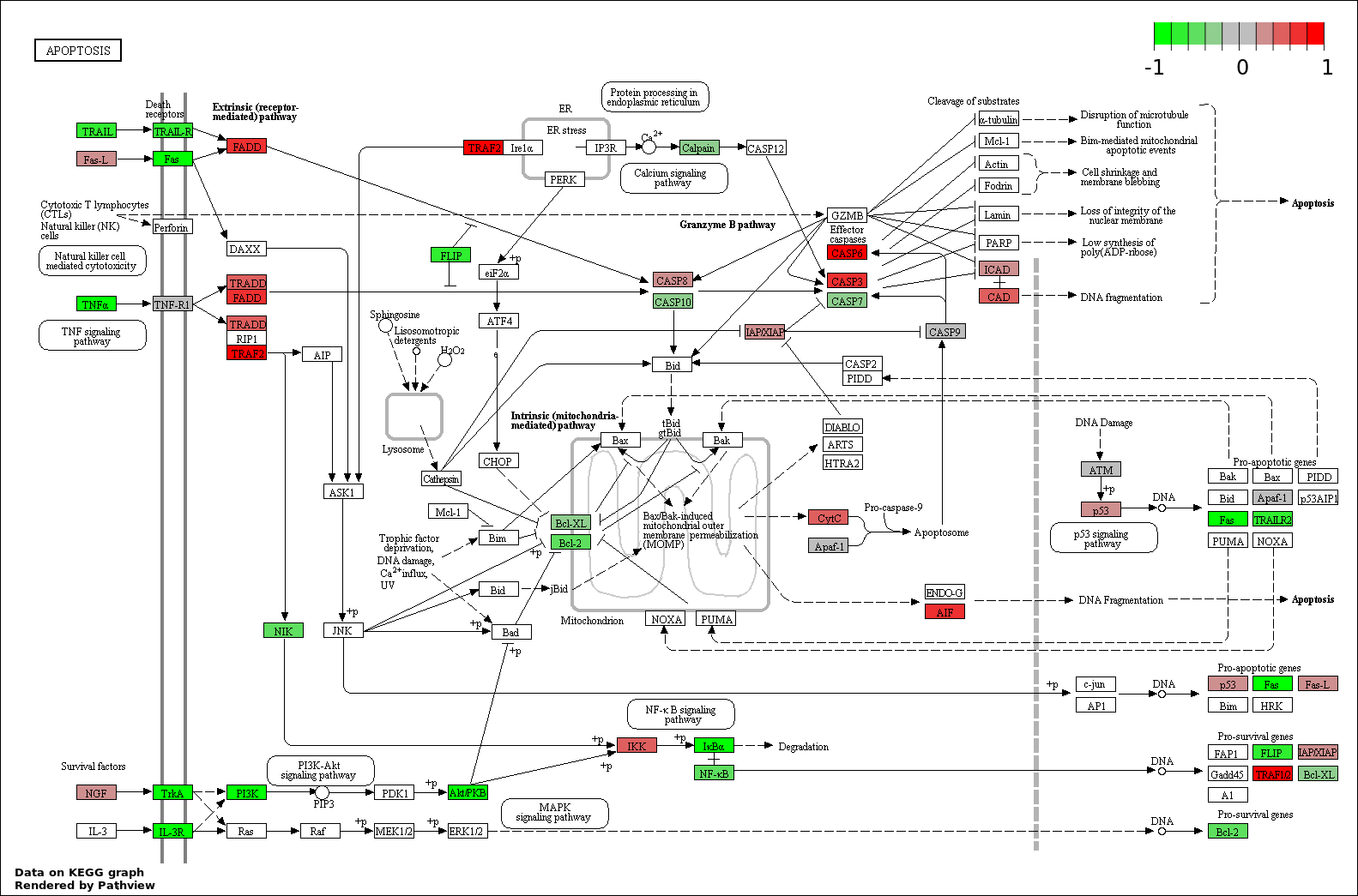
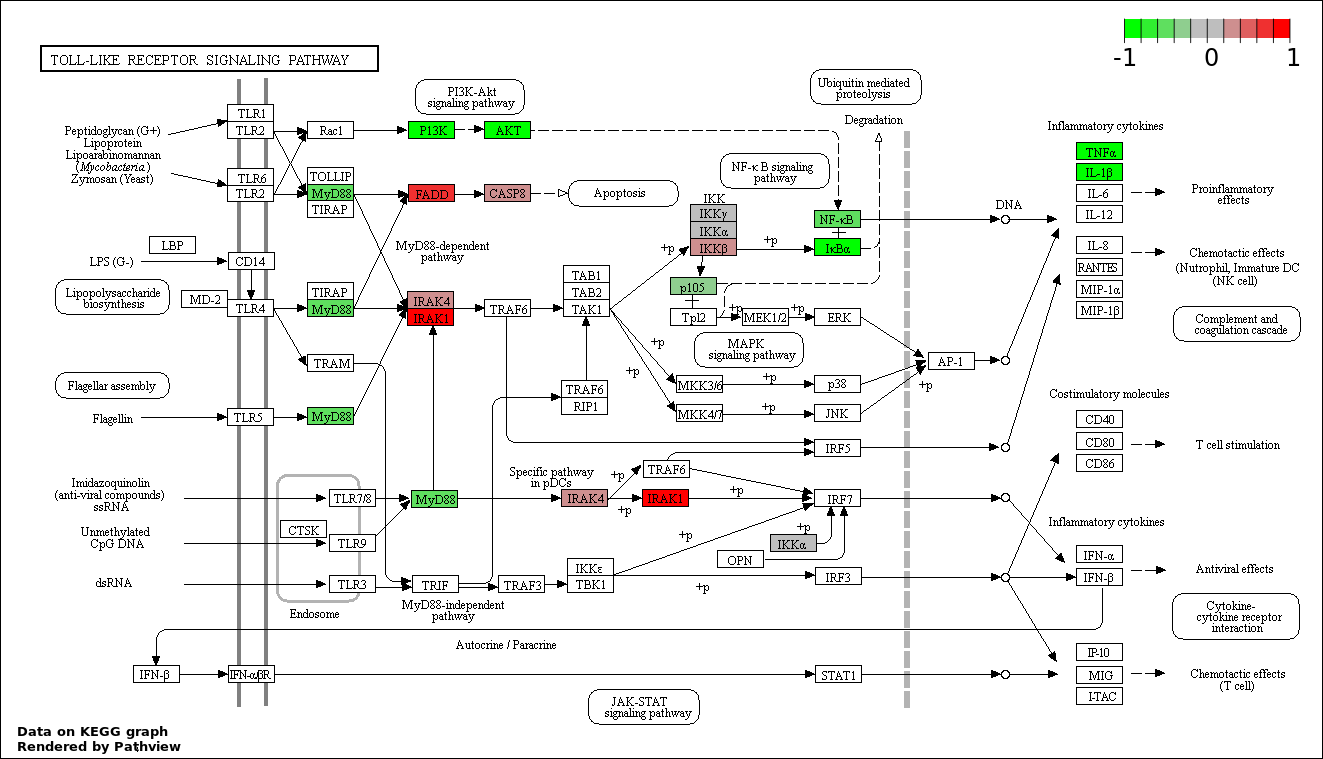
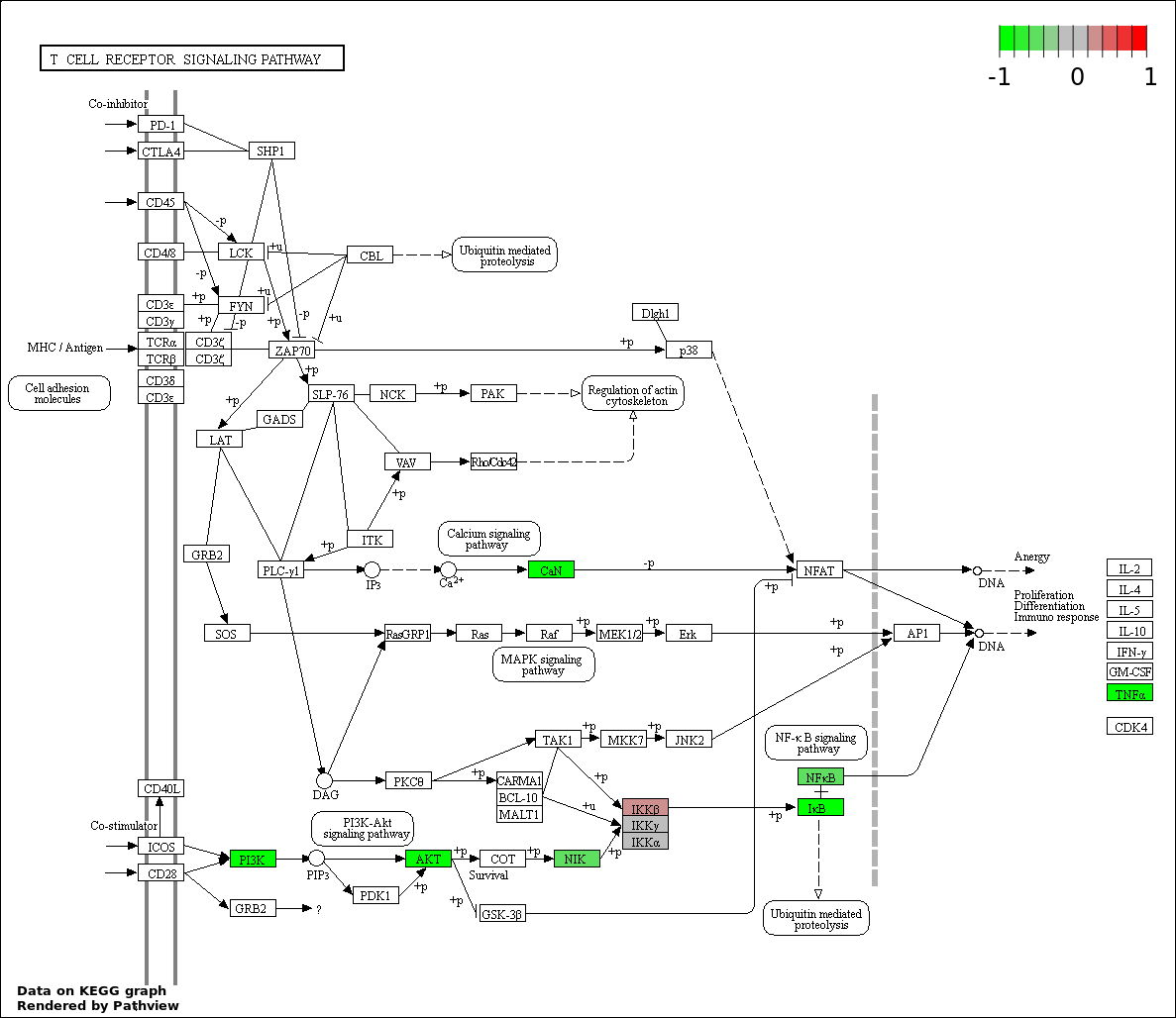
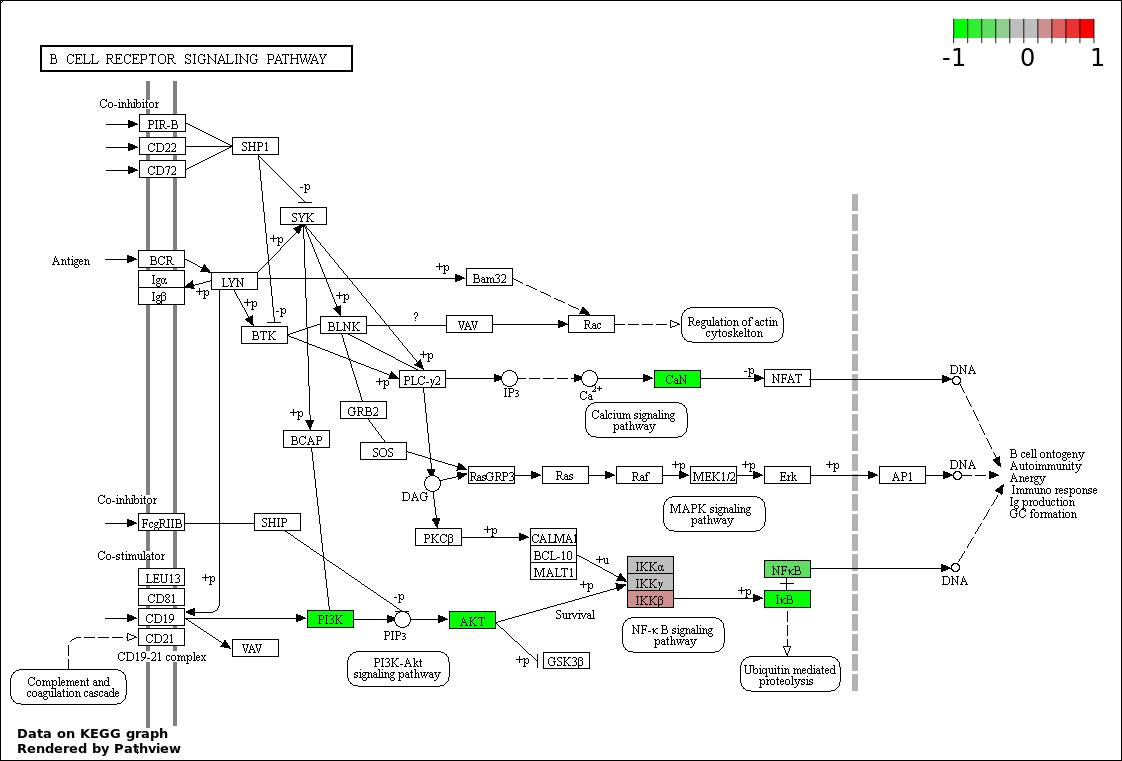
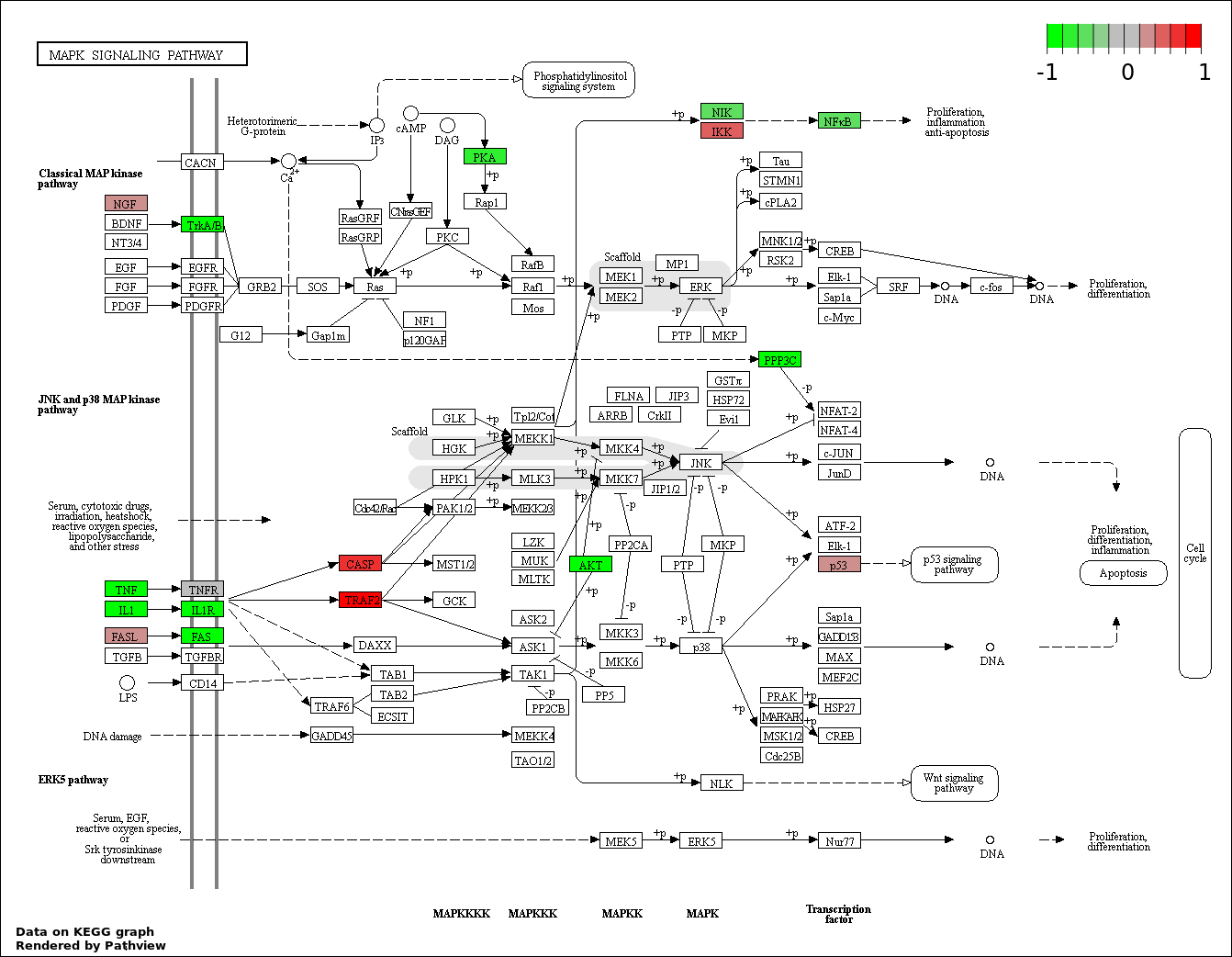
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-5p | AIFM1 | -1.37 | 0 | 0.7 | 0 | TargetScan; miRNATAP | -0.11 | 0.00018 | NA | |
| 2 | hsa-let-7b-5p | AIFM1 | -1.62 | 0 | 0.7 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 3 | hsa-miR-125a-5p | AIFM1 | -1.05 | 0 | 0.7 | 0 | miRanda | -0.08 | 0.00249 | NA | |
| 4 | hsa-miR-125b-5p | AIFM1 | -0.55 | 0.01072 | 0.7 | 0 | miRNATAP | -0.09 | 0.00028 | NA | |
| 5 | hsa-miR-145-5p | AIFM1 | -1.35 | 0 | 0.7 | 0 | miRNATAP | -0.09 | 1.0E-5 | 20332243 | Artificial overexpression of miR145 by using adenoviral vectors in prostate cancer PC-3 and DU145 cells significantly downregulated BNIP3 together with the upregulation of AIF reduced cell growth and increased cell death |
| 6 | hsa-miR-155-5p | AIFM1 | 0.81 | 0.00061 | 0.7 | 0 | miRNAWalker2 validate | -0.06 | 0.01116 | NA | |
| 7 | hsa-miR-199a-5p | AIFM1 | 1.31 | 0 | 0.7 | 0 | miRanda | -0.06 | 0.00903 | NA | |
| 8 | hsa-miR-143-3p | AKT1 | -1.21 | 1.0E-5 | -0.13 | 0.25348 | miRNAWalker2 validate | -0.06 | 0.00284 | NA | |
| 9 | hsa-let-7a-5p | AKT2 | -1.37 | 0 | 0.16 | 0.1135 | TargetScan | -0.12 | 0 | NA | |
| 10 | hsa-miR-29a-3p | AKT2 | 0.1 | 0.5732 | 0.16 | 0.1135 | MirTarget | -0.1 | 4.0E-5 | 24076586 | Furthermore a feed-back loop comprising of c-Myc miR-29 family and Akt2 were found in myeloid leukemogenesis |
| 11 | hsa-miR-29c-3p | AKT2 | 1.32 | 0 | 0.16 | 0.1135 | MirTarget | -0.07 | 5.0E-5 | NA | |
| 12 | hsa-miR-30d-3p | AKT2 | 0 | 0.98646 | 0.16 | 0.1135 | mirMAP | -0.05 | 0.01424 | NA | |
| 13 | hsa-miR-30e-3p | AKT2 | -0.1 | 0.52624 | 0.16 | 0.1135 | mirMAP | -0.07 | 0.01256 | NA | |
| 14 | hsa-miR-106a-5p | AKT3 | 1.39 | 6.0E-5 | -1.44 | 0 | miRNATAP | -0.09 | 0.01547 | NA | |
| 15 | hsa-miR-106b-5p | AKT3 | 1.47 | 0 | -1.44 | 0 | miRNATAP | -0.16 | 0.00426 | NA | |
| 16 | hsa-miR-107 | AKT3 | 0.66 | 0 | -1.44 | 0 | PITA; miRanda | -0.26 | 0.0031 | NA | |
| 17 | hsa-miR-142-3p | AKT3 | 3.98 | 0 | -1.44 | 0 | miRanda | -0.1 | 0.0026 | NA | |
| 18 | hsa-miR-146b-5p | AKT3 | 1.09 | 1.0E-5 | -1.44 | 0 | miRNAWalker2 validate | -0.15 | 0.00189 | NA | |
| 19 | hsa-miR-15a-5p | AKT3 | 1.63 | 0 | -1.44 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.41 | 0 | NA | |
| 20 | hsa-miR-16-5p | AKT3 | 0.75 | 0 | -1.44 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.27 | 0.0001 | NA | |
| 21 | hsa-miR-17-3p | AKT3 | 1.37 | 0 | -1.44 | 0 | miRNATAP | -0.15 | 0.01027 | NA | |
| 22 | hsa-miR-17-5p | AKT3 | 2.07 | 0 | -1.44 | 0 | TargetScan; miRNATAP | -0.22 | 0 | NA | |
| 23 | hsa-miR-181a-5p | AKT3 | -0.38 | 0.05621 | -1.44 | 0 | miRNATAP | -0.23 | 9.0E-5 | NA | |
| 24 | hsa-miR-181b-5p | AKT3 | 0.67 | 0.00024 | -1.44 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 25 | hsa-miR-20a-5p | AKT3 | 2.65 | 0 | -1.44 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 26 | hsa-miR-22-3p | AKT3 | 1.43 | 0 | -1.44 | 0 | miRNATAP | -0.26 | 0.00109 | NA | |
| 27 | hsa-miR-28-3p | AKT3 | 0.39 | 0.00778 | -1.44 | 0 | miRNATAP | -0.2 | 0.01226 | NA | |
| 28 | hsa-miR-29a-3p | AKT3 | 0.1 | 0.5732 | -1.44 | 0 | miRNATAP | -0.15 | 0.02016 | NA | |
| 29 | hsa-miR-29b-2-5p | AKT3 | 0.35 | 0.19484 | -1.44 | 0 | mirMAP | -0.18 | 2.0E-5 | NA | |
| 30 | hsa-miR-29b-3p | AKT3 | 3.11 | 0 | -1.44 | 0 | miRNATAP | -0.27 | 0 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 31 | hsa-miR-3065-5p | AKT3 | 0.65 | 0.09995 | -1.44 | 0 | mirMAP | -0.18 | 0 | NA | |
| 32 | hsa-miR-335-3p | AKT3 | 1.51 | 0 | -1.44 | 0 | mirMAP | -0.13 | 0.01067 | NA | |
| 33 | hsa-miR-362-5p | AKT3 | 0.66 | 0.02433 | -1.44 | 0 | PITA; TargetScan; miRNATAP | -0.23 | 0 | NA | |
| 34 | hsa-miR-424-5p | AKT3 | 1.26 | 1.0E-5 | -1.44 | 0 | miRNATAP | -0.09 | 0.02225 | 26315541 | Silencing Akt3 and E2F3 by siRNA pheno-copied the effect of ectopic miR-424 on HCC growth; Whereas overexpression of Akt3 and E2F3 attenuated the effect of miR-424 on HCC growth |
| 35 | hsa-miR-542-3p | AKT3 | 1.62 | 0 | -1.44 | 0 | miRanda | -0.19 | 1.0E-5 | NA | |
| 36 | hsa-miR-93-5p | AKT3 | 1.51 | 0 | -1.44 | 0 | miRNATAP | -0.25 | 0 | NA | |
| 37 | hsa-miR-195-3p | APAF1 | -1.33 | 0 | 0.05 | 0.6283 | mirMAP | -0.06 | 0.00936 | NA | |
| 38 | hsa-miR-23a-3p | APAF1 | 0.11 | 0.39309 | 0.05 | 0.6283 | miRNATAP | -0.13 | 0.00069 | 24992592; 24249161 | Luciferase assay was performed to verify a putative target site of miR-23a in the 3'-UTR of apoptosis protease activating factor 1 APAF1 mRNA; The expression levels of miR-23a and APAF1 in CRC cell lines SW480 and SW620 and clinical samples were assessed using reverse transcription-quantitative real-time PCR RT-qPCR and Western blot; Moreover miR-23a up-regulation was coupled with APAF1 down-regulation in CRC tissue samples;We found that the expression of miR-23a was increased and the level of apoptosis-activating factor-1 APAF-1 was decreased in 5-FU-treated colon cancer cells compared to untreated cells; APAF-1 as a target gene of miR-23a was identified and miR-23a antisense-induced increase in the activation of caspase-9 was observed |
| 39 | hsa-miR-23b-3p | APAF1 | -0.07 | 0.62059 | 0.05 | 0.6283 | miRNATAP | -0.11 | 0.00216 | NA | |
| 40 | hsa-miR-27a-3p | APAF1 | 0.43 | 0.00737 | 0.05 | 0.6283 | miRNATAP | -0.16 | 0 | NA | |
| 41 | hsa-miR-27b-3p | APAF1 | 0.24 | 0.12264 | 0.05 | 0.6283 | miRNATAP | -0.17 | 0 | NA | |
| 42 | hsa-miR-374b-5p | APAF1 | 0.47 | 0.01092 | 0.05 | 0.6283 | mirMAP | -0.07 | 0.01286 | NA | |
| 43 | hsa-miR-664a-3p | APAF1 | 0.44 | 0.02142 | 0.05 | 0.6283 | mirMAP | -0.11 | 2.0E-5 | NA | |
| 44 | hsa-miR-125a-3p | ATM | -1.05 | 0 | 0.08 | 0.53013 | miRanda | -0.06 | 0.02074 | NA | |
| 45 | hsa-miR-18a-5p | ATM | 1.37 | 1.0E-5 | 0.08 | 0.53013 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.08 | 0.0004 | 23437304; 25963391; 23857602; 23229340 | MicroRNA 18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer; Through in silico search the 3'UTR of Ataxia telangiectasia mutated ATM contains a conserved miR-18a binding site; Expression of ATM was down-regulated in CRC tumors p<0.0001 and inversely correlated with miR-18a expression r = -0.4562 p<0.01; This was further confirmed by the down-regulation of ATM protein by miR-18a; As ATM is a key enzyme in DNA damage repair we evaluated the effect of miR-18a on DNA double-strand breaks; miR-18a attenuates cellular repair of DNA double-strand breaks by directly suppressing ATM a key enzyme in DNA damage repair;However the upregulation of miR-18a suppressed the level of ataxia-telangiectasia mutated and attenuated DNA double-strand break repair after irradiation which re-sensitized the cervical cancer cells to radiotherapy by promoting apoptosis;Furthermore we used antisense oligonucleotides against micro RNAs miRNA or miRNA overexpression plasmids to study the role of miR-18a and -106a on ATM expression; Furthermore we identified that ERα activates miR-18a and -106a to downregulate ATM expression; We reveal a novel mechanism involving ERα and miR-18a and -106a regulation of ATM in breast cancer;MicroRNA 18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells; Previous studies showed that certain microRNAs including miR-18a potentially regulate ATM in cancer cells; However the mechanisms behind the modulation of ATM by miR-18a remain to be elucidated in colon cancer cells; In the present study we explored the impact of miR-18a on the autophagy process and ATM expression in HCT116 colon cancer cells; Western blotting and luciferase assays were implemented to explore the impact of miR-18a on ATM gene expression in HCT116 cells; Moreover miR-18a overexpression led to the upregulation of ATM expression and suppression of mTORC1 activity; Results of the present study pertaining to the role of miR-18a in regulating autophagy and ATM gene expression in colon cancer cells revealed a novel function for miR-18a in a critical cellular event and on a crucial gene with significant impacts in cancer development progression treatment and in other diseases |
| 46 | hsa-miR-590-5p | ATM | 2.07 | 0 | 0.08 | 0.53013 | mirMAP | -0.07 | 0.00604 | NA | |
| 47 | hsa-miR-15a-5p | BCL2 | 1.63 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.19 | 0.00401 | 26915294; 25594541; 18931683; 25623762; 22335947 | As a result transcript levels of the tumor-suppressive miR-15 and let-7 families increased which targeted and decreased the expression of the crucial prosurvival genes BCL-2 and BCL-XL respectively;MicroRNAs miRNAs encoded by the miR-15 cluster are known to induce G1 arrest and apoptosis by targeting G1 checkpoints and the anti-apoptotic B cell lymphoma 2 BCL-2 gene;MicroRNAs miRNAs are noncoding small RNAs that repress protein translation by targeting specific messenger RNAs miR-15a and miR-16-1 act as putative tumor suppressors by targeting the oncogene BCL2;miR 15a and miR 16 modulate drug resistance by targeting bcl 2 in human colon cancer cells; To investigate the reversal effect of targeted modulation of bcl-2 expression by miR-15a and miR-16 on drug resistance of human colon cancer cells;The expression of MiR-15a was significantly inhibited by Bcl-2 P < 0.05 |
| 48 | hsa-miR-17-5p | BCL2 | 2.07 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.12 | 0.02174 | 25435430 | Combined overexpression of miR-16 and miR-17 greatly reduced Beclin-1 and Bcl-2 expressions respectively; miR-17 overexpression reduced cytoprotective autophagy by targeting Beclin-1 whereas overexpression of miR-16 potentiated paclitaxel induced apoptotic cell death by inhibiting anti-apoptotic protein Bcl-2 |
| 49 | hsa-miR-200c-3p | BCL2 | 0.38 | 0.08422 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.13 | 0.02205 | NA | |
| 50 | hsa-miR-20a-3p | BCL2 | 2.52 | 0 | -0.49 | 0.06421 | mirMAP | -0.1 | 0.01974 | NA | |
| 51 | hsa-miR-20a-5p | BCL2 | 2.65 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.12 | 0.01368 | NA | |
| 52 | hsa-miR-21-5p | BCL2 | 4.38 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.2 | 1.0E-5 | 22964582; 21468550; 25994220; 25381586; 26555418; 23359184; 21376256 | Tumors harvested from these lungs have elevated levels of oncogenic miRNAs miR-21 and miR-155; are deficient for p53-regulated miRNAs; and have heightened expression of miR-34 target genes such as Met and Bcl-2;BCL-2 up-regulation could be achieved by miR-21 overexpression which prevented T24 cells from apoptosis induced by doxorubicin; Furthermore the miR-21 induced BCL-2 up-regulation could be cancelled by the PI3K inhibitor LY294002;Meanwhile miR-21 loss reduced STAT3 and Bcl-2 activation causing an increase in the apoptosis of tumour cells in CAC mice;Changes in the sensitivity of osteosarcoma cells to CDDP were examined after transfection with miR-21 mimics or anti-miR-21 or bcl-2 siRNA in combination with CDDP;The expression of Bax Bcl-2 and miR-21 in parental and paclitaxel-resistant cells was detected by RT-PCR and Western blotting;Resveratrol induces apoptosis of pancreatic cancers cells by inhibiting miR 21 regulation of BCL 2 expression; We also used Western blot to measure BCL-2 protein levels after down-regulation of miR-21 expression; Besides down-regulation of miR-21 expression can inhibit BCL-2 expression in PANC-1 CFPAC-1 and MIA Paca-2 cells; Over-expression of miR-21 expression can reverse down-regulation of BCL-2 expression and apoptosis induced by resveratrol; In this study we demonstrated that the effect of resveratrol on apoptosis is due to inhibiting miR-21 regulation of BCL-2 expression;Bcl 2 upregulation induced by miR 21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa 2 pancreatic cancer cells; However the roles and mechanisms of miRNA miR-21 in regulation of Bcl-2 in pancreatic cancer remain to be elucidated; Then luciferase activity was observed after miR-21 mimics and pRL-TK plasmids containing wild-type and mutant 3'UTRs of Bcl-2 mRNA were co-transfected; Cells transfected with miR-21 inhibitor revealed an opposite trend. There was a significant increase in luciferase activity in the cells transfected with the wild-type pRL-TK plasmid in contrast to those transfected with the mutant one indicating that miR-21 promotes Bcl-2 expression by binding directly to the 3'UTR of Bcl-2 mRNA; Upregulation of Bcl-2 directly induced by miR-21 is associated with apoptosis chemoresistance and proliferation of MIA PaCa-2 pancreatic cancer cells |
| 53 | hsa-miR-224-5p | BCL2 | 1.92 | 0 | -0.49 | 0.06421 | mirMAP | -0.14 | 0 | 24796455 | In addition the expressions of Bcl2 mRNA and protein were 1.05 ± 0.04 and 0.21 ± 0.03 in the miR-224 ASO group significantly lower than that in the control group 4.87 ± 0.96 and 0.88 ± 0.09 P < 0.01 |
| 54 | hsa-miR-24-2-5p | BCL2 | 1.44 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.16 | 0.00352 | NA | |
| 55 | hsa-miR-29a-5p | BCL2 | 1.9 | 0 | -0.49 | 0.06421 | mirMAP | -0.24 | 0 | 20041405 | Subsequent investigation characterized two antiapoptotic molecules Bcl-2 and Mcl-1 as direct targets of miR-29; Furthermore silencing of Bcl-2 and Mcl-1 phenocopied the proapoptotic effect of miR-29 whereas overexpression of these proteins attenuated the effect of miR-29 |
| 56 | hsa-miR-29b-3p | BCL2 | 3.11 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.1 | 0.02485 | 20041405 | Subsequent investigation characterized two antiapoptotic molecules Bcl-2 and Mcl-1 as direct targets of miR-29; Furthermore silencing of Bcl-2 and Mcl-1 phenocopied the proapoptotic effect of miR-29 whereas overexpression of these proteins attenuated the effect of miR-29 |
| 57 | hsa-miR-3065-5p | BCL2 | 0.65 | 0.09995 | -0.49 | 0.06421 | mirMAP | -0.12 | 0.00023 | NA | |
| 58 | hsa-miR-33a-5p | BCL2 | 2.8 | 0 | -0.49 | 0.06421 | mirMAP | -0.1 | 0.00701 | NA | |
| 59 | hsa-miR-34a-5p | BCL2 | 1.41 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.19 | 0.00135 | 22964582; 24565525; 23155233; 24444609; 20687223; 22623155; 24988056; 18803879; 19714243; 25053345; 20433755; 21399894; 23862748 | Tumors harvested from these lungs have elevated levels of oncogenic miRNAs miR-21 and miR-155; are deficient for p53-regulated miRNAs; and have heightened expression of miR-34 target genes such as Met and Bcl-2;In vitro and in vivo experiments showed that miR-34a and DOX can be efficiently encapsulated into HA-CS NPs and delivered into tumor cells or tumor tissues and enhance anti-tumor effects of DOX by suppressing the expression of non-pump resistance and anti-apoptosis proto-oncogene Bcl-2;The miR-34a expression levels in cells after irradiation at 30 and 60 Gy were 0.17- and 18.7-times the BCL2 and caspase-9 expression levels respectively;Functional analyses further indicate that restoration of miR-34a inhibits B cell lymphoma-2 Bcl-2 protein expression to withdraw the survival advantage of these resistant NSCLC cells;Thus in PC3PR cells reduced expression of miR-34a confers paclitaxel resistance via up-regulating SIRT1 and Bcl2 expression; MiR-34a and its downstream targets SIRT1 and Bcl2 play important roles in the development of paclitaxel resistance all of which can be useful biomarkers and promising therapeutic targets for the drug resistance in hormone-refractory prostate cancer;MiR 34a inhibits proliferation and migration of breast cancer through down regulation of Bcl 2 and SIRT1; In this study we aimed to determine the effect of miR-34a on the growth of breast cancer and to investigate whether its effect is achieved by targeting Bcl-2 and SIRT1; Bcl-2 and SIRT1 as the targets of miR-34a were found to be in reverse correlation with ectopic expression of miR-34a;Target analysis indicated that micro RNA miR-34a directly regulates Bcl-2 and miR-34a overexpression decreased Bcl-2 protein level in gastric cancer cells; We also found that luteolin upregulates miR-34a expression and downregulates Bcl-2 expression; Based on these results we can draw the conclusion that luteolin partly decreases Bcl-2 expression through upregulating miR-34a expression;miR-34 targets Notch HMGA2 and Bcl-2 genes involved in the self-renewal and survival of cancer stem cells; Human gastric cancer cells were transfected with miR-34 mimics or infected with the lentiviral miR-34-MIF expression system and validated by miR-34 reporter assay using Bcl-2 3'UTR reporter; Human gastric cancer Kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2 Notch and HMGA2; Bcl-2 3'UTR reporter assay showed that the transfected miR-34s were functional and confirmed that Bcl-2 is a direct target of miR-34; The mechanism of miR-34-mediated suppression of self-renewal appears to be related to the direct modulation of downstream targets Bcl-2 Notch and HMGA2 indicating that miR-34 may be involved in gastric cancer stem cell self-renewal/differentiation decision-making;Among the target proteins regulated by miR-34 are Notch pathway proteins and Bcl-2 suggesting the possibility of a role for miR-34 in the maintenance and survival of cancer stem cells; Our data support the view that miR-34 may be involved in pancreatic cancer stem cell self-renewal potentially via the direct modulation of downstream targets Bcl-2 and Notch implying that miR-34 may play an important role in pancreatic cancer stem cell self-renewal and/or cell fate determination;Manipulating miR-34a in prostate cancer cells confirms that this miRNA regulates BCL-2 and may in part regulate response to docetaxel;For instance miR-34a up-regulation corresponded with a down-regulation of BCL2 protein; Treating Par-4-overexpressing HT29 cells with a miR-34a antagomir functionally reversed both BCL2 down-regulation and apoptosis by 5-FU;Quantitative PCR and western analysis confirmed decreased expression of two genes BCL-2 and CCND1 in docetaxel-resistant cells which are both targeted by miR-34a;MicroRNA 34a targets Bcl 2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment; HCC tissues with lower miR-34a expression displayed higher expression of Bcl-2 protein than those with high expression of miR-34a; therefore an inverse correlation is evident between the miR-34a level and Bcl-2 expression; Bioinformatics and luciferase reporter assays revealed that miR-34a binds the 3'-UTR of the Bcl-2 mRNA and represses its translation; Western blotting analysis and qRT-PCR confirmed that Bcl-2 is inhibited by miR-34a overexpression; Functional analyses indicated that the restoration of miR-34a reduced cell viability promoted cell apoptosis and potentiated sorafenib-induced apoptosis and toxicity in HCC cell lines by inhibiting Bcl-2 expression |
| 60 | hsa-miR-365a-3p | BCL2 | 0.01 | 0.9536 | -0.49 | 0.06421 | miRNAWalker2 validate; miRTarBase | -0.14 | 0.01107 | NA | |
| 61 | hsa-miR-450b-5p | BCL2 | 1.69 | 0 | -0.49 | 0.06421 | mirMAP | -0.2 | 0 | NA | |
| 62 | hsa-miR-452-5p | BCL2 | 0.64 | 0.04582 | -0.49 | 0.06421 | mirMAP | -0.12 | 0.00113 | NA | |
| 63 | hsa-miR-542-3p | BCL2 | 1.62 | 0 | -0.49 | 0.06421 | mirMAP | -0.19 | 3.0E-5 | NA | |
| 64 | hsa-miR-590-3p | BCL2 | 0.84 | 0.00129 | -0.49 | 0.06421 | miRanda; mirMAP | -0.12 | 0.02705 | NA | |
| 65 | hsa-miR-590-5p | BCL2 | 2.07 | 0 | -0.49 | 0.06421 | miRanda | -0.18 | 0.00054 | NA | |
| 66 | hsa-miR-96-5p | BCL2 | 3.04 | 0 | -0.49 | 0.06421 | miRNAWalker2 validate; TargetScan | -0.14 | 0.00193 | NA | |
| 67 | hsa-miR-326 | BCL2L1 | -0.99 | 0.00335 | -0.25 | 0.07732 | PITA; miRanda; mirMAP; miRNATAP | -0.06 | 0.00125 | NA | |
| 68 | hsa-miR-330-5p | BCL2L1 | 0.17 | 0.33643 | -0.25 | 0.07732 | PITA; miRanda; miRNATAP | -0.08 | 0.03195 | NA | |
| 69 | hsa-miR-421 | BCL2L1 | 0.17 | 0.53528 | -0.25 | 0.07732 | miRanda | -0.09 | 0.00056 | NA | |
| 70 | hsa-miR-204-5p | BIRC2 | -1.02 | 0.00767 | -0.06 | 0.62026 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.08 | 0 | NA | |
| 71 | hsa-miR-29a-3p | BIRC2 | 0.1 | 0.5732 | -0.06 | 0.62026 | MirTarget | -0.06 | 0.04035 | NA | |
| 72 | hsa-miR-29b-3p | BIRC2 | 3.11 | 0 | -0.06 | 0.62026 | MirTarget | -0.06 | 0.00214 | NA | |
| 73 | hsa-miR-500a-5p | BIRC2 | 0.65 | 0.01047 | -0.06 | 0.62026 | MirTarget | -0.11 | 0 | NA | |
| 74 | hsa-miR-624-5p | BIRC2 | -0.06 | 0.82985 | -0.06 | 0.62026 | MirTarget | -0.05 | 0.02001 | NA | |
| 75 | hsa-miR-24-1-5p | BIRC3 | 0.86 | 0.00011 | 0.13 | 0.67648 | MirTarget | -0.21 | 0.00165 | NA | |
| 76 | hsa-miR-375 | BIRC3 | 0.62 | 0.1492 | 0.13 | 0.67648 | miRNAWalker2 validate | -0.2 | 0 | 23726271 | Taken together these data suggest that miR-375 sensitizes TNF-α-induced apoptosis and the reduction in the expression of the apoptosis inhibitory proteins cFLIP-L and cIAP2 plays an important role in this sensitization |
| 77 | hsa-miR-664a-3p | BIRC3 | 0.44 | 0.02142 | 0.13 | 0.67648 | mirMAP | -0.17 | 0.0216 | NA | |
| 78 | hsa-miR-98-5p | BIRC3 | 1.17 | 0 | 0.13 | 0.67648 | miRNAWalker2 validate | -0.14 | 0.0476 | NA | |
| 79 | hsa-miR-107 | CAPN2 | 0.66 | 0 | -0.31 | 0.05176 | miRanda | -0.13 | 0.02025 | NA | |
| 80 | hsa-miR-1468-5p | CAPN2 | -0.9 | 0.00072 | -0.31 | 0.05176 | MirTarget | -0.06 | 0.03764 | NA | |
| 81 | hsa-miR-16-2-3p | CAPN2 | 0.5 | 0.02636 | -0.31 | 0.05176 | mirMAP | -0.11 | 0.00157 | NA | |
| 82 | hsa-miR-20a-3p | CAPN2 | 2.52 | 0 | -0.31 | 0.05176 | MirTarget | -0.13 | 0 | NA | |
| 83 | hsa-miR-320b | CAPN2 | 0.23 | 0.37882 | -0.31 | 0.05176 | miRanda | -0.12 | 1.0E-5 | NA | |
| 84 | hsa-miR-421 | CAPN2 | 0.17 | 0.53528 | -0.31 | 0.05176 | miRanda | -0.16 | 0 | NA | |
| 85 | hsa-miR-590-3p | CAPN2 | 0.84 | 0.00129 | -0.31 | 0.05176 | miRanda | -0.1 | 0.00294 | NA | |
| 86 | hsa-miR-590-5p | CAPN2 | 2.07 | 0 | -0.31 | 0.05176 | miRanda | -0.16 | 0 | NA | |
| 87 | hsa-miR-7-5p | CAPN2 | 0.34 | 0.43506 | -0.31 | 0.05176 | miRNAWalker2 validate | -0.08 | 6.0E-5 | NA | |
| 88 | hsa-miR-129-5p | CASP10 | -0.41 | 0.34149 | -0.27 | 0.09806 | mirMAP | -0.06 | 0.00151 | NA | |
| 89 | hsa-miR-148b-5p | CASP10 | 1.39 | 0 | -0.27 | 0.09806 | mirMAP | -0.14 | 2.0E-5 | NA | |
| 90 | hsa-miR-19a-3p | CASP10 | 2.12 | 0 | -0.27 | 0.09806 | MirTarget; mirMAP | -0.1 | 6.0E-5 | NA | |
| 91 | hsa-miR-19b-1-5p | CASP10 | 1.71 | 0 | -0.27 | 0.09806 | mirMAP | -0.18 | 0 | NA | |
| 92 | hsa-miR-19b-3p | CASP10 | 2.11 | 0 | -0.27 | 0.09806 | MirTarget; mirMAP | -0.12 | 0.00012 | NA | |
| 93 | hsa-miR-200c-3p | CASP10 | 0.38 | 0.08422 | -0.27 | 0.09806 | mirMAP | -0.07 | 0.048 | NA | |
| 94 | hsa-miR-320b | CASP10 | 0.23 | 0.37882 | -0.27 | 0.09806 | miRanda | -0.09 | 0.00149 | NA | |
| 95 | hsa-miR-421 | CASP10 | 0.17 | 0.53528 | -0.27 | 0.09806 | mirMAP | -0.17 | 0 | NA | |
| 96 | hsa-miR-744-3p | CASP10 | 2.07 | 0 | -0.27 | 0.09806 | mirMAP | -0.13 | 0 | NA | |
| 97 | hsa-let-7a-5p | CASP3 | -1.37 | 0 | 0.75 | 0 | miRNAWalker2 validate; MirTarget; TargetScan; miRNATAP | -0.09 | 0.00189 | NA | |
| 98 | hsa-let-7b-5p | CASP3 | -1.62 | 0 | 0.75 | 0 | MirTarget; miRNATAP | -0.07 | 0.01162 | NA | |
| 99 | hsa-let-7c-5p | CASP3 | -2.14 | 0 | 0.75 | 0 | MirTarget | -0.07 | 0.00043 | NA | |
| 100 | hsa-miR-139-5p | CASP3 | -2.27 | 0 | 0.75 | 0 | miRanda | -0.1 | 0 | NA | |
| 101 | hsa-miR-195-3p | CASP3 | -1.33 | 0 | 0.75 | 0 | MirTarget | -0.12 | 0 | NA | |
| 102 | hsa-miR-30a-5p | CASP3 | -0.92 | 0.00076 | 0.75 | 0 | miRNATAP | -0.09 | 0 | NA | |
| 103 | hsa-miR-30c-5p | CASP3 | -0.33 | 0.1236 | 0.75 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 104 | hsa-miR-30d-5p | CASP3 | -0.92 | 4.0E-5 | 0.75 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.17 | 0 | NA | |
| 105 | hsa-miR-362-3p | CASP3 | 0.19 | 0.52808 | 0.75 | 0 | miRanda | -0.06 | 0.00859 | NA | |
| 106 | hsa-miR-374a-5p | CASP3 | -0.2 | 0.29808 | 0.75 | 0 | mirMAP | -0.07 | 0.0379 | NA | |
| 107 | hsa-miR-125a-5p | CASP6 | -1.05 | 0 | 0.9 | 0 | miRanda | -0.12 | 0.00016 | NA | |
| 108 | hsa-miR-106b-5p | CASP7 | 1.47 | 0 | -0.21 | 0.08139 | miRNAWalker2 validate | -0.06 | 0.01907 | 22986525 | MicroRNA 106b 25 cluster expression is associated with early disease recurrence and targets caspase 7 and focal adhesion in human prostate cancer; Moreover increased tumor miR-106b expression was associated with disease recurrence and the combination of high miR-106b and low CASP7 caspase-7 expressions in primary tumors was an independent predictor of early disease recurrence adjusted hazard ratio=4.1; 95% confidence interval: 1.6-12.3; The approach revealed that CASP7 is a direct target of miR-106b which was confirmed by western blot analysis and a 3'-untranslated region reporter assay; Moreover selected phenotypes induced by miR-106b knockdown in DU145 human prostate cancer cells did not develop when both miR-106b and CASP7 expression were inhibited |
| 109 | hsa-miR-664a-3p | CASP7 | 0.44 | 0.02142 | -0.21 | 0.08139 | MirTarget | -0.11 | 0.00018 | NA | |
| 110 | hsa-miR-17-5p | CASP8 | 2.07 | 0 | 0.38 | 0.00232 | MirTarget | -0.07 | 0.00207 | NA | |
| 111 | hsa-miR-19b-1-5p | CASP8 | 1.71 | 0 | 0.38 | 0.00232 | miRNAWalker2 validate | -0.07 | 0.00768 | NA | |
| 112 | hsa-miR-19b-3p | CASP8 | 2.11 | 0 | 0.38 | 0.00232 | MirTarget | -0.05 | 0.02505 | NA | |
| 113 | hsa-miR-20a-5p | CASP8 | 2.65 | 0 | 0.38 | 0.00232 | MirTarget | -0.05 | 0.02464 | NA | |
| 114 | hsa-miR-93-5p | CASP8 | 1.51 | 0 | 0.38 | 0.00232 | MirTarget | -0.06 | 0.01825 | NA | |
| 115 | hsa-miR-195-5p | CASP9 | -1.02 | 5.0E-5 | -0.03 | 0.82555 | mirMAP | -0.06 | 0.01207 | NA | |
| 116 | hsa-miR-199a-3p | CASP9 | 1.68 | 0 | -0.03 | 0.82555 | mirMAP | -0.09 | 0.00108 | 23319430 | The techniques used were the MTT assay flow cytometry real-time PCR to assess miR-199a expression as also caspase-8 and caspase-9 activity in HepG2 cells treated with Propofol |
| 117 | hsa-miR-199b-3p | CASP9 | 1.68 | 0 | -0.03 | 0.82555 | mirMAP | -0.09 | 0.00109 | NA | |
| 118 | hsa-miR-222-5p | CASP9 | 0.1 | 0.71933 | -0.03 | 0.82555 | mirMAP | -0.05 | 0.01673 | NA | |
| 119 | hsa-miR-342-5p | CASP9 | -0.38 | 0.0897 | -0.03 | 0.82555 | MirTarget | -0.07 | 0.01107 | NA | |
| 120 | hsa-miR-497-5p | CASP9 | -0.05 | 0.78621 | -0.03 | 0.82555 | mirMAP | -0.08 | 0.00786 | NA | |
| 121 | hsa-miR-107 | CFLAR | 0.66 | 0 | -0.6 | 0.00015 | miRanda | -0.15 | 0.00732 | NA | |
| 122 | hsa-miR-130b-3p | CFLAR | 1.83 | 0 | -0.6 | 0.00015 | mirMAP | -0.17 | 0 | NA | |
| 123 | hsa-miR-186-5p | CFLAR | 0.85 | 0 | -0.6 | 0.00015 | mirMAP | -0.11 | 0.03888 | NA | |
| 124 | hsa-miR-301a-3p | CFLAR | 2.7 | 0 | -0.6 | 0.00015 | mirMAP | -0.06 | 0.01637 | NA | |
| 125 | hsa-miR-320b | CFLAR | 0.23 | 0.37882 | -0.6 | 0.00015 | miRanda | -0.15 | 0 | NA | |
| 126 | hsa-miR-330-5p | CFLAR | 0.17 | 0.33643 | -0.6 | 0.00015 | miRanda | -0.11 | 0.0124 | NA | |
| 127 | hsa-miR-421 | CFLAR | 0.17 | 0.53528 | -0.6 | 0.00015 | miRanda | -0.11 | 0.00019 | NA | |
| 128 | hsa-miR-454-3p | CFLAR | 1.49 | 0 | -0.6 | 0.00015 | mirMAP | -0.11 | 0.0011 | NA | |
| 129 | hsa-miR-550a-5p | CFLAR | 0.6 | 0.03148 | -0.6 | 0.00015 | mirMAP | -0.06 | 0.01426 | NA | |
| 130 | hsa-miR-708-5p | CFLAR | 3.04 | 0 | -0.6 | 0.00015 | mirMAP | -0.1 | 2.0E-5 | NA | |
| 131 | hsa-miR-106b-5p | CHP2 | 1.47 | 0 | -0.7 | 0.27547 | MirTarget | -0.29 | 0.04606 | NA | |
| 132 | hsa-miR-582-5p | CHP2 | 1.08 | 0.00149 | -0.7 | 0.27547 | miRNATAP | -0.17 | 0.04861 | NA | |
| 133 | hsa-miR-148a-3p | CHUK | 2.31 | 0 | 0.04 | 0.74424 | MirTarget | -0.07 | 0.0015 | NA | |
| 134 | hsa-miR-15a-5p | CHUK | 1.63 | 0 | 0.04 | 0.74424 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.16 | 0 | NA | |
| 135 | hsa-miR-15b-5p | CHUK | -1.26 | 0 | 0.04 | 0.74424 | MirTarget | -0.06 | 0.03926 | NA | |
| 136 | hsa-miR-195-5p | CHUK | -1.02 | 5.0E-5 | 0.04 | 0.74424 | miRNAWalker2 validate; MirTarget | -0.06 | 0.00384 | NA | |
| 137 | hsa-miR-23b-3p | CHUK | -0.07 | 0.62059 | 0.04 | 0.74424 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.08 | 0.04582 | NA | |
| 138 | hsa-miR-29b-2-5p | CHUK | 0.35 | 0.19484 | 0.04 | 0.74424 | MirTarget | -0.12 | 0 | NA | |
| 139 | hsa-miR-339-5p | CHUK | 0.54 | 0.04881 | 0.04 | 0.74424 | miRanda | -0.1 | 0 | NA | |
| 140 | hsa-miR-342-3p | CHUK | -0.13 | 0.56103 | 0.04 | 0.74424 | miRanda | -0.18 | 0 | NA | |
| 141 | hsa-miR-362-3p | CHUK | 0.19 | 0.52808 | 0.04 | 0.74424 | miRanda | -0.07 | 0.00121 | NA | |
| 142 | hsa-miR-376c-3p | CHUK | 0.02 | 0.95781 | 0.04 | 0.74424 | MirTarget | -0.06 | 8.0E-5 | NA | |
| 143 | hsa-miR-429 | CHUK | 2.38 | 0 | 0.04 | 0.74424 | miRanda | -0.06 | 0.00053 | NA | |
| 144 | hsa-miR-497-5p | CHUK | -0.05 | 0.78621 | 0.04 | 0.74424 | MirTarget | -0.19 | 0 | NA | |
| 145 | hsa-miR-15b-3p | CSF2RB | 0.8 | 0.0004 | -1.28 | 0 | mirMAP | -0.15 | 0.00822 | NA | |
| 146 | hsa-miR-181d-5p | CSF2RB | 1.19 | 0 | -1.28 | 0 | MirTarget | -0.23 | 8.0E-5 | NA | |
| 147 | hsa-miR-19a-3p | CSF2RB | 2.12 | 0 | -1.28 | 0 | MirTarget | -0.17 | 4.0E-5 | NA | |
| 148 | hsa-miR-19b-3p | CSF2RB | 2.11 | 0 | -1.28 | 0 | MirTarget | -0.25 | 0 | NA | |
| 149 | hsa-miR-139-5p | CYCS | -2.27 | 0 | 0.53 | 0.0008 | miRanda | -0.18 | 0 | NA | |
| 150 | hsa-miR-200a-3p | CYCS | 3.15 | 0 | 0.53 | 0.0008 | mirMAP | -0.06 | 0.00378 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 54 | 1929 | 1.986e-38 | 9.239e-35 |
| 2 | INTRACELLULAR SIGNAL TRANSDUCTION | 49 | 1572 | 5.008e-36 | 7.988e-33 |
| 3 | REGULATION OF CELL DEATH | 48 | 1472 | 5.15e-36 | 7.988e-33 |
| 4 | CELL DEATH | 42 | 1001 | 4.305e-35 | 5.007e-32 |
| 5 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 49 | 1848 | 1.059e-32 | 9.852e-30 |
| 6 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 21 | 99 | 6.827e-32 | 5.294e-29 |
| 7 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 45 | 1492 | 8.67e-32 | 5.763e-29 |
| 8 | POSITIVE REGULATION OF CELL COMMUNICATION | 45 | 1532 | 2.724e-31 | 1.584e-28 |
| 9 | APOPTOTIC SIGNALING PATHWAY | 27 | 289 | 5.996e-31 | 3.1e-28 |
| 10 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 45 | 1656 | 7.799e-30 | 3.629e-27 |
| 11 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 46 | 1791 | 1.396e-29 | 5.628e-27 |
| 12 | POSITIVE REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 23 | 179 | 1.451e-29 | 5.628e-27 |
| 13 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 42 | 1381 | 2.122e-29 | 7.596e-27 |
| 14 | NEGATIVE REGULATION OF CELL DEATH | 36 | 872 | 3.364e-29 | 1.118e-26 |
| 15 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 36 | 876 | 3.948e-29 | 1.225e-26 |
| 16 | IMMUNE SYSTEM PROCESS | 47 | 1984 | 8.021e-29 | 2.333e-26 |
| 17 | REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 24 | 233 | 1.789e-28 | 4.897e-26 |
| 18 | RESPONSE TO CYTOKINE | 33 | 714 | 4.143e-28 | 1.071e-25 |
| 19 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 15 | 39 | 2.149e-27 | 5.263e-25 |
| 20 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 22 | 213 | 4.172e-26 | 9.705e-24 |
| 21 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 17 | 95 | 1.678e-24 | 3.718e-22 |
| 22 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 17 | 98 | 2.967e-24 | 6.024e-22 |
| 23 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 39 | 1518 | 2.978e-24 | 6.024e-22 |
| 24 | POSITIVE REGULATION OF CELL DEATH | 28 | 605 | 1.298e-23 | 2.516e-21 |
| 25 | ZYMOGEN ACTIVATION | 17 | 112 | 3.355e-23 | 6.245e-21 |
| 26 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 27 | 606 | 2.654e-22 | 4.749e-20 |
| 27 | REGULATION OF IMMUNE SYSTEM PROCESS | 36 | 1403 | 4.177e-22 | 7.198e-20 |
| 28 | RESPONSE TO NITROGEN COMPOUND | 30 | 859 | 7.373e-22 | 1.183e-19 |
| 29 | REGULATION OF IMMUNE RESPONSE | 30 | 858 | 7.131e-22 | 1.183e-19 |
| 30 | POSITIVE REGULATION OF IMMUNE RESPONSE | 26 | 563 | 7.682e-22 | 1.191e-19 |
| 31 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 17 | 153 | 8.596e-21 | 1.29e-18 |
| 32 | ACTIVATION OF IMMUNE RESPONSE | 23 | 427 | 8.975e-21 | 1.305e-18 |
| 33 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 17 | 154 | 9.636e-21 | 1.359e-18 |
| 34 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 21 | 321 | 1.031e-20 | 1.411e-18 |
| 35 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 32 | 1135 | 1.293e-20 | 1.718e-18 |
| 36 | REGULATION OF PEPTIDASE ACTIVITY | 22 | 392 | 2.987e-20 | 3.861e-18 |
| 37 | CYTOKINE MEDIATED SIGNALING PATHWAY | 23 | 452 | 3.225e-20 | 4.055e-18 |
| 38 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 9 | 13 | 6.301e-20 | 7.715e-18 |
| 39 | RESPONSE TO ABIOTIC STIMULUS | 30 | 1024 | 1.071e-19 | 1.278e-17 |
| 40 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 21 | 363 | 1.322e-19 | 1.538e-17 |
| 41 | POSITIVE REGULATION OF DEFENSE RESPONSE | 21 | 364 | 1.4e-19 | 1.588e-17 |
| 42 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 28 | 867 | 2.02e-19 | 2.237e-17 |
| 43 | REGULATION OF PROTEOLYSIS | 26 | 711 | 2.619e-19 | 2.835e-17 |
| 44 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 27 | 799 | 3.37e-19 | 3.564e-17 |
| 45 | RESPONSE TO BIOTIC STIMULUS | 28 | 886 | 3.577e-19 | 3.698e-17 |
| 46 | RESPONSE TO TUMOR NECROSIS FACTOR | 18 | 233 | 4.279e-19 | 4.328e-17 |
| 47 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 35 | 1618 | 5.082e-19 | 5.031e-17 |
| 48 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 17 | 200 | 8.966e-19 | 8.691e-17 |
| 49 | ACTIVATION OF INNATE IMMUNE RESPONSE | 17 | 204 | 1.259e-18 | 1.196e-16 |
| 50 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 15 | 132 | 1.416e-18 | 1.318e-16 |
| 51 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 29 | 1036 | 1.828e-18 | 1.636e-16 |
| 52 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 29 | 1036 | 1.828e-18 | 1.636e-16 |
| 53 | REGULATION OF TRANSFERASE ACTIVITY | 28 | 946 | 1.998e-18 | 1.754e-16 |
| 54 | POSITIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 9 | 17 | 2.117e-18 | 1.824e-16 |
| 55 | REGULATION OF KINASE ACTIVITY | 26 | 776 | 2.26e-18 | 1.912e-16 |
| 56 | REGULATION OF PROTEIN MODIFICATION PROCESS | 35 | 1710 | 2.958e-18 | 2.458e-16 |
| 57 | PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 14 | 109 | 3.663e-18 | 2.99e-16 |
| 58 | PROTEIN MATURATION | 18 | 265 | 4.321e-18 | 3.466e-16 |
| 59 | TOLL LIKE RECEPTOR SIGNALING PATHWAY | 13 | 85 | 5.798e-18 | 4.572e-16 |
| 60 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 22 | 505 | 6.723e-18 | 5.214e-16 |
| 61 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 17 | 228 | 8.406e-18 | 6.412e-16 |
| 62 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 18 | 279 | 1.083e-17 | 8.126e-16 |
| 63 | PHOSPHORYLATION | 30 | 1228 | 1.676e-17 | 1.238e-15 |
| 64 | RESPONSE TO BACTERIUM | 22 | 528 | 1.722e-17 | 1.252e-15 |
| 65 | RESPONSE TO ENDOGENOUS STIMULUS | 32 | 1450 | 1.793e-17 | 1.283e-15 |
| 66 | RESPONSE TO EXTERNAL STIMULUS | 35 | 1821 | 2.158e-17 | 1.521e-15 |
| 67 | REGULATION OF RESPONSE TO STRESS | 32 | 1468 | 2.567e-17 | 1.782e-15 |
| 68 | I KAPPAB KINASE NF KAPPAB SIGNALING | 12 | 70 | 2.754e-17 | 1.884e-15 |
| 69 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 17 | 246 | 3.047e-17 | 2.055e-15 |
| 70 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 10 | 34 | 3.596e-17 | 2.39e-15 |
| 71 | POSITIVE REGULATION OF KINASE ACTIVITY | 21 | 482 | 4.321e-17 | 2.793e-15 |
| 72 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 9 | 22 | 4.267e-17 | 2.793e-15 |
| 73 | POSITIVE REGULATION OF PROTEOLYSIS | 19 | 363 | 5.899e-17 | 3.76e-15 |
| 74 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 15 | 171 | 7.476e-17 | 4.701e-15 |
| 75 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 14 | 144 | 2.035e-16 | 1.262e-14 |
| 76 | INFLAMMATORY RESPONSE | 20 | 454 | 2.217e-16 | 1.357e-14 |
| 77 | PROTEIN PHOSPHORYLATION | 26 | 944 | 2.635e-16 | 1.592e-14 |
| 78 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 20 | 470 | 4.309e-16 | 2.57e-14 |
| 79 | TUMOR NECROSIS FACTOR MEDIATED SIGNALING PATHWAY | 13 | 118 | 4.962e-16 | 2.923e-14 |
| 80 | REGULATION OF INNATE IMMUNE RESPONSE | 18 | 357 | 8.375e-16 | 4.871e-14 |
| 81 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 16 | 263 | 2.164e-15 | 1.243e-13 |
| 82 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 34 | 1977 | 2.243e-15 | 1.273e-13 |
| 83 | REGULATION OF TUMOR NECROSIS FACTOR MEDIATED SIGNALING PATHWAY | 10 | 50 | 2.686e-15 | 1.506e-13 |
| 84 | REGULATION OF DEFENSE RESPONSE | 23 | 759 | 2.788e-15 | 1.544e-13 |
| 85 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 17 | 323 | 2.892e-15 | 1.583e-13 |
| 86 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 13 | 142 | 5.79e-15 | 3.097e-13 |
| 87 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 21 | 616 | 5.772e-15 | 3.097e-13 |
| 88 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 10 | 55 | 7.537e-15 | 3.985e-13 |
| 89 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 11 | 80 | 8.188e-15 | 4.281e-13 |
| 90 | REGULATION OF HYDROLASE ACTIVITY | 28 | 1327 | 1.176e-14 | 6.079e-13 |
| 91 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 13 | 152 | 1.416e-14 | 7.238e-13 |
| 92 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 17 | 365 | 2.162e-14 | 1.093e-12 |
| 93 | REGULATION OF NEURON DEATH | 15 | 252 | 2.454e-14 | 1.228e-12 |
| 94 | REGULATION OF CELL PROLIFERATION | 29 | 1496 | 2.957e-14 | 1.464e-12 |
| 95 | RESPONSE TO MECHANICAL STIMULUS | 14 | 210 | 4.086e-14 | 2.001e-12 |
| 96 | NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR SIGNALING PATHWAY | 8 | 28 | 7.791e-14 | 3.737e-12 |
| 97 | RESPONSE TO LIPID | 23 | 888 | 7.757e-14 | 3.737e-12 |
| 98 | IMMUNE RESPONSE | 25 | 1100 | 9.214e-14 | 4.375e-12 |
| 99 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 23 | 905 | 1.154e-13 | 5.425e-12 |
| 100 | POSITIVE REGULATION OF GENE EXPRESSION | 30 | 1733 | 1.757e-13 | 8.175e-12 |
| 101 | SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 8 | 33 | 3.429e-13 | 1.549e-11 |
| 102 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 8 | 33 | 3.429e-13 | 1.549e-11 |
| 103 | CYTOPLASMIC PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 8 | 33 | 3.429e-13 | 1.549e-11 |
| 104 | EXECUTION PHASE OF APOPTOSIS | 9 | 55 | 4.945e-13 | 2.212e-11 |
| 105 | NEURON APOPTOTIC PROCESS | 8 | 35 | 5.778e-13 | 2.56e-11 |
| 106 | CELLULAR RESPONSE TO STRESS | 28 | 1565 | 6.985e-13 | 3.066e-11 |
| 107 | FC RECEPTOR SIGNALING PATHWAY | 13 | 206 | 7.255e-13 | 3.155e-11 |
| 108 | PROTEOLYSIS | 25 | 1208 | 7.466e-13 | 3.217e-11 |
| 109 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 14 | 264 | 9.448e-13 | 4.033e-11 |
| 110 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 23 | 1008 | 1.084e-12 | 4.587e-11 |
| 111 | RESPONSE TO PEPTIDE | 16 | 404 | 1.649e-12 | 6.913e-11 |
| 112 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 15 | 360 | 4.325e-12 | 1.797e-10 |
| 113 | REGULATION OF NECROTIC CELL DEATH | 7 | 26 | 4.863e-12 | 2.002e-10 |
| 114 | T CELL RECEPTOR SIGNALING PATHWAY | 11 | 146 | 7.039e-12 | 2.873e-10 |
| 115 | RESPONSE TO HORMONE | 21 | 893 | 7.484e-12 | 3.002e-10 |
| 116 | NEURON DEATH | 8 | 47 | 7.448e-12 | 3.002e-10 |
| 117 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 12 | 195 | 8.024e-12 | 3.191e-10 |
| 118 | DEFENSE RESPONSE | 24 | 1231 | 8.694e-12 | 3.428e-10 |
| 119 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 25 | 1360 | 1.009e-11 | 3.944e-10 |
| 120 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 8 | 49 | 1.062e-11 | 4.117e-10 |
| 121 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 21 | 917 | 1.233e-11 | 4.74e-10 |
| 122 | MYD88 INDEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 7 | 30 | 1.487e-11 | 5.672e-10 |
| 123 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 17 | 552 | 1.653e-11 | 6.253e-10 |
| 124 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 28 | 1805 | 2.173e-11 | 8.153e-10 |
| 125 | RESPONSE TO WOUNDING | 17 | 563 | 2.253e-11 | 8.386e-10 |
| 126 | NIK NF KAPPAB SIGNALING | 9 | 83 | 2.352e-11 | 8.684e-10 |
| 127 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 10 | 121 | 2.681e-11 | 9.745e-10 |
| 128 | CELLULAR RESPONSE TO PEPTIDE | 13 | 274 | 2.677e-11 | 9.745e-10 |
| 129 | NEGATIVE REGULATION OF CELL COMMUNICATION | 23 | 1192 | 3.294e-11 | 1.188e-09 |
| 130 | INOSITOL LIPID MEDIATED SIGNALING | 10 | 124 | 3.427e-11 | 1.227e-09 |
| 131 | HOMEOSTATIC PROCESS | 24 | 1337 | 4.88e-11 | 1.734e-09 |
| 132 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 16 | 514 | 6.077e-11 | 2.142e-09 |
| 133 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 14 | 370 | 8.694e-11 | 3.042e-09 |
| 134 | REGULATION OF NEURON APOPTOTIC PROCESS | 11 | 192 | 1.374e-10 | 4.77e-09 |
| 135 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 15 | 465 | 1.593e-10 | 5.49e-09 |
| 136 | CELLULAR RESPONSE TO HORMONE STIMULUS | 16 | 552 | 1.736e-10 | 5.94e-09 |
| 137 | REGULATION OF MAP KINASE ACTIVITY | 13 | 319 | 1.767e-10 | 6.001e-09 |
| 138 | WOUND HEALING | 15 | 470 | 1.848e-10 | 6.231e-09 |
| 139 | CELLULAR COMPONENT DISASSEMBLY INVOLVED IN EXECUTION PHASE OF APOPTOSIS | 7 | 43 | 2.265e-10 | 7.581e-09 |
| 140 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 8 | 71 | 2.346e-10 | 7.796e-09 |
| 141 | RESPONSE TO INORGANIC SUBSTANCE | 15 | 479 | 2.404e-10 | 7.933e-09 |
| 142 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 21 | 1079 | 2.513e-10 | 8.234e-09 |
| 143 | CELLULAR GLUCOSE HOMEOSTASIS | 8 | 75 | 3.676e-10 | 1.196e-08 |
| 144 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 7 | 46 | 3.728e-10 | 1.205e-08 |
| 145 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 25 | 3.795e-10 | 1.218e-08 |
| 146 | RESPONSE TO INTERLEUKIN 1 | 9 | 115 | 4.573e-10 | 1.458e-08 |
| 147 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 1004 | 4.956e-10 | 1.569e-08 |
| 148 | CELLULAR RESPONSE TO BIOTIC STIMULUS | 10 | 163 | 5.149e-10 | 1.619e-08 |
| 149 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 11 | 220 | 5.871e-10 | 1.833e-08 |
| 150 | RESPONSE TO OXIDATIVE STRESS | 13 | 352 | 5.908e-10 | 1.833e-08 |
| 151 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 9 | 120 | 6.7e-10 | 2.065e-08 |
| 152 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 12 | 289 | 7.664e-10 | 2.346e-08 |
| 153 | POSITIVE REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 7 | 51 | 7.944e-10 | 2.416e-08 |
| 154 | NECROTIC CELL DEATH | 6 | 28 | 8.002e-10 | 2.418e-08 |
| 155 | INTERLEUKIN 1 MEDIATED SIGNALING PATHWAY | 5 | 13 | 8.141e-10 | 2.444e-08 |
| 156 | REGULATION OF CATABOLIC PROCESS | 17 | 731 | 1.259e-09 | 3.755e-08 |
| 157 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 9 | 129 | 1.278e-09 | 3.788e-08 |
| 158 | CELLULAR RESPONSE TO INTERLEUKIN 1 | 8 | 88 | 1.347e-09 | 3.967e-08 |
| 159 | REGULATION OF PROTEIN LOCALIZATION | 19 | 950 | 1.384e-09 | 4.051e-08 |
| 160 | LIPOPOLYSACCHARIDE MEDIATED SIGNALING PATHWAY | 6 | 31 | 1.55e-09 | 4.508e-08 |
| 161 | RESPONSE TO OXYGEN LEVELS | 12 | 311 | 1.762e-09 | 5.092e-08 |
| 162 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 6 | 32 | 1.902e-09 | 5.43e-08 |
| 163 | NEGATIVE REGULATION OF SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 6 | 32 | 1.902e-09 | 5.43e-08 |
| 164 | POSITIVE REGULATION OF MAPK CASCADE | 14 | 470 | 1.956e-09 | 5.551e-08 |
| 165 | REGULATION OF MAPK CASCADE | 16 | 660 | 2.31e-09 | 6.514e-08 |
| 166 | RESPONSE TO ACID CHEMICAL | 12 | 319 | 2.347e-09 | 6.578e-08 |
| 167 | CELL ACTIVATION | 15 | 568 | 2.486e-09 | 6.91e-08 |
| 168 | REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 8 | 95 | 2.495e-09 | 6.91e-08 |
| 169 | CHEMICAL HOMEOSTASIS | 18 | 874 | 2.586e-09 | 7.121e-08 |
| 170 | T CELL HOMEOSTASIS | 6 | 34 | 2.806e-09 | 7.681e-08 |
| 171 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 11 | 258 | 3.145e-09 | 8.557e-08 |
| 172 | ACTIVATION OF PROTEIN KINASE A ACTIVITY | 5 | 17 | 3.869e-09 | 1.047e-07 |
| 173 | AGING | 11 | 264 | 3.998e-09 | 1.075e-07 |
| 174 | NEGATIVE REGULATION OF CATABOLIC PROCESS | 10 | 203 | 4.342e-09 | 1.161e-07 |
| 175 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 8 | 103 | 4.765e-09 | 1.267e-07 |
| 176 | POSITIVE REGULATION OF PROTEIN IMPORT | 8 | 104 | 5.146e-09 | 1.361e-07 |
| 177 | POSITIVE REGULATION OF NEURON DEATH | 7 | 67 | 5.69e-09 | 1.496e-07 |
| 178 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 8 | 106 | 5.99e-09 | 1.566e-07 |
| 179 | POSITIVE REGULATION OF TRANSPORT | 18 | 936 | 7.553e-09 | 1.963e-07 |
| 180 | REGULATION OF LIPID METABOLIC PROCESS | 11 | 282 | 7.939e-09 | 2.052e-07 |
| 181 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 17 | 829 | 8.284e-09 | 2.118e-07 |
| 182 | REGULATION OF INTRACELLULAR TRANSPORT | 15 | 621 | 8.267e-09 | 2.118e-07 |
| 183 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 10 | 218 | 8.606e-09 | 2.188e-07 |
| 184 | RESPONSE TO AMINO ACID | 8 | 112 | 9.276e-09 | 2.346e-07 |
| 185 | REGULATION OF NIK NF KAPPAB SIGNALING | 6 | 42 | 1.069e-08 | 2.689e-07 |
| 186 | LEUKOCYTE DIFFERENTIATION | 11 | 292 | 1.139e-08 | 2.848e-07 |
| 187 | CELLULAR RESPONSE TO CARBOHYDRATE STIMULUS | 7 | 74 | 1.153e-08 | 2.869e-07 |
| 188 | REGULATION OF INFLAMMATORY RESPONSE | 11 | 294 | 1.222e-08 | 3.024e-07 |
| 189 | NECROPTOTIC PROCESS | 5 | 21 | 1.258e-08 | 3.096e-07 |
| 190 | GLUCOSE HOMEOSTASIS | 9 | 170 | 1.454e-08 | 3.543e-07 |
| 191 | CARBOHYDRATE HOMEOSTASIS | 9 | 170 | 1.454e-08 | 3.543e-07 |
| 192 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 12 | 381 | 1.705e-08 | 4.133e-07 |
| 193 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 6 | 47 | 2.156e-08 | 5.198e-07 |
| 194 | HEMOSTASIS | 11 | 311 | 2.179e-08 | 5.226e-07 |
| 195 | RESPONSE TO TOXIC SUBSTANCE | 10 | 241 | 2.236e-08 | 5.336e-07 |
| 196 | REGULATION OF EXECUTION PHASE OF APOPTOSIS | 5 | 24 | 2.604e-08 | 6.181e-07 |
| 197 | REGULATION OF PROTEIN IMPORT | 9 | 183 | 2.758e-08 | 6.514e-07 |
| 198 | IMMUNE SYSTEM DEVELOPMENT | 14 | 582 | 2.926e-08 | 6.877e-07 |
| 199 | LYMPHOCYTE HOMEOSTASIS | 6 | 50 | 3.162e-08 | 7.394e-07 |
| 200 | RESPONSE TO NICOTINE | 6 | 51 | 3.573e-08 | 8.313e-07 |
| 201 | LEUKOCYTE CELL CELL ADHESION | 10 | 255 | 3.813e-08 | 8.826e-07 |
| 202 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 7 | 88 | 3.901e-08 | 8.987e-07 |
| 203 | RESPONSE TO REACTIVE OXYGEN SPECIES | 9 | 191 | 3.992e-08 | 9.15e-07 |
| 204 | RESPONSE TO RADIATION | 12 | 413 | 4.142e-08 | 9.447e-07 |
| 205 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 9 | 193 | 4.367e-08 | 9.912e-07 |
| 206 | RESPONSE TO ALKALOID | 8 | 137 | 4.525e-08 | 1.018e-06 |
| 207 | REGULATION OF ORGANELLE ORGANIZATION | 19 | 1178 | 4.53e-08 | 1.018e-06 |
| 208 | LYMPHOCYTE ACTIVATION | 11 | 342 | 5.746e-08 | 1.285e-06 |
| 209 | REGULATION OF NECROPTOTIC PROCESS | 4 | 11 | 5.9e-08 | 1.307e-06 |
| 210 | POSITIVE REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 4 | 11 | 5.9e-08 | 1.307e-06 |
| 211 | RESPONSE TO DRUG | 12 | 431 | 6.598e-08 | 1.455e-06 |
| 212 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 29 | 7.169e-08 | 1.559e-06 |
| 213 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 29 | 7.169e-08 | 1.559e-06 |
| 214 | POSITIVE REGULATION OF NIK NF KAPPAB SIGNALING | 5 | 29 | 7.169e-08 | 1.559e-06 |
| 215 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 18 | 1087 | 7.46e-08 | 1.614e-06 |
| 216 | REGULATION OF CELL ADHESION | 14 | 629 | 7.662e-08 | 1.65e-06 |
| 217 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 23 | 1784 | 7.787e-08 | 1.67e-06 |
| 218 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 9 | 207 | 7.969e-08 | 1.701e-06 |
| 219 | PEPTIDYL SERINE MODIFICATION | 8 | 148 | 8.252e-08 | 1.745e-06 |
| 220 | RESPONSE TO TEMPERATURE STIMULUS | 8 | 148 | 8.252e-08 | 1.745e-06 |
| 221 | REGULATION OF ENDOTHELIAL CELL DEVELOPMENT | 4 | 12 | 8.824e-08 | 1.831e-06 |
| 222 | LIPID PHOSPHORYLATION | 7 | 99 | 8.854e-08 | 1.831e-06 |
| 223 | REGULATION OF ESTABLISHMENT OF ENDOTHELIAL BARRIER | 4 | 12 | 8.824e-08 | 1.831e-06 |
| 224 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 59 | 8.731e-08 | 1.831e-06 |
| 225 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 12 | 8.824e-08 | 1.831e-06 |
| 226 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 9 | 211 | 9.386e-08 | 1.932e-06 |
| 227 | REGULATION OF GLUCOSE TRANSPORT | 7 | 100 | 9.492e-08 | 1.946e-06 |
| 228 | REGULATION OF GLUCOSE IMPORT | 6 | 60 | 9.672e-08 | 1.965e-06 |
| 229 | LEUKOCYTE HOMEOSTASIS | 6 | 60 | 9.672e-08 | 1.965e-06 |
| 230 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 10 | 282 | 9.794e-08 | 1.981e-06 |
| 231 | RENAL SYSTEM PROCESS | 7 | 102 | 1.089e-07 | 2.193e-06 |
| 232 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 22 | 1672 | 1.145e-07 | 2.297e-06 |
| 233 | MYD88 DEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 5 | 32 | 1.205e-07 | 2.396e-06 |
| 234 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 5 | 32 | 1.205e-07 | 2.396e-06 |
| 235 | RESPONSE TO COBALT ION | 4 | 13 | 1.271e-07 | 2.517e-06 |
| 236 | REGULATION OF CYTOKINE PRODUCTION | 13 | 563 | 1.555e-07 | 3.067e-06 |
| 237 | REGULATION OF CELLULAR LOCALIZATION | 19 | 1277 | 1.609e-07 | 3.159e-06 |
| 238 | PROTEIN KINASE B SIGNALING | 5 | 34 | 1.656e-07 | 3.21e-06 |
| 239 | RENAL WATER HOMEOSTASIS | 5 | 34 | 1.656e-07 | 3.21e-06 |
| 240 | CELLULAR RESPONSE TO ALKALOID | 5 | 34 | 1.656e-07 | 3.21e-06 |
| 241 | TOLL LIKE RECEPTOR 9 SIGNALING PATHWAY | 4 | 14 | 1.774e-07 | 3.426e-06 |
| 242 | REGULATION OF CYTOPLASMIC TRANSPORT | 12 | 481 | 2.16e-07 | 4.136e-06 |
| 243 | REGULATION OF PROTEIN TARGETING | 10 | 307 | 2.154e-07 | 4.136e-06 |
| 244 | RESPONSE TO CARBOHYDRATE | 8 | 168 | 2.193e-07 | 4.165e-06 |
| 245 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 8 | 168 | 2.193e-07 | 4.165e-06 |
| 246 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 36 | 2.23e-07 | 4.218e-06 |
| 247 | REGULATION OF CELL ACTIVATION | 12 | 484 | 2.309e-07 | 4.349e-06 |
| 248 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 9 | 235 | 2.345e-07 | 4.4e-06 |
| 249 | T CELL APOPTOTIC PROCESS | 4 | 15 | 2.413e-07 | 4.509e-06 |
| 250 | REGULATION OF MEMBRANE PERMEABILITY | 6 | 70 | 2.459e-07 | 4.577e-06 |
| 251 | NEGATIVE REGULATION OF NEURON DEATH | 8 | 171 | 2.512e-07 | 4.656e-06 |
| 252 | CELLULAR RESPONSE TO GLUCAGON STIMULUS | 5 | 38 | 2.952e-07 | 5.45e-06 |
| 253 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 11 | 404 | 3.065e-07 | 5.636e-06 |
| 254 | RESPONSE TO VIRUS | 9 | 247 | 3.57e-07 | 6.539e-06 |
| 255 | REGULATION OF CELL DIFFERENTIATION | 20 | 1492 | 3.781e-07 | 6.899e-06 |
| 256 | REGULATION OF RESPONSE TO WOUNDING | 11 | 413 | 3.814e-07 | 6.932e-06 |
| 257 | LEUKOCYTE ACTIVATION | 11 | 414 | 3.906e-07 | 7.072e-06 |
| 258 | REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 4 | 17 | 4.183e-07 | 7.486e-06 |
| 259 | ACTIVATION OF NF KAPPAB INDUCING KINASE ACTIVITY | 4 | 17 | 4.183e-07 | 7.486e-06 |
| 260 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 17 | 4.183e-07 | 7.486e-06 |
| 261 | REGULATION OF TRANSPORT | 22 | 1804 | 4.291e-07 | 7.649e-06 |
| 262 | RESPONSE TO UV | 7 | 126 | 4.633e-07 | 8.229e-06 |
| 263 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 5 | 42 | 4.944e-07 | 8.748e-06 |
| 264 | LEUKOCYTE MIGRATION | 9 | 259 | 5.315e-07 | 9.368e-06 |
| 265 | LYMPHOCYTE APOPTOTIC PROCESS | 4 | 18 | 5.363e-07 | 9.416e-06 |
| 266 | INSULIN RECEPTOR SIGNALING PATHWAY | 6 | 80 | 5.472e-07 | 9.572e-06 |
| 267 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 5 | 43 | 5.579e-07 | 9.722e-06 |
| 268 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 19 | 1395 | 6.262e-07 | 1.087e-05 |
| 269 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 9 | 272 | 7.999e-07 | 1.384e-05 |
| 270 | CELL DEVELOPMENT | 19 | 1426 | 8.736e-07 | 1.505e-05 |
| 271 | RESPONSE TO ANTIBIOTIC | 5 | 47 | 8.787e-07 | 1.509e-05 |
| 272 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 17 | 1152 | 9.294e-07 | 1.59e-05 |
| 273 | RESPONSE TO GLUCAGON | 5 | 48 | 9.78e-07 | 1.667e-05 |
| 274 | CELLULAR RESPONSE TO LIPID | 11 | 457 | 1.033e-06 | 1.754e-05 |
| 275 | NEGATIVE REGULATION OF LIPID CATABOLIC PROCESS | 4 | 21 | 1.04e-06 | 1.76e-05 |
| 276 | SINGLE ORGANISM CELL ADHESION | 11 | 459 | 1.078e-06 | 1.817e-05 |
| 277 | LYMPHOCYTE DIFFERENTIATION | 8 | 209 | 1.151e-06 | 1.934e-05 |
| 278 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 10 | 370 | 1.184e-06 | 1.982e-05 |
| 279 | RESPONSE TO GAMMA RADIATION | 5 | 50 | 1.203e-06 | 2.006e-05 |
| 280 | CELLULAR HOMEOSTASIS | 13 | 676 | 1.232e-06 | 2.047e-05 |
| 281 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 10 | 372 | 1.243e-06 | 2.059e-05 |
| 282 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 6 | 92 | 1.253e-06 | 2.068e-05 |
| 283 | CELLULAR CHEMICAL HOMEOSTASIS | 12 | 570 | 1.301e-06 | 2.14e-05 |
| 284 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 13 | 684 | 1.404e-06 | 2.3e-05 |
| 285 | REGULATION OF LIPID CATABOLIC PROCESS | 5 | 52 | 1.467e-06 | 2.395e-05 |
| 286 | REGULATION OF CELL CELL ADHESION | 10 | 380 | 1.506e-06 | 2.45e-05 |
| 287 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 23 | 1.53e-06 | 2.472e-05 |
| 288 | LEUKOCYTE APOPTOTIC PROCESS | 4 | 23 | 1.53e-06 | 2.472e-05 |
| 289 | REGULATION OF MITOCHONDRION ORGANIZATION | 8 | 218 | 1.579e-06 | 2.543e-05 |
| 290 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 7 | 152 | 1.642e-06 | 2.635e-05 |
| 291 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 9 | 297 | 1.657e-06 | 2.649e-05 |
| 292 | IMMUNE EFFECTOR PROCESS | 11 | 486 | 1.878e-06 | 2.992e-05 |
| 293 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 9 | 307 | 2.175e-06 | 3.454e-05 |
| 294 | NEGATIVE REGULATION OF HYDROLASE ACTIVITY | 10 | 397 | 2.229e-06 | 3.527e-05 |
| 295 | MULTICELLULAR ORGANISMAL WATER HOMEOSTASIS | 5 | 58 | 2.542e-06 | 4.009e-05 |
| 296 | DEVELOPMENTAL PROGRAMMED CELL DEATH | 4 | 26 | 2.561e-06 | 4.026e-05 |
| 297 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 8 | 233 | 2.593e-06 | 4.063e-05 |
| 298 | REGULATION OF BODY FLUID LEVELS | 11 | 506 | 2.77e-06 | 4.324e-05 |
| 299 | REGULATION OF ENDOTHELIAL CELL DIFFERENTIATION | 4 | 27 | 2.998e-06 | 4.65e-05 |
| 300 | DNA CATABOLIC PROCESS | 4 | 27 | 2.998e-06 | 4.65e-05 |
| 301 | POSITIVE REGULATION OF CELL CELL ADHESION | 8 | 243 | 3.541e-06 | 5.474e-05 |
| 302 | PROTEIN HETEROOLIGOMERIZATION | 6 | 113 | 4.173e-06 | 6.43e-05 |
| 303 | RESPONSE TO METAL ION | 9 | 333 | 4.222e-06 | 6.483e-05 |
| 304 | RESPONSE TO CORTICOSTEROID | 7 | 176 | 4.349e-06 | 6.657e-05 |
| 305 | PROTEIN OLIGOMERIZATION | 10 | 434 | 4.913e-06 | 7.496e-05 |
| 306 | RESPONSE TO KETONE | 7 | 182 | 5.424e-06 | 8.248e-05 |
| 307 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 6 | 122 | 6.501e-06 | 9.821e-05 |
| 308 | WATER HOMEOSTASIS | 5 | 70 | 6.483e-06 | 9.821e-05 |
| 309 | MYELOID CELL DIFFERENTIATION | 7 | 189 | 6.949e-06 | 0.0001046 |
| 310 | GLYCEROLIPID METABOLIC PROCESS | 9 | 356 | 7.241e-06 | 0.0001087 |
| 311 | PROTEIN AUTOPHOSPHORYLATION | 7 | 192 | 7.703e-06 | 0.0001153 |
| 312 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 6 | 126 | 7.828e-06 | 0.0001167 |
| 313 | NEGATIVE REGULATION OF NECROTIC CELL DEATH | 3 | 11 | 8.179e-06 | 0.0001216 |
| 314 | RESPONSE TO ALCOHOL | 9 | 362 | 8.281e-06 | 0.0001227 |
| 315 | PHOSPHOLIPID METABOLIC PROCESS | 9 | 364 | 8.655e-06 | 0.0001278 |
| 316 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 11 | 573 | 9.026e-06 | 0.0001329 |
| 317 | POSITIVE REGULATION OF CELL PROLIFERATION | 13 | 814 | 9.361e-06 | 0.0001374 |
| 318 | B CELL ACTIVATION | 6 | 132 | 1.022e-05 | 0.0001496 |
| 319 | RESPONSE TO GROWTH FACTOR | 10 | 475 | 1.082e-05 | 0.0001578 |
| 320 | REGULATION OF VITAMIN METABOLIC PROCESS | 3 | 12 | 1.088e-05 | 0.0001581 |
| 321 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 14 | 957 | 1.099e-05 | 0.0001593 |
| 322 | RESPONSE TO INSULIN | 7 | 205 | 1.181e-05 | 0.0001706 |
| 323 | NEGATIVE REGULATION OF LIPID METABOLIC PROCESS | 5 | 80 | 1.251e-05 | 0.0001802 |
| 324 | MITOCHONDRION ORGANIZATION | 11 | 594 | 1.264e-05 | 0.000181 |
| 325 | ACTIVATION OF MAPK ACTIVITY | 6 | 137 | 1.265e-05 | 0.000181 |
| 326 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 138 | 1.318e-05 | 0.0001881 |
| 327 | LIPID MODIFICATION | 7 | 210 | 1.38e-05 | 0.0001964 |
| 328 | JNK CASCADE | 5 | 82 | 1.411e-05 | 0.0001978 |
| 329 | HEPATOCYTE APOPTOTIC PROCESS | 3 | 13 | 1.41e-05 | 0.0001978 |
| 330 | REGULATION OF PROTEIN MATURATION | 5 | 82 | 1.411e-05 | 0.0001978 |
| 331 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 12 | 720 | 1.411e-05 | 0.0001978 |
| 332 | REGULATION OF BICELLULAR TIGHT JUNCTION ASSEMBLY | 3 | 13 | 1.41e-05 | 0.0001978 |
| 333 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 11 | 602 | 1.432e-05 | 0.0002001 |
| 334 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 4 | 40 | 1.505e-05 | 0.0002096 |
| 335 | PLATELET ACTIVATION | 6 | 142 | 1.551e-05 | 0.0002154 |
| 336 | CELL CELL ADHESION | 11 | 608 | 1.57e-05 | 0.0002174 |
| 337 | CELLULAR RESPONSE TO OXYGEN LEVELS | 6 | 143 | 1.614e-05 | 0.0002229 |
| 338 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 10 | 498 | 1.628e-05 | 0.0002241 |
| 339 | PROTEIN COMPLEX BIOGENESIS | 15 | 1132 | 1.652e-05 | 0.0002261 |
| 340 | PROTEIN COMPLEX ASSEMBLY | 15 | 1132 | 1.652e-05 | 0.0002261 |
| 341 | RESPONSE TO IONIZING RADIATION | 6 | 145 | 1.747e-05 | 0.0002384 |
| 342 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 5 | 86 | 1.781e-05 | 0.0002423 |
| 343 | REGULATION OF CAMP DEPENDENT PROTEIN KINASE ACTIVITY | 3 | 14 | 1.79e-05 | 0.0002428 |
| 344 | CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 146 | 1.816e-05 | 0.0002457 |
| 345 | INNATE IMMUNE RESPONSE | 11 | 619 | 1.854e-05 | 0.00025 |
| 346 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 5 | 88 | 1.991e-05 | 0.0002678 |
| 347 | RESPONSE TO HEAT | 5 | 89 | 2.104e-05 | 0.0002821 |
| 348 | POSITIVE REGULATION OF CELL ACTIVATION | 8 | 311 | 2.134e-05 | 0.0002854 |
| 349 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 4 | 44 | 2.21e-05 | 0.0002946 |
| 350 | APOPTOTIC DNA FRAGMENTATION | 3 | 15 | 2.231e-05 | 0.0002958 |
| 351 | NEUROTROPHIN TRK RECEPTOR SIGNALING PATHWAY | 3 | 15 | 2.231e-05 | 0.0002958 |
| 352 | NEGATIVE REGULATION OF CELLULAR CATABOLIC PROCESS | 6 | 156 | 2.642e-05 | 0.0003492 |
| 353 | REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 5 | 94 | 2.742e-05 | 0.0003614 |
| 354 | REGULATION OF LIPID TRANSPORT | 5 | 95 | 2.886e-05 | 0.0003793 |
| 355 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 4 | 48 | 3.132e-05 | 0.0004093 |
| 356 | REGULATION OF LIPID KINASE ACTIVITY | 4 | 48 | 3.132e-05 | 0.0004093 |
| 357 | NEGATIVE REGULATION OF PROTEOLYSIS | 8 | 329 | 3.188e-05 | 0.0004156 |
| 358 | NEGATIVE REGULATION OF ANOIKIS | 3 | 17 | 3.316e-05 | 0.000431 |
| 359 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 13 | 926 | 3.634e-05 | 0.000471 |
| 360 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 9 | 437 | 3.666e-05 | 0.0004738 |
| 361 | NEGATIVE REGULATION OF PEPTIDASE ACTIVITY | 7 | 245 | 3.713e-05 | 0.0004786 |
| 362 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 6 | 167 | 3.874e-05 | 0.000498 |
| 363 | CELL PROLIFERATION | 11 | 672 | 3.935e-05 | 0.000503 |
| 364 | RESPONSE TO EXTRACELLULAR STIMULUS | 9 | 441 | 3.935e-05 | 0.000503 |
| 365 | NEGATIVE REGULATION OF ORGANIC ACID TRANSPORT | 3 | 18 | 3.969e-05 | 0.0005046 |
| 366 | INOSITOL PHOSPHATE MEDIATED SIGNALING | 3 | 18 | 3.969e-05 | 0.0005046 |
| 367 | REGULATION OF AUTOPHAGY | 7 | 249 | 4.115e-05 | 0.0005217 |
| 368 | NEGATIVE REGULATION OF KINASE ACTIVITY | 7 | 250 | 4.221e-05 | 0.0005337 |
| 369 | REGULATION OF T CELL MEDIATED IMMUNITY | 4 | 52 | 4.308e-05 | 0.0005433 |
| 370 | CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM | 7 | 252 | 4.44e-05 | 0.0005583 |
| 371 | RESPONSE TO MUSCLE STRETCH | 3 | 19 | 4.701e-05 | 0.0005771 |
| 372 | REGULATION OF NITRIC OXIDE BIOSYNTHETIC PROCESS | 4 | 53 | 4.647e-05 | 0.0005771 |
| 373 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 9 | 450 | 4.602e-05 | 0.0005771 |
| 374 | NEGATIVE REGULATION OF MEIOTIC CELL CYCLE | 3 | 19 | 4.701e-05 | 0.0005771 |
| 375 | REGULATION OF CELL CYCLE | 13 | 949 | 4.68e-05 | 0.0005771 |
| 376 | CELLULAR RESPONSE TO AMINO ACID STIMULUS | 4 | 53 | 4.647e-05 | 0.0005771 |
| 377 | POSITIVE REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 19 | 4.701e-05 | 0.0005771 |
| 378 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 4 | 53 | 4.647e-05 | 0.0005771 |
| 379 | DNA CATABOLIC PROCESS ENDONUCLEOLYTIC | 3 | 19 | 4.701e-05 | 0.0005771 |
| 380 | STRIATED MUSCLE CELL DIFFERENTIATION | 6 | 173 | 4.719e-05 | 0.0005778 |
| 381 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 11 | 689 | 4.934e-05 | 0.0006025 |
| 382 | CELLULAR RESPONSE TO ACID CHEMICAL | 6 | 175 | 5.031e-05 | 0.0006105 |
| 383 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 8 | 351 | 5.038e-05 | 0.0006105 |
| 384 | HOMEOSTASIS OF NUMBER OF CELLS | 6 | 175 | 5.031e-05 | 0.0006105 |
| 385 | MITOCHONDRIAL TRANSPORT | 6 | 177 | 5.36e-05 | 0.0006478 |
| 386 | RESPONSE TO HYDROGEN PEROXIDE | 5 | 109 | 5.587e-05 | 0.0006734 |
| 387 | RESPONSE TO NITRIC OXIDE | 3 | 21 | 6.417e-05 | 0.0007715 |
| 388 | REGULATION OF MYELOID CELL DIFFERENTIATION | 6 | 183 | 6.45e-05 | 0.0007735 |
| 389 | POSITIVE REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 4 | 58 | 6.638e-05 | 0.0007931 |
| 390 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 6 | 184 | 6.648e-05 | 0.0007931 |
| 391 | REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 5 | 114 | 6.919e-05 | 0.0008233 |
| 392 | REGULATION OF MONOOXYGENASE ACTIVITY | 4 | 60 | 7.586e-05 | 0.0009005 |
| 393 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 14 | 1142 | 7.685e-05 | 0.0009099 |
| 394 | PROTEIN DEPHOSPHORYLATION | 6 | 190 | 7.939e-05 | 0.0009375 |
| 395 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 4 | 61 | 8.096e-05 | 0.0009536 |
| 396 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 5 | 118 | 8.151e-05 | 0.0009554 |
| 397 | POSITIVE REGULATION OF CELL ADHESION | 8 | 376 | 8.151e-05 | 0.0009554 |
| 398 | NEUROTROPHIN SIGNALING PATHWAY | 3 | 23 | 8.499e-05 | 0.0009936 |
| 399 | RESPONSE TO LIGHT STIMULUS | 7 | 280 | 8.613e-05 | 0.001004 |
| 400 | REGULATION OF ANOIKIS | 3 | 24 | 9.687e-05 | 0.001127 |
| 401 | RESPONSE TO STEROID HORMONE | 9 | 497 | 9.851e-05 | 0.001143 |
| 402 | REGULATION OF ADAPTIVE IMMUNE RESPONSE | 5 | 123 | 9.92e-05 | 0.001145 |
| 403 | T CELL DIFFERENTIATION | 5 | 123 | 9.92e-05 | 0.001145 |
| 404 | REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 4 | 65 | 0.0001038 | 0.001196 |
| 405 | CELLULAR EXTRAVASATION | 3 | 25 | 0.0001098 | 0.001255 |
| 406 | APOPTOTIC NUCLEAR CHANGES | 3 | 25 | 0.0001098 | 0.001255 |
| 407 | EPITHELIAL CELL APOPTOTIC PROCESS | 3 | 25 | 0.0001098 | 0.001255 |
| 408 | POSITIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 4 | 66 | 0.0001102 | 0.001257 |
| 409 | GLAND DEVELOPMENT | 8 | 395 | 0.0001147 | 0.001305 |
| 410 | EMBRYO DEVELOPMENT | 12 | 894 | 0.000115 | 0.001305 |
| 411 | CELLULAR RESPONSE TO DRUG | 4 | 67 | 0.0001169 | 0.001323 |
| 412 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 5 | 128 | 0.0001197 | 0.001346 |
| 413 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 5 | 128 | 0.0001197 | 0.001346 |
| 414 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 5 | 128 | 0.0001197 | 0.001346 |
| 415 | REGULATION OF LEUKOCYTE PROLIFERATION | 6 | 206 | 0.0001238 | 0.001382 |
| 416 | NEGATIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 4 | 68 | 0.0001239 | 0.001382 |
| 417 | NEGATIVE REGULATION OF LIPID TRANSPORT | 3 | 26 | 0.0001238 | 0.001382 |
| 418 | NUCLEAR IMPORT | 5 | 129 | 0.0001242 | 0.001382 |
| 419 | CELLULAR COMPONENT DISASSEMBLY | 9 | 515 | 0.0001289 | 0.001431 |
| 420 | POSITIVE REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 4 | 69 | 0.0001311 | 0.001452 |
| 421 | TISSUE DEVELOPMENT | 16 | 1518 | 0.0001315 | 0.001453 |
| 422 | POSITIVE REGULATION OF TYPE I INTERFERON PRODUCTION | 4 | 70 | 0.0001387 | 0.001527 |
| 423 | REGULATION OF FATTY ACID TRANSPORT | 3 | 27 | 0.0001389 | 0.001527 |
| 424 | CELLULAR LIPID METABOLIC PROCESS | 12 | 913 | 0.0001401 | 0.001537 |
| 425 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 16 | 1527 | 0.0001409 | 0.001542 |
| 426 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 5 | 135 | 0.0001537 | 0.001679 |
| 427 | POSITIVE REGULATION OF ACUTE INFLAMMATORY RESPONSE | 3 | 28 | 0.0001551 | 0.00169 |
| 428 | POSITIVE REGULATION OF ADAPTIVE IMMUNE RESPONSE | 4 | 73 | 0.0001632 | 0.001774 |
| 429 | CELLULAR RESPONSE TO RADIATION | 5 | 137 | 0.0001647 | 0.001786 |
| 430 | RESPONSE TO ESTROGEN | 6 | 218 | 0.0001685 | 0.001823 |
| 431 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 4 | 74 | 0.000172 | 0.00185 |
| 432 | PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 3 | 29 | 0.0001725 | 0.00185 |
| 433 | MUSCLE ADAPTATION | 3 | 29 | 0.0001725 | 0.00185 |
| 434 | POSITIVE REGULATION OF MONOOXYGENASE ACTIVITY | 3 | 29 | 0.0001725 | 0.00185 |
| 435 | POSITIVE REGULATION OF LOCOMOTION | 8 | 420 | 0.0001746 | 0.001867 |
| 436 | REGULATION OF HEMOPOIESIS | 7 | 314 | 0.0001751 | 0.001867 |
| 437 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 10 | 662 | 0.0001758 | 0.001867 |
| 438 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 10 | 662 | 0.0001758 | 0.001867 |
| 439 | GRANULOCYTE MIGRATION | 4 | 75 | 0.0001812 | 0.001918 |
| 440 | LIPID BIOSYNTHETIC PROCESS | 9 | 539 | 0.0001813 | 0.001918 |
| 441 | MACROMOLECULAR COMPLEX ASSEMBLY | 15 | 1398 | 0.0001825 | 0.001925 |
| 442 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 9 | 541 | 0.0001864 | 0.001954 |
| 443 | REGULATION OF IMMUNE EFFECTOR PROCESS | 8 | 424 | 0.0001863 | 0.001954 |
| 444 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 9 | 541 | 0.0001864 | 0.001954 |
| 445 | NEGATIVE REGULATION OF B CELL ACTIVATION | 3 | 30 | 0.0001912 | 0.001999 |
| 446 | EPITHELIUM DEVELOPMENT | 12 | 945 | 0.000193 | 0.002014 |
| 447 | LYMPHOCYTE COSTIMULATION | 4 | 78 | 0.0002109 | 0.002193 |
| 448 | POSITIVE REGULATION OF INTERLEUKIN 2 PRODUCTION | 3 | 31 | 0.0002111 | 0.002193 |
| 449 | NEGATIVE REGULATION OF CELL CYCLE | 8 | 433 | 0.0002148 | 0.002226 |
| 450 | RESPONSE TO ESTRADIOL | 5 | 146 | 0.0002215 | 0.002291 |
| 451 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 9 | 554 | 0.0002224 | 0.002294 |
| 452 | POSITIVE REGULATION OF T CELL MEDIATED IMMUNITY | 3 | 32 | 0.0002323 | 0.002392 |
| 453 | REGULATION OF CELL CYCLE PROCESS | 9 | 558 | 0.0002345 | 0.002409 |
| 454 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 6 | 232 | 0.0002359 | 0.002417 |
| 455 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 11 | 823 | 0.0002366 | 0.00242 |
| 456 | REGULATION OF CELLULAR RESPONSE TO STRESS | 10 | 691 | 0.0002481 | 0.002532 |
| 457 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 33 | 0.0002549 | 0.002589 |
| 458 | NEGATIVE REGULATION OF ANION TRANSPORT | 3 | 33 | 0.0002549 | 0.002589 |
| 459 | MUSCLE CELL DIFFERENTIATION | 6 | 237 | 0.0002645 | 0.002681 |
| 460 | SYSTEM PROCESS | 17 | 1785 | 0.0002651 | 0.002682 |
| 461 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 12 | 983 | 0.0002773 | 0.002799 |
| 462 | PEPTIDYL AMINO ACID MODIFICATION | 11 | 841 | 0.0002849 | 0.002863 |
| 463 | REGULATION OF DNA METABOLIC PROCESS | 7 | 340 | 0.0002843 | 0.002863 |
| 464 | REPRODUCTION | 14 | 1297 | 0.0002913 | 0.002922 |
| 465 | PROTEIN IMPORT | 5 | 155 | 0.0002922 | 0.002924 |
| 466 | POSITIVE REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 4 | 85 | 0.0002935 | 0.00293 |
| 467 | REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 5 | 156 | 0.0003011 | 0.002993 |
| 468 | PROTEIN LOCALIZATION TO NUCLEUS | 5 | 156 | 0.0003011 | 0.002993 |
| 469 | NEGATIVE REGULATION OF PROTEIN PROCESSING | 3 | 35 | 0.0003041 | 0.003011 |
| 470 | NEGATIVE REGULATION OF PROTEIN MATURATION | 3 | 35 | 0.0003041 | 0.003011 |
| 471 | NEGATIVE REGULATION OF TRANSPORT | 8 | 458 | 0.0003135 | 0.003097 |
| 472 | REGULATION OF JNK CASCADE | 5 | 159 | 0.0003287 | 0.00324 |
| 473 | B CELL DIFFERENTIATION | 4 | 89 | 0.0003499 | 0.003435 |
| 474 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 4 | 89 | 0.0003499 | 0.003435 |
| 475 | RESPONSE TO NERVE GROWTH FACTOR | 3 | 37 | 0.0003591 | 0.003517 |
| 476 | REGULATION OF OXIDOREDUCTASE ACTIVITY | 4 | 90 | 0.0003652 | 0.003562 |
| 477 | CALCIUM MEDIATED SIGNALING | 4 | 90 | 0.0003652 | 0.003562 |
| 478 | NEGATIVE REGULATION OF GENE EXPRESSION | 15 | 1493 | 0.0003713 | 0.003614 |
| 479 | GAMETE GENERATION | 9 | 595 | 0.0003757 | 0.003649 |
| 480 | ESTABLISHMENT OF LOCALIZATION IN CELL | 16 | 1676 | 0.000406 | 0.003936 |
| 481 | WNT SIGNALING PATHWAY CALCIUM MODULATING PATHWAY | 3 | 39 | 0.0004201 | 0.004047 |
| 482 | ERBB2 SIGNALING PATHWAY | 3 | 39 | 0.0004201 | 0.004047 |
| 483 | SPLEEN DEVELOPMENT | 3 | 39 | 0.0004201 | 0.004047 |
| 484 | BIOLOGICAL ADHESION | 12 | 1032 | 0.0004309 | 0.004143 |
| 485 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 5 | 170 | 0.0004466 | 0.004285 |
| 486 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 4 | 95 | 0.0004486 | 0.004295 |
| 487 | REGULATION OF MEIOTIC CELL CYCLE | 3 | 40 | 0.0004529 | 0.004318 |
| 488 | REGULATION OF ACTIVATED T CELL PROLIFERATION | 3 | 40 | 0.0004529 | 0.004318 |
| 489 | MYELOID LEUKOCYTE DIFFERENTIATION | 4 | 96 | 0.0004668 | 0.004432 |
| 490 | FEMALE GAMETE GENERATION | 4 | 96 | 0.0004668 | 0.004432 |
| 491 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 10 | 750 | 0.0004743 | 0.004495 |
| 492 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 9 | 616 | 0.0004831 | 0.004569 |
| 493 | REGULATION OF LIPID STORAGE | 3 | 41 | 0.0004873 | 0.004581 |
| 494 | MITOTIC SPINDLE ASSEMBLY | 3 | 41 | 0.0004873 | 0.004581 |
| 495 | MICROTUBULE CYTOSKELETON ORGANIZATION INVOLVED IN MITOSIS | 3 | 41 | 0.0004873 | 0.004581 |
| 496 | SINGLE ORGANISM CELLULAR LOCALIZATION | 11 | 898 | 0.0004963 | 0.004656 |
| 497 | MEMBRANE ORGANIZATION | 11 | 899 | 0.000501 | 0.00469 |
| 498 | MYELOID LEUKOCYTE MIGRATION | 4 | 99 | 0.0005245 | 0.0049 |
| 499 | MYELOID LEUKOCYTE MEDIATED IMMUNITY | 3 | 43 | 0.0005612 | 0.005222 |
| 500 | REGULATION OF POLYSACCHARIDE METABOLIC PROCESS | 3 | 43 | 0.0005612 | 0.005222 |
| 501 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 10 | 767 | 0.0005647 | 0.005245 |
| 502 | MULTICELLULAR ORGANISM REPRODUCTION | 10 | 768 | 0.0005705 | 0.005288 |
| 503 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 7 | 387 | 0.0006167 | 0.005704 |
| 504 | REGULATION OF INTERLEUKIN 6 PRODUCTION | 4 | 104 | 0.0006317 | 0.00582 |
| 505 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 4 | 104 | 0.0006317 | 0.00582 |
| 506 | POSITIVE REGULATION OF INTERLEUKIN 8 PRODUCTION | 3 | 45 | 0.0006418 | 0.005855 |
| 507 | THYMOCYTE AGGREGATION | 3 | 45 | 0.0006418 | 0.005855 |
| 508 | T CELL DIFFERENTIATION IN THYMUS | 3 | 45 | 0.0006418 | 0.005855 |
| 509 | MACROMOLECULE CATABOLIC PROCESS | 11 | 926 | 0.000641 | 0.005855 |
| 510 | REGULATION OF ALCOHOL BIOSYNTHETIC PROCESS | 3 | 45 | 0.0006418 | 0.005855 |
| 511 | MUSCLE SYSTEM PROCESS | 6 | 282 | 0.0006643 | 0.006049 |
| 512 | REGULATION OF PROTEIN CATABOLIC PROCESS | 7 | 393 | 0.0006752 | 0.006136 |
| 513 | PEPTIDYL THREONINE MODIFICATION | 3 | 46 | 0.0006847 | 0.006211 |
| 514 | DEPHOSPHORYLATION | 6 | 286 | 0.000715 | 0.006472 |
| 515 | REGULATION OF CELL CYCLE ARREST | 4 | 108 | 0.0007279 | 0.006564 |
| 516 | REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 4 | 108 | 0.0007279 | 0.006564 |
| 517 | POSITIVE REGULATION OF OXIDOREDUCTASE ACTIVITY | 3 | 47 | 0.0007294 | 0.006565 |
| 518 | REGULATION OF CERAMIDE BIOSYNTHETIC PROCESS | 2 | 11 | 0.0007466 | 0.006681 |
| 519 | POSITIVE REGULATION OF HAIR CYCLE | 2 | 11 | 0.0007466 | 0.006681 |
| 520 | REGULATION OF FEVER GENERATION | 2 | 11 | 0.0007466 | 0.006681 |
| 521 | REGULATION OF NF KAPPAB IMPORT INTO NUCLEUS | 3 | 48 | 0.000776 | 0.006904 |
| 522 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 3 | 48 | 0.000776 | 0.006904 |
| 523 | REGULATION OF INTERLEUKIN 2 PRODUCTION | 3 | 48 | 0.000776 | 0.006904 |
| 524 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 10 | 801 | 0.000789 | 0.007006 |
| 525 | REGULATION OF TYPE I INTERFERON PRODUCTION | 4 | 111 | 0.0008067 | 0.007149 |
| 526 | REGULATION OF TOLL LIKE RECEPTOR SIGNALING PATHWAY | 3 | 49 | 0.0008244 | 0.007279 |
| 527 | REGULATION OF STEROID BIOSYNTHETIC PROCESS | 3 | 49 | 0.0008244 | 0.007279 |
| 528 | REPRODUCTIVE SYSTEM DEVELOPMENT | 7 | 408 | 0.0008411 | 0.007412 |
| 529 | LOCOMOTION | 12 | 1114 | 0.0008482 | 0.00746 |
| 530 | POSITIVE REGULATION OF INFLAMMATORY RESPONSE | 4 | 113 | 0.0008624 | 0.007571 |
| 531 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 5 | 197 | 0.0008702 | 0.007626 |
| 532 | REGULATION OF ORGANIC ACID TRANSPORT | 3 | 50 | 0.0008746 | 0.007636 |
| 533 | POSITIVE REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 3 | 50 | 0.0008746 | 0.007636 |
| 534 | REGULATION OF CHEMOKINE BIOSYNTHETIC PROCESS | 2 | 12 | 0.0008938 | 0.00773 |
| 535 | I KAPPAB PHOSPHORYLATION | 2 | 12 | 0.0008938 | 0.00773 |
| 536 | REPLICATIVE SENESCENCE | 2 | 12 | 0.0008938 | 0.00773 |
| 537 | MYELIN MAINTENANCE | 2 | 12 | 0.0008938 | 0.00773 |
| 538 | POSITIVE REGULATION OF INTERLEUKIN 2 BIOSYNTHETIC PROCESS | 2 | 12 | 0.0008938 | 0.00773 |
| 539 | REGULATION OF INTERLEUKIN 12 PRODUCTION | 3 | 51 | 0.0009268 | 0.008001 |
| 540 | CELLULAR RESPONSE TO IONIZING RADIATION | 3 | 52 | 0.0009809 | 0.008421 |
| 541 | POSITIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 3 | 52 | 0.0009809 | 0.008421 |
| 542 | ACTIVATION OF MAPKK ACTIVITY | 3 | 52 | 0.0009809 | 0.008421 |
| 543 | PLASMA MEMBRANE ORGANIZATION | 5 | 203 | 0.0009952 | 0.008528 |
| 544 | NEGATIVE REGULATION OF PHOSPHORYLATION | 7 | 422 | 0.001024 | 0.008755 |
| 545 | NEGATIVE REGULATION OF AUTOPHAGY | 3 | 53 | 0.001037 | 0.008853 |
| 546 | REGULATION OF SPHINGOLIPID BIOSYNTHETIC PROCESS | 2 | 13 | 0.001054 | 0.008882 |
| 547 | REGULATION OF MEMBRANE LIPID METABOLIC PROCESS | 2 | 13 | 0.001054 | 0.008882 |
| 548 | POSITIVE REGULATION OF STEROID BIOSYNTHETIC PROCESS | 2 | 13 | 0.001054 | 0.008882 |
| 549 | REGULATION OF HISTONE PHOSPHORYLATION | 2 | 13 | 0.001054 | 0.008882 |
| 550 | GLUCOSE METABOLIC PROCESS | 4 | 119 | 0.001046 | 0.008882 |
| 551 | REGULATION OF PROTEIN POLYUBIQUITINATION | 2 | 13 | 0.001054 | 0.008882 |
| 552 | POSITIVE REGULATION OF MACROPHAGE DIFFERENTIATION | 2 | 13 | 0.001054 | 0.008882 |
| 553 | CYTOKINE PRODUCTION | 4 | 120 | 0.001079 | 0.009075 |
| 554 | CELL MOTILITY | 10 | 835 | 0.001082 | 0.009075 |
| 555 | LOCALIZATION OF CELL | 10 | 835 | 0.001082 | 0.009075 |
| 556 | NEGATIVE REGULATION OF REPRODUCTIVE PROCESS | 3 | 54 | 0.001095 | 0.009131 |
| 557 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 3 | 54 | 0.001095 | 0.009131 |
| 558 | REGULATION OF CELL DEVELOPMENT | 10 | 836 | 0.001092 | 0.009131 |
| 559 | REGULATION OF B CELL ACTIVATION | 4 | 121 | 0.001112 | 0.00926 |
| 560 | NEGATIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 5 | 209 | 0.001133 | 0.009397 |
| 561 | GERM CELL DEVELOPMENT | 5 | 209 | 0.001133 | 0.009397 |
| 562 | REGULATION OF B CELL PROLIFERATION | 3 | 55 | 0.001155 | 0.009564 |
| 563 | LIPID METABOLIC PROCESS | 12 | 1158 | 0.001186 | 0.009804 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | TUMOR NECROSIS FACTOR RECEPTOR SUPERFAMILY BINDING | 11 | 47 | 1.5e-17 | 1.393e-14 |
| 2 | CYTOKINE RECEPTOR BINDING | 17 | 271 | 1.554e-16 | 7.219e-14 |
| 3 | KINASE ACTIVITY | 24 | 842 | 2.336e-15 | 7.235e-13 |
| 4 | ENZYME BINDING | 32 | 1737 | 3.216e-15 | 7.47e-13 |
| 5 | CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC PROCESS | 7 | 15 | 4.916e-14 | 9.134e-12 |
| 6 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 24 | 992 | 8.568e-14 | 1.327e-11 |
| 7 | DEATH RECEPTOR BINDING | 7 | 18 | 2.409e-13 | 3.198e-11 |
| 8 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 8 | 43 | 3.476e-12 | 4.036e-10 |
| 9 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 9 | 70 | 4.836e-12 | 4.992e-10 |
| 10 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 16 | 445 | 7.092e-12 | 6.589e-10 |
| 11 | PROTEIN KINASE ACTIVITY | 18 | 640 | 1.705e-11 | 1.131e-09 |
| 12 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 8 | 51 | 1.49e-11 | 1.131e-09 |
| 13 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 13 | 264 | 1.681e-11 | 1.131e-09 |
| 14 | TUMOR NECROSIS FACTOR RECEPTOR BINDING | 7 | 30 | 1.487e-11 | 1.131e-09 |
| 15 | KINASE BINDING | 17 | 606 | 7.101e-11 | 4.398e-09 |
| 16 | PROTEIN HETERODIMERIZATION ACTIVITY | 15 | 468 | 1.742e-10 | 1.009e-08 |
| 17 | PROTEASE BINDING | 9 | 104 | 1.846e-10 | 1.009e-08 |
| 18 | IDENTICAL PROTEIN BINDING | 22 | 1209 | 3.028e-10 | 1.481e-08 |
| 19 | DEATH RECEPTOR ACTIVITY | 6 | 24 | 2.893e-10 | 1.481e-08 |
| 20 | ADENYL NUCLEOTIDE BINDING | 24 | 1514 | 6.224e-10 | 2.891e-08 |
| 21 | CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 8 | 86 | 1.119e-09 | 4.949e-08 |
| 22 | RECEPTOR BINDING | 22 | 1476 | 1.233e-08 | 5.206e-07 |
| 23 | PROTEIN DIMERIZATION ACTIVITY | 19 | 1149 | 3.048e-08 | 1.229e-06 |
| 24 | KINASE REGULATOR ACTIVITY | 9 | 186 | 3.175e-08 | 1.229e-06 |
| 25 | RIBONUCLEOTIDE BINDING | 24 | 1860 | 3.606e-08 | 1.34e-06 |
| 26 | ENZYME REGULATOR ACTIVITY | 17 | 959 | 6.971e-08 | 2.491e-06 |
| 27 | INTERLEUKIN 1 RECEPTOR BINDING | 4 | 16 | 3.208e-07 | 1.064e-05 |
| 28 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 4 | 16 | 3.208e-07 | 1.064e-05 |
| 29 | PROTEIN PHOSPHATASE BINDING | 7 | 120 | 3.323e-07 | 1.065e-05 |
| 30 | MOLECULAR FUNCTION REGULATOR | 19 | 1353 | 3.928e-07 | 1.216e-05 |
| 31 | CYSTEINE TYPE PEPTIDASE ACTIVITY | 8 | 184 | 4.394e-07 | 1.317e-05 |
| 32 | PROTEIN DOMAIN SPECIFIC BINDING | 13 | 624 | 5.022e-07 | 1.414e-05 |
| 33 | CYSTEINE TYPE ENDOPEPTIDASE REGULATOR ACTIVITY INVOLVED IN APOPTOTIC PROCESS | 5 | 42 | 4.944e-07 | 1.414e-05 |
| 34 | PHOSPHATASE BINDING | 7 | 162 | 2.512e-06 | 6.863e-05 |
| 35 | SIGNAL TRANSDUCER ACTIVITY | 20 | 1731 | 3.845e-06 | 0.0001021 |
| 36 | PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 4 | 30 | 4.642e-06 | 0.0001198 |
| 37 | INSULIN RECEPTOR SUBSTRATE BINDING | 3 | 11 | 8.179e-06 | 0.0002054 |
| 38 | PROTEIN KINASE A BINDING | 4 | 42 | 1.832e-05 | 0.0004365 |
| 39 | NEUROTROPHIN RECEPTOR BINDING | 3 | 14 | 1.79e-05 | 0.0004365 |
| 40 | PROTEIN KINASE A CATALYTIC SUBUNIT BINDING | 3 | 15 | 2.231e-05 | 0.0005182 |
| 41 | PROTEIN COMPLEX BINDING | 13 | 935 | 4.016e-05 | 0.00091 |
| 42 | PROTEIN HOMODIMERIZATION ACTIVITY | 11 | 722 | 7.508e-05 | 0.001661 |
| 43 | CAMP BINDING | 3 | 23 | 8.499e-05 | 0.001755 |
| 44 | CYSTEINE TYPE ENDOPEPTIDASE INHIBITOR ACTIVITY INVOLVED IN APOPTOTIC PROCESS | 3 | 23 | 8.499e-05 | 0.001755 |
| 45 | ENZYME INHIBITOR ACTIVITY | 8 | 378 | 8.458e-05 | 0.001755 |
| 46 | PROTEIN SERINE THREONINE PHOSPHATASE ACTIVITY | 4 | 64 | 9.773e-05 | 0.001974 |
| 47 | GROWTH FACTOR RECEPTOR BINDING | 5 | 129 | 0.0001242 | 0.002455 |
| 48 | PEPTIDASE REGULATOR ACTIVITY | 6 | 214 | 0.0001524 | 0.002941 |
| 49 | PROTEIN PHOSPHATASE 2A BINDING | 3 | 28 | 0.0001551 | 0.002941 |
| 50 | MACROMOLECULAR COMPLEX BINDING | 15 | 1399 | 0.0001839 | 0.003417 |
| 51 | ENDOPEPTIDASE ACTIVITY | 8 | 448 | 0.0002703 | 0.004924 |
| 52 | CYCLIC NUCLEOTIDE BINDING | 3 | 36 | 0.0003308 | 0.005911 |
| 53 | HEAT SHOCK PROTEIN BINDING | 4 | 89 | 0.0003499 | 0.00602 |
| 54 | KINASE INHIBITOR ACTIVITY | 4 | 89 | 0.0003499 | 0.00602 |
| 55 | PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 5 | 178 | 0.0005507 | 0.009302 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 17 | 237 | 1.622e-17 | 5.562e-15 |
| 2 | MEMBRANE MICRODOMAIN | 18 | 288 | 1.905e-17 | 5.562e-15 |
| 3 | CATALYTIC COMPLEX | 26 | 1038 | 2.544e-15 | 4.722e-13 |
| 4 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 8 | 20 | 3.234e-15 | 4.722e-13 |
| 5 | MEMBRANE PROTEIN COMPLEX | 23 | 1020 | 1.384e-12 | 1.616e-10 |
| 6 | TRANSFERASE COMPLEX | 18 | 703 | 7.927e-11 | 7.715e-09 |
| 7 | PROTEIN KINASE COMPLEX | 8 | 90 | 1.615e-09 | 1.347e-07 |
| 8 | CILIARY BASE | 5 | 23 | 2.067e-08 | 1.509e-06 |
| 9 | MEMBRANE REGION | 19 | 1134 | 2.471e-08 | 1.603e-06 |
| 10 | PLASMA MEMBRANE PROTEIN COMPLEX | 13 | 510 | 4.952e-08 | 2.892e-06 |
| 11 | CD40 RECEPTOR COMPLEX | 4 | 11 | 5.9e-08 | 3.132e-06 |
| 12 | CYTOSOLIC PART | 9 | 223 | 1.504e-07 | 7.319e-06 |
| 13 | RECEPTOR COMPLEX | 9 | 327 | 3.642e-06 | 0.0001636 |
| 14 | EXTRINSIC COMPONENT OF MEMBRANE | 8 | 252 | 4.63e-06 | 0.0001931 |
| 15 | IKAPPAB KINASE COMPLEX | 3 | 11 | 8.179e-06 | 0.0003184 |
| 16 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 18 | 1649 | 2.855e-05 | 0.001042 |
| 17 | PHOSPHATASE COMPLEX | 4 | 48 | 3.132e-05 | 0.001076 |
| 18 | PLASMA MEMBRANE RECEPTOR COMPLEX | 6 | 175 | 5.031e-05 | 0.001632 |
| 19 | MITOCHONDRION | 16 | 1633 | 0.0003035 | 0.008963 |
| 20 | PLASMA MEMBRANE RAFT | 4 | 86 | 0.0003069 | 0.008963 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04210_Apoptosis | 75 | 89 | 5.759e-198 | 1.019e-195 | |
| 2 | hsa04380_Osteoclast_differentiation | 27 | 128 | 4.502e-41 | 3.984e-39 | |
| 3 | hsa04620_Toll.like_receptor_signaling_pathway | 24 | 102 | 1.092e-37 | 6.442e-36 | |
| 4 | hsa04660_T_cell_receptor_signaling_pathway | 24 | 108 | 5.065e-37 | 2.236e-35 | |
| 5 | hsa04662_B_cell_receptor_signaling_pathway | 22 | 75 | 6.316e-37 | 2.236e-35 | |
| 6 | hsa04010_MAPK_signaling_pathway | 29 | 268 | 3.07e-35 | 9.057e-34 | |
| 7 | hsa04722_Neurotrophin_signaling_pathway | 24 | 127 | 3.679e-35 | 9.302e-34 | |
| 8 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 22 | 136 | 1.34e-30 | 2.965e-29 | |
| 9 | hsa04370_VEGF_signaling_pathway | 17 | 76 | 2.635e-26 | 5.182e-25 | |
| 10 | hsa04062_Chemokine_signaling_pathway | 20 | 189 | 5.913e-24 | 1.047e-22 | |
| 11 | hsa04910_Insulin_signaling_pathway | 18 | 138 | 2.745e-23 | 4.417e-22 | |
| 12 | hsa04914_Progesterone.mediated_oocyte_maturation | 14 | 87 | 1.316e-19 | 1.941e-18 | |
| 13 | hsa04920_Adipocytokine_signaling_pathway | 13 | 68 | 2.586e-19 | 3.52e-18 | |
| 14 | hsa04621_NOD.like_receptor_signaling_pathway | 12 | 59 | 2.992e-18 | 3.783e-17 | |
| 15 | hsa04973_Carbohydrate_digestion_and_absorption | 11 | 44 | 6.663e-18 | 7.862e-17 | |
| 16 | hsa04510_Focal_adhesion | 16 | 200 | 2.779e-17 | 3.074e-16 | |
| 17 | hsa04622_RIG.I.like_receptor_signaling_pathway | 12 | 71 | 3.304e-17 | 3.44e-16 | |
| 18 | hsa04150_mTOR_signaling_pathway | 11 | 52 | 5.126e-17 | 5.04e-16 | |
| 19 | hsa04664_Fc_epsilon_RI_signaling_pathway | 12 | 79 | 1.291e-16 | 1.203e-15 | |
| 20 | hsa04630_Jak.STAT_signaling_pathway | 13 | 155 | 1.828e-14 | 1.618e-13 | |
| 21 | hsa04012_ErbB_signaling_pathway | 11 | 87 | 2.144e-14 | 1.807e-13 | |
| 22 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 11 | 95 | 5.838e-14 | 4.697e-13 | |
| 23 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 8 | 42 | 2.838e-12 | 2.184e-11 | |
| 24 | hsa04115_p53_signaling_pathway | 9 | 69 | 4.227e-12 | 3.117e-11 | |
| 25 | hsa04720_Long.term_potentiation | 8 | 70 | 2.088e-10 | 1.478e-09 | |
| 26 | hsa04070_Phosphatidylinositol_signaling_system | 8 | 78 | 5.063e-10 | 3.447e-09 | |
| 27 | hsa04623_Cytosolic_DNA.sensing_pathway | 7 | 56 | 1.568e-09 | 1.028e-08 | |
| 28 | hsa04310_Wnt_signaling_pathway | 9 | 151 | 5.153e-09 | 3.257e-08 | |
| 29 | hsa04114_Oocyte_meiosis | 8 | 114 | 1.067e-08 | 6.514e-08 | |
| 30 | hsa04670_Leukocyte_transendothelial_migration | 8 | 117 | 1.311e-08 | 7.733e-08 | |
| 31 | hsa04020_Calcium_signaling_pathway | 8 | 177 | 3.27e-07 | 1.867e-06 | |
| 32 | hsa04810_Regulation_of_actin_cytoskeleton | 8 | 214 | 1.375e-06 | 7.604e-06 | |
| 33 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 6.198e-05 | 0.0003324 | |
| 34 | hsa04360_Axon_guidance | 5 | 130 | 0.0001288 | 0.0006704 | |
| 35 | hsa04640_Hematopoietic_cell_lineage | 4 | 88 | 0.0003351 | 0.001695 | |
| 36 | hsa04962_Vasopressin.regulated_water_reabsorption | 3 | 44 | 0.0006006 | 0.002953 | |
| 37 | hsa04742_Taste_transduction | 3 | 52 | 0.0009809 | 0.004693 | |
| 38 | hsa04340_Hedgehog_signaling_pathway | 3 | 56 | 0.001217 | 0.00567 | |
| 39 | hsa04976_Bile_secretion | 3 | 71 | 0.002411 | 0.01094 | |
| 40 | hsa04971_Gastric_acid_secretion | 3 | 74 | 0.002712 | 0.012 | |
| 41 | hsa04970_Salivary_secretion | 3 | 89 | 0.004565 | 0.01971 | |
| 42 | hsa04540_Gap_junction | 3 | 90 | 0.00471 | 0.01985 | |
| 43 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.006488 | 0.0261 | |
| 44 | hsa04916_Melanogenesis | 3 | 101 | 0.006488 | 0.0261 | |
| 45 | hsa04270_Vascular_smooth_muscle_contraction | 3 | 116 | 0.009482 | 0.03729 | |
| 46 | hsa04530_Tight_junction | 3 | 133 | 0.01371 | 0.05274 | |
| 47 | hsa04120_Ubiquitin_mediated_proteolysis | 3 | 139 | 0.01541 | 0.05805 | |
| 48 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 3 | 168 | 0.0253 | 0.0933 | |
| 49 | hsa04110_Cell_cycle | 2 | 128 | 0.08341 | 0.3013 | |
| 50 | hsa04740_Olfactory_transduction | 3 | 388 | 0.1783 | 0.619 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-629-3p | 23 | PIK3R1 | Sponge network | -2.108 | 0 | -1.285 | 0 | 0.544 |
| 2 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | PIK3R1 | Sponge network | -2.791 | 0 | -1.285 | 0 | 0.529 |
| 3 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 22 | PIK3R1 | Sponge network | -2.039 | 0 | -1.285 | 0 | 0.52 |
| 4 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PIK3R1 | Sponge network | -4.19 | 0 | -1.285 | 0 | 0.51 |
| 5 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | AKT3 | Sponge network | -2.68 | 0 | -1.44 | 0 | 0.497 |
| 6 | RP11-720L2.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | -2.305 | 0 | -1.285 | 0 | 0.496 |
| 7 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | PIK3R1 | Sponge network | -2.856 | 0 | -1.285 | 0 | 0.494 |
| 8 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -1.892 | 0 | -1.285 | 0 | 0.486 |
| 9 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-93-5p | 14 | AKT3 | Sponge network | -2.856 | 0 | -1.44 | 0 | 0.485 |
| 10 | RP11-815I9.4 |
hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-5p | 10 | PRKAR2A | Sponge network | -0.881 | 0.0002 | -0.542 | 0.00821 | 0.477 |
| 11 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -2.952 | 0 | -1.285 | 0 | 0.458 |
| 12 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PIK3R1 | Sponge network | -4.222 | 0 | -1.285 | 0 | 0.438 |
| 13 | AC007743.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -2.595 | 0 | -1.285 | 0 | 0.436 |
| 14 | RP11-815I9.4 |
hsa-miR-106a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-590-5p | 12 | PIK3R1 | Sponge network | -0.881 | 0.0002 | -1.285 | 0 | 0.43 |
| 15 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-589-3p | 15 | PIK3R1 | Sponge network | -2.09 | 0 | -1.285 | 0 | 0.428 |
| 16 | RP11-456K23.1 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | BCL2 | Sponge network | -1.488 | 0 | -0.487 | 0.06421 | 0.423 |
| 17 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | PIK3R1 | Sponge network | -2.062 | 0 | -1.285 | 0 | 0.418 |
| 18 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -1.193 | 0.00359 | -1.285 | 0 | 0.418 |
| 19 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-424-5p | 13 | PIK3R1 | Sponge network | -2.68 | 0 | -1.285 | 0 | 0.417 |
| 20 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -1.488 | 0 | -1.285 | 0 | 0.416 |
| 21 | RP11-354E11.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | -2.138 | 0 | -1.285 | 0 | 0.41 |
| 22 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-374a-3p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-93-5p | 17 | PIK3R1 | Sponge network | -3.04 | 0 | -1.285 | 0 | 0.408 |
| 23 | LIPE-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-766-3p | 15 | PRKAR2A | Sponge network | -0.734 | 0.00039 | -0.542 | 0.00821 | 0.408 |
| 24 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 28 | PIK3R1 | Sponge network | -1.745 | 0 | -1.285 | 0 | 0.407 |
| 25 | CTD-2353F22.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p | 10 | PIK3R1 | Sponge network | -1.501 | 0.00083 | -1.285 | 0 | 0.407 |
| 26 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-28-3p;hsa-miR-335-3p | 11 | AKT3 | Sponge network | -1.761 | 0 | -1.44 | 0 | 0.397 |
| 27 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | PIK3R1 | Sponge network | -2.028 | 0 | -1.285 | 0 | 0.395 |
| 28 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PIK3R1 | Sponge network | -2.142 | 0 | -1.285 | 0 | 0.391 |
| 29 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-28-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | AKT3 | Sponge network | -1.488 | 0 | -1.44 | 0 | 0.389 |
| 30 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-28-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-542-3p | 14 | AKT3 | Sponge network | -2.108 | 0 | -1.44 | 0 | 0.386 |
| 31 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-93-5p | 13 | AKT3 | Sponge network | -1.892 | 0 | -1.44 | 0 | 0.386 |
| 32 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-429;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | -0.873 | 0.00072 | -1.285 | 0 | 0.38 |
| 33 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -2.159 | 0 | -1.285 | 0 | 0.378 |
| 34 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | PIK3R1 | Sponge network | -2.807 | 0 | -1.285 | 0 | 0.374 |
| 35 | LINC00092 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | -2.383 | 0 | -1.285 | 0 | 0.369 |
| 36 | RP11-378A13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | -1.713 | 0 | -1.285 | 0 | 0.367 |
| 37 | RP11-462G12.2 |
hsa-miR-130b-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | ENDOD1 | Sponge network | -0.602 | 0.22954 | 0.057 | 0.80229 | 0.366 |
| 38 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | AKT3 | Sponge network | -1.745 | 0 | -1.44 | 0 | 0.366 |
| 39 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-582-5p;hsa-miR-590-5p | 15 | PIK3R1 | Sponge network | -1.583 | 0 | -1.285 | 0 | 0.364 |
| 40 | AC011899.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | PIK3R1 | Sponge network | -2.611 | 0 | -1.285 | 0 | 0.364 |
| 41 | AC079630.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -3.758 | 0 | -1.285 | 0 | 0.359 |
| 42 | RP5-1042I8.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -0.733 | 0.00018 | -1.285 | 0 | 0.358 |
| 43 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -3.16 | 2.0E-5 | -1.285 | 0 | 0.358 |
| 44 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p | 20 | PIK3R1 | Sponge network | -1.761 | 0 | -1.285 | 0 | 0.354 |
| 45 | RP11-815I9.4 |
hsa-miR-106a-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 10 | AKT3 | Sponge network | -0.881 | 0.0002 | -1.44 | 0 | 0.35 |
| 46 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-660-5p | 12 | IRAK3 | Sponge network | -1.892 | 0 | -1.317 | 1.0E-5 | 0.348 |
| 47 | LINC00961 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -2.724 | 0 | -1.285 | 0 | 0.346 |
| 48 | RBMS3-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -2.307 | 0.0011 | -1.285 | 0 | 0.345 |
| 49 | AC010226.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -1.081 | 2.0E-5 | -1.285 | 0 | 0.344 |
| 50 | AC003991.3 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | ENDOD1 | Sponge network | -0.787 | 0.08132 | 0.057 | 0.80229 | 0.343 |
| 51 | LINC00987 |
hsa-miR-106a-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -0.927 | 0 | -1.285 | 0 | 0.343 |
| 52 | BZRAP1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.785 | 0.00723 | -1.285 | 0 | 0.34 |
| 53 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | PIK3R1 | Sponge network | -0.758 | 0.00105 | -1.285 | 0 | 0.339 |
| 54 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | PIK3R1 | Sponge network | -2.783 | 0 | -1.285 | 0 | 0.338 |
| 55 | AC004947.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | PIK3R1 | Sponge network | -3.94 | 0 | -1.285 | 0 | 0.332 |
| 56 | CTC-523E23.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | PIK3R1 | Sponge network | -1.636 | 0.00051 | -1.285 | 0 | 0.33 |
| 57 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -1.297 | 0 | -1.285 | 0 | 0.329 |
| 58 | CTD-2003C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -3.403 | 0 | -1.285 | 0 | 0.328 |
| 59 | LINC00883 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | -1.466 | 0 | -1.285 | 0 | 0.327 |
| 60 | CTC-429P9.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.908 | 0.00061 | -1.285 | 0 | 0.326 |
| 61 | RP11-389C8.2 |
hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | BCL2 | Sponge network | -2.039 | 0 | -0.487 | 0.06421 | 0.325 |
| 62 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-28-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-542-3p;hsa-miR-93-5p | 12 | AKT3 | Sponge network | -2.039 | 0 | -1.44 | 0 | 0.324 |
| 63 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-542-3p;hsa-miR-93-5p | 11 | AKT3 | Sponge network | -3.04 | 0 | -1.44 | 0 | 0.323 |
| 64 | AC020571.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-3065-5p;hsa-miR-335-3p;hsa-miR-362-5p | 10 | AKT3 | Sponge network | 0.248 | 0.40984 | -1.44 | 0 | 0.321 |
| 65 | RP11-293M10.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.199 | 0.00063 | -1.285 | 0 | 0.32 |
| 66 | RP11-354E11.2 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | ENDOD1 | Sponge network | -2.138 | 0 | 0.057 | 0.80229 | 0.32 |
| 67 | RP11-434D9.1 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -4.573 | 0 | -1.285 | 0 | 0.319 |
| 68 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | PIK3R1 | Sponge network | -0.427 | 0.1559 | -1.285 | 0 | 0.318 |
| 69 | CTC-559E9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.737 | 0.0562 | -1.285 | 0 | 0.318 |
| 70 | RP11-789C1.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -4.664 | 0.00012 | -1.285 | 0 | 0.317 |
| 71 | LINC00607 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | PIK3R1 | Sponge network | -2.277 | 0 | -1.285 | 0 | 0.316 |
| 72 | RP11-517P14.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -0.795 | 6.0E-5 | -1.285 | 0 | 0.315 |
| 73 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -1.312 | 0 | -1.285 | 0 | 0.314 |
| 74 | RP11-1223D19.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 11 | ENDOD1 | Sponge network | -0.862 | 0.05389 | 0.057 | 0.80229 | 0.314 |
| 75 | RP1-151F17.2 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-5p | 10 | PIK3R1 | Sponge network | -0.793 | 0.00074 | -1.285 | 0 | 0.313 |
| 76 | RP11-736K20.5 | hsa-miR-15a-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -1.84 | 0 | -1.285 | 0 | 0.312 |
| 77 | FZD10-AS1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-28-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | AKT3 | Sponge network | -1.071 | 0.02419 | -1.44 | 0 | 0.309 |
| 78 | CTD-2619J13.17 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.819 | 0.00282 | -1.285 | 0 | 0.307 |
| 79 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-28-3p;hsa-miR-424-5p;hsa-miR-93-5p | 12 | AKT3 | Sponge network | -4.19 | 0 | -1.44 | 0 | 0.305 |
| 80 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -0.428 | 0.21699 | -1.285 | 0 | 0.303 |
| 81 | RP11-462G12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | PIK3R1 | Sponge network | -1.071 | 0.01175 | -1.285 | 0 | 0.302 |
| 82 | LINC01088 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -1.385 | 0.01604 | -1.285 | 0 | 0.3 |
| 83 | CTD-2269F5.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | AKT3 | Sponge network | -1.576 | 0.00334 | -1.44 | 0 | 0.3 |
| 84 | AC016735.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -2.711 | 0.00282 | -1.285 | 0 | 0.298 |
| 85 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -1.062 | 0 | -1.285 | 0 | 0.298 |
| 86 | TBX5-AS1 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-365a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-5p | 14 | BCL2 | Sponge network | -2.108 | 0 | -0.487 | 0.06421 | 0.297 |
| 87 | LINC00702 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | BCL2 | Sponge network | -2.856 | 0 | -0.487 | 0.06421 | 0.297 |
| 88 | RP11-88I21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -8.789 | 0 | -1.285 | 0 | 0.297 |
| 89 | RP11-1008C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | -1.249 | 0 | -1.285 | 0 | 0.297 |
| 90 | AC034187.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -1.974 | 0 | -1.285 | 0 | 0.297 |
| 91 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-93-5p | 12 | AKT3 | Sponge network | -2.791 | 0 | -1.44 | 0 | 0.293 |
| 92 | RP11-365O16.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -2.765 | 0.00017 | -1.285 | 0 | 0.293 |
| 93 | FENDRR |
hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-3200-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p | 10 | IRAK3 | Sponge network | -4.222 | 0 | -1.317 | 1.0E-5 | 0.291 |
| 94 | CASC2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 21 | PIK3R1 | Sponge network | -1.086 | 0 | -1.285 | 0 | 0.29 |
| 95 | RP11-532F6.3 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | BCL2 | Sponge network | -2.028 | 0 | -0.487 | 0.06421 | 0.29 |
| 96 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -0.161 | 0.52228 | -1.285 | 0 | 0.29 |
| 97 | RP11-416I2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 10 | PPP3CA | Sponge network | 3.177 | 1.0E-5 | -0.314 | 0.01652 | 0.289 |
| 98 | LINC00702 |
hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p | 12 | IRAK3 | Sponge network | -2.856 | 0 | -1.317 | 1.0E-5 | 0.285 |
| 99 | RP11-268F1.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -6.474 | 1.0E-5 | -1.285 | 0 | 0.285 |
| 100 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | PIK3R1 | Sponge network | -0.761 | 0.05061 | -1.285 | 0 | 0.285 |
| 101 | AF131215.9 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | PIK3R1 | Sponge network | -1.808 | 0 | -1.285 | 0 | 0.282 |
| 102 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-642a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PPP3CA | Sponge network | -1.297 | 0 | -0.314 | 0.01652 | 0.281 |
| 103 | RP11-352D13.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PIK3R1 | Sponge network | -4.634 | 0 | -1.285 | 0 | 0.28 |
| 104 | LIPE-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -0.734 | 0.00039 | -1.285 | 0 | 0.28 |
| 105 | SRGAP3-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-450b-5p | 10 | PIK3R1 | Sponge network | -5.086 | 5.0E-5 | -1.285 | 0 | 0.279 |
| 106 | RP11-401P9.4 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 10 | BCL2 | Sponge network | -3.04 | 0 | -0.487 | 0.06421 | 0.279 |
| 107 | RP11-476D10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -4.519 | 0 | -1.285 | 0 | 0.279 |
| 108 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | PIK3R1 | Sponge network | -0.568 | 0.02011 | -1.285 | 0 | 0.277 |
| 109 | CTD-2135D7.5 | hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -3.162 | 0 | -1.285 | 0 | 0.277 |
| 110 | CTC-441N14.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-582-5p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -2.273 | 0.01231 | -1.285 | 0 | 0.276 |
| 111 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 10 | AKT3 | Sponge network | -2.783 | 0 | -1.44 | 0 | 0.276 |
| 112 | LINC00443 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-542-3p | 15 | PIK3R1 | Sponge network | -3.704 | 0.0003 | -1.285 | 0 | 0.276 |
| 113 | AC008268.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -5.661 | 0 | -1.285 | 0 | 0.276 |
| 114 | LIPE-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-642a-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | PPP3CA | Sponge network | -0.734 | 0.00039 | -0.314 | 0.01652 | 0.274 |
| 115 | RP11-1008C21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | -1.826 | 3.0E-5 | -1.285 | 0 | 0.272 |
| 116 | SNHG18 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -1.073 | 0.00533 | -1.285 | 0 | 0.27 |
| 117 | C1orf132 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -0.86 | 0.02429 | -1.285 | 0 | 0.269 |
| 118 | LINC00551 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -2.179 | 0.00466 | -1.285 | 0 | 0.269 |
| 119 | CTD-2013N24.2 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | BCL2 | Sponge network | -1.745 | 0 | -0.487 | 0.06421 | 0.268 |
| 120 | RP11-890B15.3 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -0.389 | 0.02728 | -1.285 | 0 | 0.268 |
| 121 | TBX5-AS1 |
hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-3200-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-5p | 12 | IRAK3 | Sponge network | -2.108 | 0 | -1.317 | 1.0E-5 | 0.267 |
| 122 | LINC00657 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-766-3p | 10 | PRKAR2A | Sponge network | -0.222 | 0.34002 | -0.542 | 0.00821 | 0.266 |
| 123 | LINC01024 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -0.659 | 0.02711 | -1.285 | 0 | 0.266 |
| 124 | CTC-366B18.4 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -0.652 | 0.01265 | -1.285 | 0 | 0.265 |
| 125 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-93-5p | 11 | AKT3 | Sponge network | -2.952 | 0 | -1.44 | 0 | 0.265 |
| 126 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-129-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | IL1RAP | Sponge network | -0.427 | 0.1559 | 0 | 0.99919 | 0.265 |
| 127 | LINC00511 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7c-5p;hsa-miR-139-5p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-374a-5p | 10 | CASP3 | Sponge network | 2.848 | 0 | 0.75 | 0 | 0.264 |
| 128 | RP1-78O14.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-629-3p | 12 | PIK3R1 | Sponge network | -4.409 | 0 | -1.285 | 0 | 0.264 |
| 129 | AC096670.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -1.939 | 7.0E-5 | -1.285 | 0 | 0.263 |
| 130 | AC007743.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-5p | 13 | PRKAR2A | Sponge network | -2.595 | 0 | -0.542 | 0.00821 | 0.263 |
| 131 | RP11-815I9.4 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-590-5p | 10 | BCL2 | Sponge network | -0.881 | 0.0002 | -0.487 | 0.06421 | 0.262 |
| 132 | RP5-839B4.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-93-5p | 16 | PIK3R1 | Sponge network | -5.037 | 0 | -1.285 | 0 | 0.262 |
| 133 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 11 | AKT3 | Sponge network | -2.142 | 0 | -1.44 | 0 | 0.262 |
| 134 | DIO3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | -1.936 | 0.00085 | -1.285 | 0 | 0.262 |
| 135 | CTC-523E23.1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -1.223 | 0.0005 | -1.285 | 0 | 0.262 |
| 136 | RP4-668J24.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-629-3p | 12 | PIK3R1 | Sponge network | -2.397 | 0.00331 | -1.285 | 0 | 0.261 |
| 137 | RP11-347J14.7 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -5.394 | 0 | -1.285 | 0 | 0.261 |
| 138 | RP11-582J16.4 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -2.665 | 0.00014 | -1.285 | 0 | 0.26 |
| 139 | LINC00261 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | PIK3R1 | Sponge network | -2.566 | 0.00025 | -1.285 | 0 | 0.259 |
| 140 | RP11-23P13.6 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-629-3p | 12 | PIK3R1 | Sponge network | -0.705 | 0.00072 | -1.285 | 0 | 0.256 |
| 141 | GAS6-AS2 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 10 | BCL2 | Sponge network | -1.761 | 0 | -0.487 | 0.06421 | 0.256 |
| 142 | AGAP11 | hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | -2.127 | 0 | -1.285 | 0 | 0.255 |
| 143 | LINC00619 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p | 12 | PIK3R1 | Sponge network | -2.307 | 0.02217 | -1.285 | 0 | 0.255 |
| 144 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | PIK3R1 | Sponge network | 0.053 | 0.85755 | -1.285 | 0 | 0.255 |
| 145 | CTA-221G9.11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -0.645 | 0.06404 | -1.285 | 0 | 0.254 |
| 146 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -0.672 | 0.02084 | -1.285 | 0 | 0.254 |
| 147 | RP11-1024P17.1 |
hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-3200-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-3p | 10 | IRAK3 | Sponge network | -2.062 | 0 | -1.317 | 1.0E-5 | 0.254 |
| 148 | RP11-473I1.9 |
hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 10 | PPP3CA | Sponge network | -0.037 | 0.8641 | -0.314 | 0.01652 | 0.252 |
| 149 | RP11-677M14.3 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.866 | 0 | -1.285 | 0 | 0.251 |
| 150 | MIR497HG |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | BCL2 | Sponge network | -2.142 | 0 | -0.487 | 0.06421 | 0.251 |
| 151 | AC012501.2 | hsa-miR-139-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-342-3p | 10 | PPP3R1 | Sponge network | 2.091 | 0.06305 | -0.021 | 0.82881 | 0.251 |