This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-21-5p | AKT2 | 1.75 | 0 | -0.01 | 0.9227 | miRNAWalker2 validate | -0.15 | 0.00352 | NA | |
| 2 | hsa-miR-29b-3p | AKT2 | -0.23 | 0.36746 | -0.01 | 0.9227 | MirTarget | -0.14 | 0.00022 | 26512921; 26564501; 24076586 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3;The expression of miR-29b was significantly upregualted by cisplatin treatmentwhile its target gene AKT2 was downregulated;Furthermore a feed-back loop comprising of c-Myc miR-29 family and Akt2 were found in myeloid leukemogenesis |
| 3 | hsa-miR-106b-5p | AKT3 | 1.71 | 0 | -0.75 | 0.06936 | miRNATAP | -0.37 | 7.0E-5 | NA | |
| 4 | hsa-miR-107 | AKT3 | 0.9 | 5.0E-5 | -0.75 | 0.06936 | PITA; miRanda | -0.6 | 0 | NA | |
| 5 | hsa-miR-15b-5p | AKT3 | 1.62 | 0 | -0.75 | 0.06936 | miRNATAP | -0.36 | 0.00041 | NA | |
| 6 | hsa-miR-16-5p | AKT3 | 1.01 | 1.0E-5 | -0.75 | 0.06936 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.47 | 0.00031 | NA | |
| 7 | hsa-miR-17-3p | AKT3 | 1.31 | 0 | -0.75 | 0.06936 | miRNATAP | -0.52 | 0 | NA | |
| 8 | hsa-miR-17-5p | AKT3 | 1.66 | 0 | -0.75 | 0.06936 | TargetScan; miRNATAP | -0.28 | 0.0027 | NA | |
| 9 | hsa-miR-29a-3p | AKT3 | -0.11 | 0.61501 | -0.75 | 0.06936 | miRNATAP | -0.41 | 0.00212 | NA | |
| 10 | hsa-miR-29b-3p | AKT3 | -0.23 | 0.36746 | -0.75 | 0.06936 | miRNATAP | -0.34 | 0.00361 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 11 | hsa-miR-32-3p | AKT3 | 0.58 | 0.11837 | -0.75 | 0.06936 | mirMAP | -0.32 | 0.00023 | NA | |
| 12 | hsa-miR-320b | AKT3 | 1.11 | 0.0005 | -0.75 | 0.06936 | PITA; miRanda; miRNATAP | -0.26 | 0.0039 | NA | |
| 13 | hsa-miR-335-3p | AKT3 | 2.52 | 0 | -0.75 | 0.06936 | mirMAP | -0.22 | 0.00074 | NA | |
| 14 | hsa-miR-33a-3p | AKT3 | 0.35 | 0.32171 | -0.75 | 0.06936 | mirMAP | -0.38 | 2.0E-5 | NA | |
| 15 | hsa-miR-362-3p | AKT3 | -0.03 | 0.91378 | -0.75 | 0.06936 | miRanda | -0.38 | 4.0E-5 | NA | |
| 16 | hsa-miR-362-5p | AKT3 | -0.35 | 0.35491 | -0.75 | 0.06936 | PITA; TargetScan; miRNATAP | -0.23 | 0.00253 | NA | |
| 17 | hsa-miR-501-3p | AKT3 | 0.73 | 0.02704 | -0.75 | 0.06936 | miRNATAP | -0.39 | 1.0E-5 | NA | |
| 18 | hsa-miR-502-3p | AKT3 | -0.16 | 0.55747 | -0.75 | 0.06936 | miRNATAP | -0.5 | 0 | NA | |
| 19 | hsa-miR-505-3p | AKT3 | 0.83 | 0.00112 | -0.75 | 0.06936 | mirMAP | -0.35 | 0.00235 | 22051041 | We also find that Akt3 correlate inversely with miR-505 modulates drug sensitivity in MCF7-ADR |
| 20 | hsa-miR-548v | AKT3 | -0.16 | 0.70867 | -0.75 | 0.06936 | miRNATAP | -0.27 | 0.00031 | NA | |
| 21 | hsa-miR-577 | AKT3 | 0.91 | 0.22561 | -0.75 | 0.06936 | mirMAP | -0.23 | 0 | NA | |
| 22 | hsa-miR-616-5p | AKT3 | 0.83 | 0.03478 | -0.75 | 0.06936 | mirMAP | -0.27 | 0.00041 | NA | |
| 23 | hsa-miR-193b-3p | CASP9 | 0.28 | 0.45126 | -0.31 | 0.11004 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 24 | hsa-miR-577 | HSPB1 | 0.91 | 0.22561 | 0.99 | 0.06197 | PITA | -0.31 | 0 | NA | |
| 25 | hsa-miR-149-5p | KDR | 0.71 | 0.29685 | -0.7 | 0.04936 | miRNATAP | -0.22 | 0 | NA | |
| 26 | hsa-miR-23a-3p | KDR | 1.11 | 0 | -0.7 | 0.04936 | mirMAP | -0.55 | 1.0E-5 | NA | |
| 27 | hsa-let-7a-3p | KRAS | 0.5 | 0.04111 | 0.04 | 0.88822 | mirMAP; miRNATAP | -0.27 | 0.00252 | 24727325; 20603437; 24890702; 23324806; 21994416; 23167843; 27620744; 20177422; 25183481; 22584434; 18922928 | Association study of the let 7 miRNA complementary site variant in the 3' untranslated region of the KRAS gene in stage III colon cancer NCCTG N0147 Clinical Trial; A let-7 microRNA-complementary site LCS6 polymorphism in the 3' untranslated region of the KRAS gene has been shown to disrupt let-7 binding and upregulate KRAS expression;A let 7 microRNA binding site polymorphism in 3' untranslated region of KRAS gene predicts response in wild type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy; A polymorphism in a let-7 microRNA complementary site lcs6 in the KRAS 3' untranslated region UTR is associated with an increased cancer risk in non-small-cell lung cancer and reduced overall survival OS in oral cancers; the presence of KRAS let-7 lcs6 polymorphism was evaluated in 130 mCRC patients who were enrolled in a phase II study of cetuximab monotherapy IMCL-0144; KRAS let-7 lcs6 polymorphism was found to be related to object response rate ORR in mCRC patients whose tumors had KRASwt;Let 7 microRNA binding site polymorphism in the 3'UTR of KRAS and colorectal cancer outcome: a systematic review and meta analysis; There is a small but growing body of literature regarding the predictive utility of a Let-7 microRNA-binding-site polymorphism in the 3'-untranslated region UTR of KRAS KRAS-LCS6 for colorectal cancer outcome although the results are conflicting; A PubMed search was conducted to identify all studies reporting on KRAS let-7 microRNA-binding site polymorphism LCS6; rs61764370 and colorectal cancer outcome;The LCS6 polymorphism in the binding site of let 7 microRNA to the KRAS 3' untranslated region: its role in the efficacy of anti EGFR based therapy in metastatic colorectal cancer patients; The lethal-7 let-7 family of microRNAs regulates KRAS activity; We studied the association of the KRAS let-7 LCS6 polymorphism with the response in 100 refractory mCRC patients treated with anti-EGFR antibodies; The KRAS let-7 LCS6 polymorphism was genotyped using the BioMark system in blood and tumor DNA samples; The KRAS let-7 LCS6 G-allele showed a statistically significant association with nonresponse to anti-EGFR-based treatment: 31.9% of patients with the T/T genotype presented a complete or a partial response versus no patients with T/G or G/G genotypes P=0.004;A let 7 microRNA SNP in the KRAS 3'UTR is prognostic in early stage colorectal cancer; Recently a SNP in a lethal-7 let-7 miRNA complementary site LCS6 in the KRAS 3'untranslated region was suggested to affect survival in metastatic CRC;Let 7 miRNA binding site polymorphism in the KRAS 3'UTR; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5 fluorouracil and oxaliplatin +/ cetuximab; Recent studies have reported associations between a variant allele in a let-7 microRNA complementary site LCS6 within the 3'untranslated region 3'UTR of KRAS rs61764370 and clinical outcome in metastatic colorectal cancer mCRC patients receiving cetuximab;A let 7 microRNA binding site polymorphism in the KRAS 3'UTR is associated with increased risk and reduced survival for gallbladder cancer in North Indian population; The let-7 microRNAs play a key role in regulating KRAS expression and a polymorphism in 3' untranslated region rs61764370 T/G of KRAS leads to its higher expression;Genetic modulation of the Let 7 microRNA binding to KRAS 3' untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab irinotecan; There is increasing evidence that the Let-7 microRNA miRNA exerts an effect as a tumor suppressor by targeting the KRAS mRNA; The Let-7 complementary site LCS6 T>G variant in the KRAS 3'-untranslated region weakens Let-7 binding;A let 7 microRNA binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy;High let 7a microRNA levels in KRAS mutated colorectal carcinomas may rescue anti EGFR therapy effects in patients with chemotherapy refractory metastatic disease; Preclinical and experimental data in vivo indicate that Lethal-7 Let-7 microRNA downregulates KRAS with antitumor effects in the presence of activating KRAS mutations; In 59 patients harboring KRAS mutations Let-7a levels were analyzed for association with overall survival OS and progression-free survival PFS times; An exploratory subgroup analysis was performed using the rs61764370 LCS6 T>G polymorphism that experimentally impairs Let-7 binding to KRAS mRNA; In patients with KRAS mutations Let-7a analysis may serve to identify subgroups of patients who may still benefit from EGFR inhibition and this may open up new perspectives for alternative treatment strategies;A SNP in a let 7 microRNA complementary site in the KRAS 3' untranslated region increases non small cell lung cancer risk; The purpose of this study was to identify single nucleotide polymorphisms SNP that could modify let-7 binding and to assess the effect of such SNPs on target gene regulation and risk for non-small cell lung cancer NSCLC let-7 complementary sites LCS were sequenced in the KRAS 3' untranslated region from 74 NSCLC cases to identify mutations and SNPs that correlated with NSCLC; The LCS6 variant allele in a KRAS miRANA complementary site is significantly associated with increased risk for NSCLC among moderate smokers and represents a new paradigm for let-7 miRNAs in lung cancer susceptibility |
| 28 | hsa-miR-143-3p | KRAS | -0.66 | 0.04832 | 0.04 | 0.88822 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.18 | 0.00744 | 22804917; 27725862; 19137007; 26581910; 23173124; 26824186; 21197560 | In KRAS mutated tumours increased miR-200b and decreased miR-143 expression were associated with a good PFS;The expression of miR-143 is often down-regulated and it might play an important role by targeting KRAS in colorectal cancer CRC; Dual-luciferase reporter assays were performed to validate whether KRAS was regulated by miR-143; Dual-luciferase assays indicated that KRAS was a direct target of miR-143 as subsequently demonstrated by qPCR and western blot analysis showing that infection of SW480 cells with Ad-ZD55-miR-143 resulted in the down-regulation of KRAS at both mRNA and protein levels;Role of miR 143 targeting KRAS in colorectal tumorigenesis; In particular among a panel of presumed targets generated by in silico analysis that may interact with these aberrantly expressed miRNAs KRAS oncogene has been further experimentally validated as the target of miR-143; First an inverse correlation between KRAS protein and miR-143 in vivo was found; Second KRAS expression in Lovo cells was significantly abolished by treatment with miR-143 mimic whereas miR-143 inhibitor increased KRAS protein level; Third luciferase reporter assay confirmed that miR-143 directly recognize the 3'-untranslated region of KRAS transcripts; Finally inhibition of KRAS expression by miR-143 inhibits constitutive phosphorylation of ERK1/2; Taken together the present study provides the first evidences that miR-143 is significant in suppressing colorectal cancer cell growth through inhibition of KRAS translation;MicroRNA 143 replenishment re sensitizes colorectal cancer cells harboring mutant but not wild type KRAS to paclitaxel treatment; Our results showed that miR-143 but not let-7b increased sensitization of KRAS mutant tumor cells to paclitaxel; Furthermore transfection of miR-143 but not let-7b mimic negatively regulated the expression of mutant but not wild-type KRAS; Combination of miR-143 mimic and paclitaxel induced the onset of apoptosis and reverted in vitro metastatic properties migration and invasion in KRAS mutant tumor cells; MiR-143 thus can be used as a chemosensitizer for the treatment of KRAS mutant tumors and warrants further investigations in in vitro and pre-clinical in vivo models;In turn the level of miR-143 in CRC cells decreasing the amount of MACC1 metastasis-associated in colon cancer-1 and oncogenic KRAS protein may be utilized as a prognostic marker;Stable miR-143 or miR-145 overexpression increased cell sensitivity to cetuximab resulting in a significant increase of cetuximab-mediated ADCC independently of KRAS status;miR 143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS; The expression level of miR-143 and its target gene KRAS were measured by realtime PCR and western blotting respectively; Our results revealed an inverse correlation of expression between miR-143 and KRAS protein in prostate cancer samples Pearson's correlation scatter plots: R = -0.707 P < 0.05; These findings suggest that miR-143 plays an important role in prostate cancer proliferation migration and chemosensitivity by suppressing KRAS and subsequent inactivation of MAPK pathway which provides a potential development of a new approach for the treatment of prostate cancer |
| 29 | hsa-miR-193b-3p | KRAS | 0.28 | 0.45126 | 0.04 | 0.88822 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.2 | 0.00063 | 25905463 | Deregulation of the MiR 193b KRAS Axis Contributes to Impaired Cell Growth in Pancreatic Cancer; Mechanistically KRAS was verified as a direct effector of miR-193b through which the AKT and ERK pathways were modulated and cell growth of PDAC cells was suppressed; Taken together our findings indicate that miR-193b-mediated deregulation of the KRAS axis is involved in pancreatic carcinogenesis and suggest that miR-193b could be a potentially effective target for PDAC therapy |
| 30 | hsa-miR-224-3p | KRAS | 1.52 | 0.0065 | 0.04 | 0.88822 | mirMAP | -0.11 | 0.00837 | 25919696 | MicroRNA 224 is associated with colorectal cancer progression and response to 5 fluorouracil based chemotherapy by KRAS dependent and independent mechanisms; MicroRNA-224 was differentially expressed in dysplastic colorectal disease and in isogeneic KRAS WT and mutant HCT116 cells; 5-FU chemosensitivity was significantly increased in miR-224 knockdown cells and in NIH3T3 cells expressing KRAS and BRAF mutant proteins |
| 31 | hsa-miR-27b-3p | KRAS | 0.08 | 0.72527 | 0.04 | 0.88822 | miRNATAP | -0.31 | 0.00105 | NA | |
| 32 | hsa-miR-101-3p | MAP2K1 | -0.45 | 0.02834 | 0.35 | 0.02947 | miRNAWalker2 validate | -0.21 | 0.00016 | NA | |
| 33 | hsa-miR-140-3p | MAP2K1 | -0.3 | 0.12754 | 0.35 | 0.02947 | PITA | -0.25 | 2.0E-5 | NA | |
| 34 | hsa-miR-30c-5p | MAP2K1 | -0.98 | 5.0E-5 | 0.35 | 0.02947 | miRNAWalker2 validate | -0.19 | 5.0E-5 | NA | |
| 35 | hsa-miR-497-5p | MAP2K1 | -0.8 | 0.0036 | 0.35 | 0.02947 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 36 | hsa-miR-126-5p | MAPK1 | 0.42 | 0.07532 | -0.5 | 0.00057 | mirMAP | -0.12 | 0.00476 | NA | |
| 37 | hsa-miR-140-3p | MAPK1 | -0.3 | 0.12754 | -0.5 | 0.00057 | PITA; miRNATAP | -0.15 | 0.00713 | NA | |
| 38 | hsa-miR-140-5p | MAPK1 | 0.21 | 0.303 | -0.5 | 0.00057 | miRanda | -0.17 | 0.00108 | NA | |
| 39 | hsa-miR-142-5p | MAPK1 | 1.56 | 1.0E-5 | -0.5 | 0.00057 | mirMAP | -0.12 | 2.0E-5 | NA | |
| 40 | hsa-miR-16-5p | MAPK1 | 1.01 | 1.0E-5 | -0.5 | 0.00057 | mirMAP | -0.13 | 0.00527 | NA | |
| 41 | hsa-miR-17-3p | MAPK1 | 1.31 | 0 | -0.5 | 0.00057 | mirMAP | -0.11 | 0.00661 | NA | |
| 42 | hsa-miR-32-5p | MAPK1 | 0.42 | 0.10646 | -0.5 | 0.00057 | mirMAP | -0.13 | 0.00101 | NA | |
| 43 | hsa-miR-320a | MAPK1 | 0.59 | 0.0119 | -0.5 | 0.00057 | miRNAWalker2 validate; PITA; miRanda; miRNATAP | -0.12 | 0.00441 | NA | |
| 44 | hsa-miR-320b | MAPK1 | 1.11 | 0.0005 | -0.5 | 0.00057 | PITA; miRanda; miRNATAP | -0.12 | 0.00017 | NA | |
| 45 | hsa-miR-3614-3p | MAPK1 | 0.82 | 0.01436 | -0.5 | 0.00057 | mirMAP | -0.14 | 0 | NA | |
| 46 | hsa-miR-361-3p | MAPK11 | -0.13 | 0.56605 | -0.07 | 0.83024 | mirMAP | -0.41 | 0.00017 | NA | |
| 47 | hsa-miR-103a-3p | NFAT5 | 0.84 | 0 | -0.04 | 0.82158 | MirTarget | -0.26 | 0.0018 | NA | |
| 48 | hsa-miR-106b-5p | NFAT5 | 1.71 | 0 | -0.04 | 0.82158 | MirTarget; miRNATAP | -0.16 | 0.0002 | NA | |
| 49 | hsa-miR-107 | NFAT5 | 0.9 | 5.0E-5 | -0.04 | 0.82158 | MirTarget | -0.25 | 4.0E-5 | NA | |
| 50 | hsa-miR-148b-3p | NFAT5 | 0.3 | 0.17466 | -0.04 | 0.82158 | miRNATAP | -0.26 | 4.0E-5 | NA | |
| 51 | hsa-miR-15b-3p | NFAT5 | 1.76 | 0 | -0.04 | 0.82158 | mirMAP | -0.15 | 0.00028 | NA | |
| 52 | hsa-miR-15b-5p | NFAT5 | 1.62 | 0 | -0.04 | 0.82158 | MirTarget | -0.16 | 0.00111 | NA | |
| 53 | hsa-miR-16-5p | NFAT5 | 1.01 | 1.0E-5 | -0.04 | 0.82158 | MirTarget | -0.19 | 0.00261 | NA | |
| 54 | hsa-miR-17-3p | NFAT5 | 1.31 | 0 | -0.04 | 0.82158 | mirMAP | -0.19 | 0.00021 | NA | |
| 55 | hsa-miR-212-3p | NFAT5 | 0.96 | 0.00133 | -0.04 | 0.82158 | miRNATAP | -0.13 | 0.00454 | NA | |
| 56 | hsa-miR-24-3p | NFAT5 | 1.09 | 0 | -0.04 | 0.82158 | miRNAWalker2 validate; miRNATAP | -0.18 | 0.00301 | NA | |
| 57 | hsa-miR-26b-3p | NFAT5 | -0.01 | 0.96577 | -0.04 | 0.82158 | MirTarget; miRNATAP | -0.16 | 0.0057 | NA | |
| 58 | hsa-miR-32-5p | NFAT5 | 0.42 | 0.10646 | -0.04 | 0.82158 | miRNATAP | -0.18 | 0.00113 | NA | |
| 59 | hsa-miR-320b | NFAT5 | 1.11 | 0.0005 | -0.04 | 0.82158 | mirMAP | -0.15 | 0.00047 | NA | |
| 60 | hsa-miR-330-3p | NFAT5 | 0.8 | 0.00747 | -0.04 | 0.82158 | mirMAP | -0.14 | 0.00306 | NA | |
| 61 | hsa-miR-33a-3p | NFAT5 | 0.35 | 0.32171 | -0.04 | 0.82158 | mirMAP | -0.12 | 0.00278 | NA | |
| 62 | hsa-miR-33a-5p | NFAT5 | 0.41 | 0.32143 | -0.04 | 0.82158 | MirTarget | -0.11 | 0.00088 | NA | |
| 63 | hsa-miR-340-5p | NFAT5 | -0.16 | 0.52991 | -0.04 | 0.82158 | MirTarget | -0.14 | 0.00802 | NA | |
| 64 | hsa-miR-424-3p | NFAT5 | 0.86 | 0.02346 | -0.04 | 0.82158 | miRNATAP | -0.14 | 0.00037 | NA | |
| 65 | hsa-miR-652-3p | NFAT5 | 1.03 | 0.00034 | -0.04 | 0.82158 | miRNAWalker2 validate | -0.14 | 0.00292 | NA | |
| 66 | hsa-miR-421 | NFATC1 | 1.81 | 0 | -0.65 | 0.05297 | miRanda | -0.22 | 0.0008 | NA | |
| 67 | hsa-miR-127-3p | NFATC2 | 0.21 | 0.46101 | -1.6 | 0.00281 | miRanda | -0.44 | 0.00108 | NA | |
| 68 | hsa-miR-16-2-3p | NFATC2 | 1.8 | 0 | -1.6 | 0.00281 | mirMAP | -0.41 | 0.00072 | NA | |
| 69 | hsa-miR-182-5p | NFATC2 | 0.89 | 0.03106 | -1.6 | 0.00281 | mirMAP | -0.57 | 0 | NA | |
| 70 | hsa-miR-185-5p | NFATC2 | 1.14 | 0 | -1.6 | 0.00281 | MirTarget | -0.5 | 0.00829 | NA | |
| 71 | hsa-miR-193b-5p | NFATC2 | 0.61 | 0.13924 | -1.6 | 0.00281 | MirTarget | -0.68 | 0 | NA | |
| 72 | hsa-miR-214-3p | NFATC2 | 1.01 | 0.00625 | -1.6 | 0.00281 | MirTarget; miRNATAP | -0.35 | 0.00075 | NA | |
| 73 | hsa-miR-218-5p | NFATC2 | -0.99 | 0.01083 | -1.6 | 0.00281 | MirTarget | -0.28 | 0.00423 | NA | |
| 74 | hsa-miR-2355-3p | NFATC2 | 0.98 | 0.02293 | -1.6 | 0.00281 | mirMAP | -0.45 | 0 | NA | |
| 75 | hsa-miR-2355-5p | NFATC2 | 0.59 | 0.12059 | -1.6 | 0.00281 | MirTarget | -0.53 | 0 | NA | |
| 76 | hsa-miR-24-3p | NFATC2 | 1.09 | 0 | -1.6 | 0.00281 | MirTarget | -1.45 | 0 | NA | |
| 77 | hsa-miR-27b-5p | NFATC2 | 0.68 | 0.01497 | -1.6 | 0.00281 | mirMAP | -0.42 | 0.00212 | NA | |
| 78 | hsa-miR-31-3p | NFATC2 | 0.47 | 0.50687 | -1.6 | 0.00281 | MirTarget | -0.2 | 0.00027 | NA | |
| 79 | hsa-miR-33a-3p | NFATC2 | 0.35 | 0.32171 | -1.6 | 0.00281 | mirMAP | -0.34 | 0.00353 | NA | |
| 80 | hsa-miR-452-5p | NFATC2 | 2.09 | 0.00026 | -1.6 | 0.00281 | MirTarget; miRNATAP | -0.58 | 0 | NA | |
| 81 | hsa-miR-493-5p | NFATC2 | 0.96 | 0.00325 | -1.6 | 0.00281 | miRNATAP | -0.39 | 0.00071 | NA | |
| 82 | hsa-miR-9-5p | NFATC2 | -0.2 | 0.76147 | -1.6 | 0.00281 | MirTarget | -0.27 | 1.0E-5 | NA | |
| 83 | hsa-miR-30d-5p | NFATC3 | -0.55 | 0.01401 | 0.28 | 0.07069 | MirTarget; miRNATAP | -0.15 | 0.00355 | NA | |
| 84 | hsa-miR-362-3p | NFATC3 | -0.03 | 0.91378 | 0.28 | 0.07069 | miRanda | -0.13 | 0.00024 | NA | |
| 85 | hsa-miR-1254 | NFATC4 | 0.94 | 0.06055 | -0.98 | 0.00292 | MirTarget | -0.22 | 0.00012 | NA | |
| 86 | hsa-miR-130b-5p | NFATC4 | 0.7 | 0.05101 | -0.98 | 0.00292 | mirMAP | -0.2 | 0.00216 | NA | |
| 87 | hsa-miR-15b-3p | NFATC4 | 1.76 | 0 | -0.98 | 0.00292 | mirMAP | -0.4 | 0 | NA | |
| 88 | hsa-miR-185-3p | NFATC4 | 1.34 | 3.0E-5 | -0.98 | 0.00292 | MirTarget | -0.31 | 1.0E-5 | NA | |
| 89 | hsa-miR-29b-3p | NFATC4 | -0.23 | 0.36746 | -0.98 | 0.00292 | miRNATAP | -0.27 | 0.00392 | NA | |
| 90 | hsa-miR-320a | NFATC4 | 0.59 | 0.0119 | -0.98 | 0.00292 | miRanda; mirMAP | -0.35 | 0.00047 | NA | |
| 91 | hsa-miR-320b | NFATC4 | 1.11 | 0.0005 | -0.98 | 0.00292 | miRanda; mirMAP | -0.33 | 0 | NA | |
| 92 | hsa-miR-320c | NFATC4 | 0.46 | 0.24061 | -0.98 | 0.00292 | miRanda; mirMAP | -0.23 | 0.00089 | NA | |
| 93 | hsa-miR-361-3p | NFATC4 | -0.13 | 0.56605 | -0.98 | 0.00292 | mirMAP | -0.34 | 0.00205 | NA | |
| 94 | hsa-miR-429 | NFATC4 | 1.4 | 0.009 | -0.98 | 0.00292 | miRNATAP | -0.21 | 0 | NA | |
| 95 | hsa-miR-484 | NFATC4 | 0.71 | 0.00234 | -0.98 | 0.00292 | MirTarget; miRNATAP | -0.49 | 0 | NA | |
| 96 | hsa-miR-625-5p | NFATC4 | 0.54 | 0.2038 | -0.98 | 0.00292 | mirMAP | -0.16 | 0.00461 | NA | |
| 97 | hsa-miR-629-3p | NFATC4 | 1.4 | 0.0001 | -0.98 | 0.00292 | mirMAP | -0.28 | 1.0E-5 | NA | |
| 98 | hsa-miR-940 | NFATC4 | 1.64 | 0.00069 | -0.98 | 0.00292 | miRNATAP | -0.19 | 0.00086 | NA | |
| 99 | hsa-miR-542-3p | NOS3 | 0.76 | 0.00211 | -0.5 | 0.18108 | miRanda | -0.31 | 0.00446 | NA | |
| 100 | hsa-let-7c-5p | NRAS | -0.5 | 0.20685 | 0.61 | 0.01077 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.12 | 0.00352 | NA | |
| 101 | hsa-miR-145-5p | NRAS | -1.75 | 2.0E-5 | 0.61 | 0.01077 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.11 | 0.00563 | 26973415 | miR-145 expression was significantly downregulated in colon cancer tissues with its expression in normal colonic tissues being 4-5-fold higher two sample t test P < 0.05 whereas N-ras expression showed the opposite trend |
| 102 | hsa-miR-26a-5p | NRAS | -0.38 | 0.04425 | 0.61 | 0.01077 | mirMAP; miRNATAP | -0.25 | 0.00537 | 26458859 | MiR 26a enhances the sensitivity of gastric cancer cells to cisplatin by targeting NRAS and E2F2; Our results suggest that miR-26a can improve the sensitivity of GC cells to cisplatin-based chemotherapies through targeting NRAS and E2F2 and provide the first evidence of the potential utility of miR-26a as a sensitizer in chemotherapy for GC |
| 103 | hsa-miR-28-5p | NRAS | -0.24 | 0.16732 | 0.61 | 0.01077 | MirTarget; PITA; miRanda; miRNATAP | -0.31 | 0.00228 | NA | |
| 104 | hsa-miR-29c-3p | NRAS | -1.62 | 0 | 0.61 | 0.01077 | miRNATAP | -0.18 | 0.00053 | NA | |
| 105 | hsa-miR-126-5p | PIK3CA | 0.42 | 0.07532 | 0.18 | 0.37992 | mirMAP | -0.23 | 0.00023 | NA | |
| 106 | hsa-miR-146a-5p | PIK3CA | 1.82 | 2.0E-5 | 0.18 | 0.37992 | mirMAP | -0.13 | 8.0E-5 | NA | |
| 107 | hsa-miR-148a-5p | PIK3CA | -0.69 | 0.0793 | 0.18 | 0.37992 | mirMAP | -0.16 | 1.0E-5 | NA | |
| 108 | hsa-miR-148b-3p | PIK3CA | 0.3 | 0.17466 | 0.18 | 0.37992 | miRNAWalker2 validate | -0.27 | 6.0E-5 | NA | |
| 109 | hsa-miR-186-5p | PIK3CA | 0.15 | 0.43471 | 0.18 | 0.37992 | mirMAP | -0.28 | 0.00025 | NA | |
| 110 | hsa-miR-29b-1-5p | PIK3CA | 0.61 | 0.11636 | 0.18 | 0.37992 | mirMAP | -0.11 | 0.00515 | NA | |
| 111 | hsa-miR-320b | PIK3CA | 1.11 | 0.0005 | 0.18 | 0.37992 | miRanda | -0.13 | 0.00356 | NA | |
| 112 | hsa-miR-335-5p | PIK3CA | 1.6 | 6.0E-5 | 0.18 | 0.37992 | miRNAWalker2 validate | -0.14 | 7.0E-5 | NA | |
| 113 | hsa-miR-338-5p | PIK3CA | -1.2 | 0.01003 | 0.18 | 0.37992 | mirMAP | -0.13 | 3.0E-5 | NA | |
| 114 | hsa-miR-501-5p | PIK3CA | 0.27 | 0.45478 | 0.18 | 0.37992 | mirMAP | -0.18 | 0 | NA | |
| 115 | hsa-miR-7-1-3p | PIK3CA | 0.71 | 0.04123 | 0.18 | 0.37992 | mirMAP | -0.18 | 1.0E-5 | NA | |
| 116 | hsa-miR-130a-3p | PIK3CB | 0.15 | 0.63374 | 0.25 | 0.18038 | miRNATAP | -0.14 | 0.001 | NA | |
| 117 | hsa-miR-1468-5p | PIK3CD | -1.88 | 3.0E-5 | 0.39 | 0.25308 | MirTarget | -0.22 | 3.0E-5 | NA | |
| 118 | hsa-miR-30b-3p | PIK3CD | -0.27 | 0.40085 | 0.39 | 0.25308 | MirTarget | -0.43 | 0 | NA | |
| 119 | hsa-miR-30b-5p | PIK3CD | -0.43 | 0.05936 | 0.39 | 0.25308 | MirTarget | -0.48 | 1.0E-5 | NA | |
| 120 | hsa-miR-30d-5p | PIK3CD | -0.55 | 0.01401 | 0.39 | 0.25308 | MirTarget; miRNATAP | -0.67 | 0 | NA | |
| 121 | hsa-miR-616-5p | PIK3CD | 0.83 | 0.03478 | 0.39 | 0.25308 | MirTarget | -0.17 | 0.00628 | NA | |
| 122 | hsa-miR-7-5p | PIK3CD | 1.64 | 0.01244 | 0.39 | 0.25308 | miRTarBase; MirTarget; miRNATAP | -0.21 | 0 | 22234835 | We first screened and identified a novel miR-7 target phosphoinositide 3-kinase catalytic subunit delta PIK3CD; A correlation between miR-7 and PIK3CD expression was also confirmed in clinical samples of HCC; By targeting PIK3CD mTOR and p70S6K miR-7 efficiently regulates the PI3K/Akt pathway |
| 123 | hsa-miR-103a-3p | PIK3R1 | 0.84 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.32 | 0.00394 | NA | |
| 124 | hsa-miR-106b-5p | PIK3R1 | 1.71 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.25 | 2.0E-5 | NA | |
| 125 | hsa-miR-107 | PIK3R1 | 0.9 | 5.0E-5 | -1.09 | 2.0E-5 | MirTarget; PITA; miRanda; miRNATAP | -0.24 | 0.00469 | NA | |
| 126 | hsa-miR-128-3p | PIK3R1 | 0.98 | 6.0E-5 | -1.09 | 2.0E-5 | MirTarget | -0.25 | 0.00086 | 25962360 | miR 128 3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients; Mechanistically miR-128-3p was confirmed to regulate PIK3R1 p85α expression thereby suppressing phosphatidylinositol 3-kinase PI3K/AKT pathway activation using qRT-PCR and western blot analysis; Hence we conclude that miR-128-3p which is frequently downregulated in HCC inhibits HCC progression by regulating PIK3R1 and PI3K/AKT activation and is a prognostic marker for HCC patients |
| 127 | hsa-miR-144-3p | PIK3R1 | -0.13 | 0.79825 | -1.09 | 2.0E-5 | mirMAP | -0.14 | 0.00013 | NA | |
| 128 | hsa-miR-15b-5p | PIK3R1 | 1.62 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.32 | 0 | NA | |
| 129 | hsa-miR-16-2-3p | PIK3R1 | 1.8 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.27 | 0 | NA | |
| 130 | hsa-miR-16-5p | PIK3R1 | 1.01 | 1.0E-5 | -1.09 | 2.0E-5 | MirTarget | -0.41 | 0 | NA | |
| 131 | hsa-miR-17-5p | PIK3R1 | 1.66 | 0 | -1.09 | 2.0E-5 | MirTarget; TargetScan; miRNATAP | -0.19 | 0.00107 | NA | |
| 132 | hsa-miR-182-5p | PIK3R1 | 0.89 | 0.03106 | -1.09 | 2.0E-5 | miRNATAP | -0.16 | 0.0004 | NA | |
| 133 | hsa-miR-185-5p | PIK3R1 | 1.14 | 0 | -1.09 | 2.0E-5 | miRNATAP | -0.42 | 0 | NA | |
| 134 | hsa-miR-188-5p | PIK3R1 | 1.35 | 0.00038 | -1.09 | 2.0E-5 | MirTarget | -0.16 | 0.00078 | NA | |
| 135 | hsa-miR-200b-3p | PIK3R1 | 0.97 | 0.0595 | -1.09 | 2.0E-5 | mirMAP | -0.1 | 0.0041 | NA | |
| 136 | hsa-miR-200c-3p | PIK3R1 | 1.28 | 0.0037 | -1.09 | 2.0E-5 | mirMAP | -0.14 | 0.00064 | NA | |
| 137 | hsa-miR-20a-5p | PIK3R1 | 1.45 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.18 | 0.00561 | NA | |
| 138 | hsa-miR-21-5p | PIK3R1 | 1.75 | 0 | -1.09 | 2.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.31 | 0.00155 | 26676464 | PIK3R1 targeting by miR 21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT and predicts clinical outcome of breast cancer; Next we identified the PIK3R1 as a direct target of miR-21 and showed that it was negatively regulated by miR-21; Taken together we provide novel evidence that miR-21 knockdown suppresses cell growth migration and invasion partly by inhibiting PI3K/AKT activation via direct targeting PIK3R1 and reversing EMT in breast cancer |
| 139 | hsa-miR-212-3p | PIK3R1 | 0.96 | 0.00133 | -1.09 | 2.0E-5 | MirTarget | -0.23 | 0.00022 | NA | |
| 140 | hsa-miR-22-5p | PIK3R1 | 0.8 | 0.0008 | -1.09 | 2.0E-5 | mirMAP | -0.27 | 0.0005 | NA | |
| 141 | hsa-miR-221-3p | PIK3R1 | 1.33 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.31 | 1.0E-5 | NA | |
| 142 | hsa-miR-222-3p | PIK3R1 | 1.39 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.28 | 1.0E-5 | NA | |
| 143 | hsa-miR-3065-5p | PIK3R1 | -0.24 | 0.63312 | -1.09 | 2.0E-5 | MirTarget; mirMAP; miRNATAP | -0.12 | 0.00106 | NA | |
| 144 | hsa-miR-32-3p | PIK3R1 | 0.58 | 0.11837 | -1.09 | 2.0E-5 | mirMAP | -0.17 | 0.00228 | NA | |
| 145 | hsa-miR-320b | PIK3R1 | 1.11 | 0.0005 | -1.09 | 2.0E-5 | miRNATAP | -0.16 | 0.00545 | NA | |
| 146 | hsa-miR-324-3p | PIK3R1 | 1.05 | 0.00039 | -1.09 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.18 | 0.00348 | NA | |
| 147 | hsa-miR-330-3p | PIK3R1 | 0.8 | 0.00747 | -1.09 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.26 | 2.0E-5 | NA | |
| 148 | hsa-miR-335-3p | PIK3R1 | 2.52 | 0 | -1.09 | 2.0E-5 | mirMAP | -0.15 | 0.0005 | NA | |
| 149 | hsa-miR-424-5p | PIK3R1 | 1.09 | 0.00042 | -1.09 | 2.0E-5 | MirTarget | -0.29 | 0 | NA | |
| 150 | hsa-miR-429 | PIK3R1 | 1.4 | 0.009 | -1.09 | 2.0E-5 | mirMAP; miRNATAP | -0.13 | 0.00023 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | INTRACELLULAR SIGNAL TRANSDUCTION | 34 | 1572 | 8.618e-29 | 4.01e-25 |
| 2 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 21 | 323 | 3.258e-26 | 7.579e-23 |
| 3 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 34 | 1929 | 7.869e-26 | 1.22e-22 |
| 4 | FC RECEPTOR SIGNALING PATHWAY | 18 | 206 | 1.076e-24 | 1.252e-21 |
| 5 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 16 | 142 | 9.49e-24 | 8.831e-21 |
| 6 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 13 | 74 | 5.324e-22 | 4.129e-19 |
| 7 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 20 | 498 | 1.026e-20 | 6.818e-18 |
| 8 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 30 | 1977 | 4.519e-20 | 2.628e-17 |
| 9 | PLATELET ACTIVATION | 14 | 142 | 5.938e-20 | 3.07e-17 |
| 10 | REGULATION OF IMMUNE RESPONSE | 22 | 858 | 1.018e-18 | 4.738e-16 |
| 11 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 20 | 689 | 5.974e-18 | 2.527e-15 |
| 12 | REGULATION OF IMMUNE SYSTEM PROCESS | 25 | 1403 | 9.312e-18 | 3.611e-15 |
| 13 | CELL ACTIVATION | 18 | 568 | 9.325e-17 | 3.337e-14 |
| 14 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 25 | 1656 | 4.673e-16 | 1.505e-13 |
| 15 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 20 | 867 | 5.034e-16 | 1.505e-13 |
| 16 | ACTIVATION OF IMMUNE RESPONSE | 16 | 427 | 5.176e-16 | 1.505e-13 |
| 17 | PHOSPHORYLATION | 22 | 1228 | 1.921e-15 | 5.257e-13 |
| 18 | HEMOSTASIS | 14 | 311 | 3.905e-15 | 1.009e-12 |
| 19 | REGULATION OF BODY FLUID LEVELS | 16 | 506 | 7.332e-15 | 1.796e-12 |
| 20 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 10 | 95 | 9.998e-15 | 2.326e-12 |
| 21 | POSITIVE REGULATION OF CELL COMMUNICATION | 23 | 1532 | 1.472e-14 | 3.262e-12 |
| 22 | REGULATION OF KINASE ACTIVITY | 18 | 776 | 2.097e-14 | 4.436e-12 |
| 23 | POSITIVE REGULATION OF IMMUNE RESPONSE | 16 | 563 | 3.828e-14 | 7.745e-12 |
| 24 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 23 | 1618 | 4.731e-14 | 9.172e-12 |
| 25 | ERBB SIGNALING PATHWAY | 9 | 79 | 1.156e-13 | 2.151e-11 |
| 26 | POSITIVE REGULATION OF LOCOMOTION | 14 | 420 | 2.414e-13 | 4.32e-11 |
| 27 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 8 | 55 | 3.831e-13 | 6.474e-11 |
| 28 | CALCIUM MEDIATED SIGNALING | 9 | 90 | 3.896e-13 | 6.474e-11 |
| 29 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 23 | 1805 | 4.803e-13 | 7.707e-11 |
| 30 | PROTEIN PHOSPHORYLATION | 18 | 944 | 5.917e-13 | 9.177e-11 |
| 31 | REGULATION OF TRANSFERASE ACTIVITY | 18 | 946 | 6.132e-13 | 9.204e-11 |
| 32 | LOCOMOTION | 19 | 1114 | 7.799e-13 | 1.134e-10 |
| 33 | PHOSPHOLIPID METABOLIC PROCESS | 13 | 364 | 8.385e-13 | 1.182e-10 |
| 34 | WOUND HEALING | 14 | 470 | 1.109e-12 | 1.474e-10 |
| 35 | VASCULATURE DEVELOPMENT | 14 | 469 | 1.077e-12 | 1.474e-10 |
| 36 | INOSITOL PHOSPHATE MEDIATED SIGNALING | 6 | 18 | 1.668e-12 | 2.157e-10 |
| 37 | SECOND MESSENGER MEDIATED SIGNALING | 10 | 160 | 2.028e-12 | 2.55e-10 |
| 38 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 17 | 876 | 2.395e-12 | 2.933e-10 |
| 39 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 18 | 1036 | 2.831e-12 | 3.293e-10 |
| 40 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 18 | 1036 | 2.831e-12 | 3.293e-10 |
| 41 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 40 | 3.156e-12 | 3.582e-10 |
| 42 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 11 | 235 | 3.331e-12 | 3.69e-10 |
| 43 | IMMUNE SYSTEM PROCESS | 23 | 1984 | 3.481e-12 | 3.767e-10 |
| 44 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 22 | 1791 | 4.228e-12 | 4.471e-10 |
| 45 | REGULATION OF TRANSPORT | 22 | 1804 | 4.885e-12 | 5.051e-10 |
| 46 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 19 | 1275 | 8.379e-12 | 8.476e-10 |
| 47 | LEUKOCYTE MIGRATION | 11 | 259 | 9.558e-12 | 9.462e-10 |
| 48 | REGULATION OF CELL DEATH | 20 | 1472 | 1.021e-11 | 9.896e-10 |
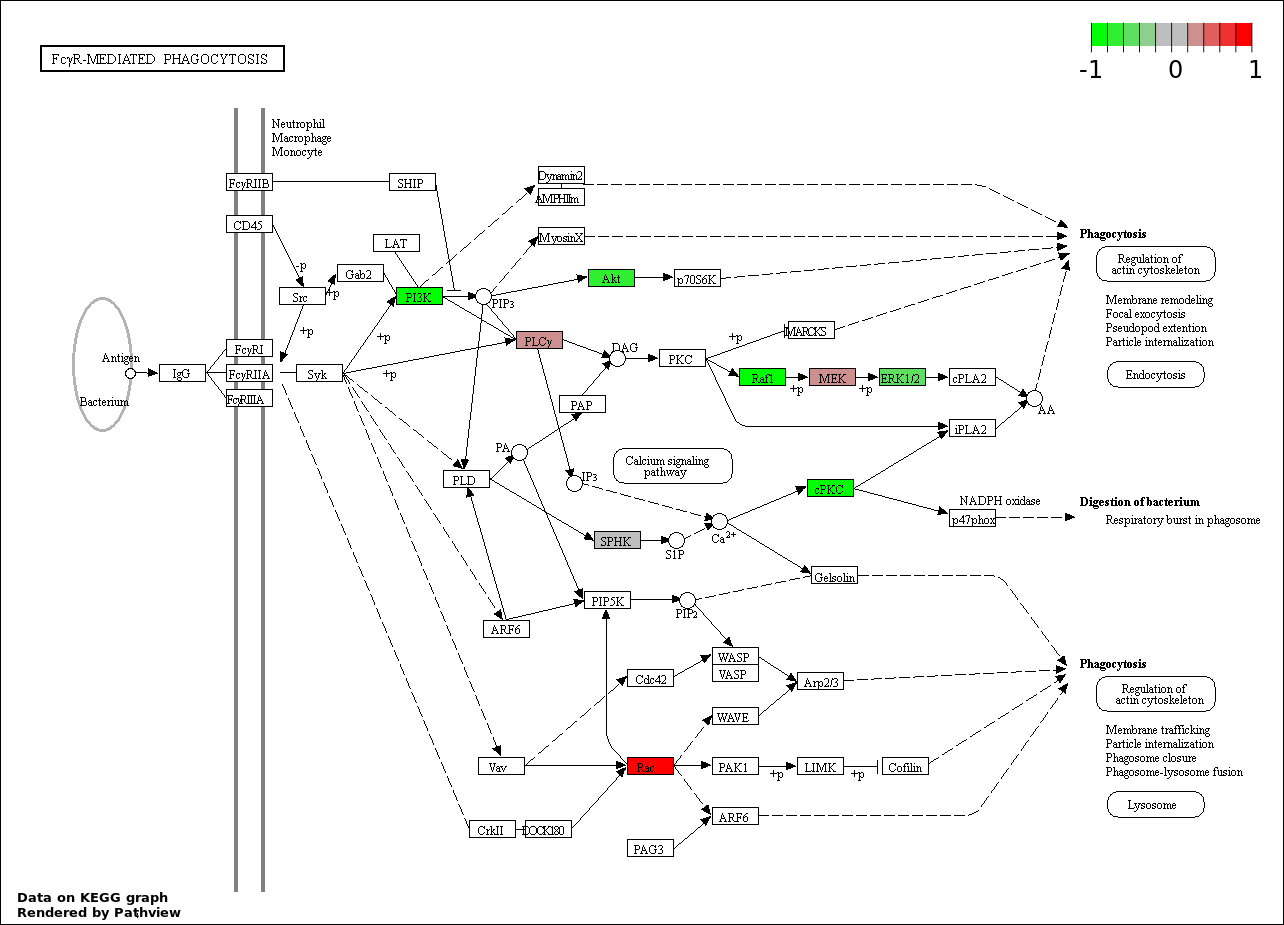
| 49 | PHAGOCYTOSIS | 10 | 190 | 1.129e-11 | 1.072e-09 |
| 50 | RESPONSE TO WOUNDING | 14 | 563 | 1.25e-11 | 1.141e-09 |
| 51 | REGULATION OF LIPID KINASE ACTIVITY | 7 | 48 | 1.23e-11 | 1.141e-09 |
| 52 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 10 | 193 | 1.319e-11 | 1.18e-09 |
| 53 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 10 | 195 | 1.461e-11 | 1.282e-09 |
| 54 | CELL MOTILITY | 16 | 835 | 1.535e-11 | 1.299e-09 |
| 55 | LOCALIZATION OF CELL | 16 | 835 | 1.535e-11 | 1.299e-09 |
| 56 | POSITIVE REGULATION OF MAPK CASCADE | 13 | 470 | 2.081e-11 | 1.729e-09 |
| 57 | POSITIVE REGULATION OF GENE EXPRESSION | 21 | 1733 | 2.157e-11 | 1.76e-09 |
| 58 | POSITIVE REGULATION OF KINASE ACTIVITY | 13 | 482 | 2.848e-11 | 2.285e-09 |
| 59 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 10 | 211 | 3.19e-11 | 2.516e-09 |
| 60 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 11 | 297 | 4.173e-11 | 3.237e-09 |
| 61 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 15 | 771 | 6.325e-11 | 4.824e-09 |
| 62 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 7 | 61 | 7.132e-11 | 5.353e-09 |
| 63 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 15 | 788 | 8.584e-11 | 6.241e-09 |
| 64 | CIRCULATORY SYSTEM DEVELOPMENT | 15 | 788 | 8.584e-11 | 6.241e-09 |
| 65 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 15 | 799 | 1.042e-10 | 7.459e-09 |
| 66 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 19 | 1492 | 1.272e-10 | 8.97e-09 |
| 67 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 17 | 1135 | 1.418e-10 | 9.85e-09 |
| 68 | REGULATION OF CELL SUBSTRATE ADHESION | 9 | 173 | 1.492e-10 | 1.021e-08 |
| 69 | REGULATION OF PROTEIN MODIFICATION PROCESS | 20 | 1710 | 1.535e-10 | 1.035e-08 |
| 70 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 19 | 1518 | 1.71e-10 | 1.137e-08 |
| 71 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 8 | 120 | 2.375e-10 | 1.556e-08 |
| 72 | GLYCEROLIPID METABOLIC PROCESS | 11 | 356 | 2.88e-10 | 1.861e-08 |
| 73 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 16 | 1021 | 3.022e-10 | 1.927e-08 |
| 74 | NEGATIVE REGULATION OF CELL DEATH | 15 | 872 | 3.516e-10 | 2.211e-08 |
| 75 | IMMUNE EFFECTOR PROCESS | 12 | 486 | 5.144e-10 | 3.191e-08 |
| 76 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 8 | 133 | 5.42e-10 | 3.318e-08 |
| 77 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 13 | 616 | 5.843e-10 | 3.531e-08 |
| 78 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 20 | 1848 | 6.113e-10 | 3.647e-08 |
| 79 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 8 | 138 | 7.279e-10 | 4.287e-08 |
| 80 | ANGIOGENESIS | 10 | 293 | 7.926e-10 | 4.61e-08 |
| 81 | REGULATION OF RESPONSE TO STRESS | 18 | 1468 | 8.743e-10 | 5.022e-08 |
| 82 | T CELL RECEPTOR SIGNALING PATHWAY | 8 | 146 | 1.141e-09 | 6.472e-08 |
| 83 | REGULATION OF CELL MATRIX ADHESION | 7 | 90 | 1.164e-09 | 6.525e-08 |
| 84 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 6 | 49 | 1.193e-09 | 6.61e-08 |
| 85 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 15 | 957 | 1.267e-09 | 6.934e-08 |
| 86 | REGULATION OF MAPK CASCADE | 13 | 660 | 1.351e-09 | 7.311e-08 |
| 87 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 23 | 1.497e-09 | 8.005e-08 |
| 88 | POSITIVE REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 6 | 51 | 1.532e-09 | 8.099e-08 |
| 89 | LIPID BIOSYNTHETIC PROCESS | 12 | 539 | 1.663e-09 | 8.694e-08 |
| 90 | HOMEOSTATIC PROCESS | 17 | 1337 | 1.764e-09 | 9.119e-08 |
| 91 | REGULATION OF MAP KINASE ACTIVITY | 10 | 319 | 1.802e-09 | 9.212e-08 |
| 92 | LIPID PHOSPHORYLATION | 7 | 99 | 2.284e-09 | 1.155e-07 |
| 93 | POSITIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 5 | 25 | 2.355e-09 | 1.166e-07 |
| 94 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 5 | 25 | 2.355e-09 | 1.166e-07 |
| 95 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 17 | 1381 | 2.885e-09 | 1.413e-07 |
| 96 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 7 | 103 | 3.02e-09 | 1.464e-07 |
| 97 | REGULATION OF EPITHELIAL CELL MIGRATION | 8 | 166 | 3.157e-09 | 1.514e-07 |
| 98 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 11 | 450 | 3.376e-09 | 1.603e-07 |
| 99 | RESPONSE TO EXTERNAL STIMULUS | 19 | 1821 | 3.694e-09 | 1.736e-07 |
| 100 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 59 | 3.781e-09 | 1.759e-07 |
| 101 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 11 | 470 | 5.305e-09 | 2.444e-07 |
| 102 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 7 | 114 | 6.158e-09 | 2.809e-07 |
| 103 | BLOOD VESSEL MORPHOGENESIS | 10 | 364 | 6.383e-09 | 2.884e-07 |
| 104 | REGULATION OF CYTOPLASMIC TRANSPORT | 11 | 481 | 6.742e-09 | 3.016e-07 |
| 105 | CELLULAR LIPID METABOLIC PROCESS | 14 | 913 | 6.871e-09 | 3.045e-07 |
| 106 | REGULATION OF INTRACELLULAR TRANSPORT | 12 | 621 | 8.139e-09 | 3.573e-07 |
| 107 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 6 | 67 | 8.263e-09 | 3.593e-07 |
| 108 | CELLULAR RESPONSE TO PEPTIDE | 9 | 274 | 8.585e-09 | 3.699e-07 |
| 109 | POSITIVE REGULATION OF TRANSPORT | 14 | 936 | 9.426e-09 | 4.024e-07 |
| 110 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 9 | 279 | 1.004e-08 | 4.249e-07 |
| 111 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 12 | 1.092e-08 | 4.578e-07 |
| 112 | REGULATION OF LIPID METABOLIC PROCESS | 9 | 282 | 1.102e-08 | 4.578e-07 |
| 113 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 8 | 207 | 1.785e-08 | 7.349e-07 |
| 114 | ERBB2 SIGNALING PATHWAY | 5 | 39 | 2.494e-08 | 1.018e-06 |
| 115 | RESPONSE TO NITROGEN COMPOUND | 13 | 859 | 3.149e-08 | 1.274e-06 |
| 116 | PEPTIDYL SERINE MODIFICATION | 7 | 148 | 3.779e-08 | 1.516e-06 |
| 117 | ORGANOPHOSPHATE METABOLIC PROCESS | 13 | 885 | 4.471e-08 | 1.764e-06 |
| 118 | REGULATION OF VASCULATURE DEVELOPMENT | 8 | 233 | 4.474e-08 | 1.764e-06 |
| 119 | RESPONSE TO ENDOGENOUS STIMULUS | 16 | 1450 | 4.621e-08 | 1.807e-06 |
| 120 | IMMUNE SYSTEM DEVELOPMENT | 11 | 582 | 4.769e-08 | 1.849e-06 |
| 121 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 17 | 5.208e-08 | 2.003e-06 |
| 122 | REGULATION OF VESICLE MEDIATED TRANSPORT | 10 | 462 | 6.072e-08 | 2.316e-06 |
| 123 | REGULATION OF CELLULAR LOCALIZATION | 15 | 1277 | 6.152e-08 | 2.327e-06 |
| 124 | TAXIS | 10 | 464 | 6.322e-08 | 2.372e-06 |
| 125 | REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 6 | 95 | 6.867e-08 | 2.541e-06 |
| 126 | REGULATION OF CELL DIFFERENTIATION | 16 | 1492 | 6.881e-08 | 2.541e-06 |
| 127 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 9 | 360 | 8.984e-08 | 3.275e-06 |
| 128 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 5 | 50 | 9.01e-08 | 3.275e-06 |
| 129 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 8 | 258 | 9.817e-08 | 3.541e-06 |
| 130 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 5 | 51 | 9.972e-08 | 3.569e-06 |
| 131 | TISSUE HOMEOSTASIS | 7 | 171 | 1.019e-07 | 3.621e-06 |
| 132 | REGULATION OF CELL ADHESION | 11 | 629 | 1.047e-07 | 3.691e-06 |
| 133 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 4 | 20 | 1.055e-07 | 3.691e-06 |
| 134 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 9 | 370 | 1.134e-07 | 3.925e-06 |
| 135 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 14 | 1142 | 1.139e-07 | 3.925e-06 |
| 136 | POSITIVE REGULATION OF PROTEIN IMPORT | 6 | 104 | 1.181e-07 | 4.04e-06 |
| 137 | B CELL RECEPTOR SIGNALING PATHWAY | 5 | 54 | 1.336e-07 | 4.537e-06 |
| 138 | LIPID METABOLIC PROCESS | 14 | 1158 | 1.352e-07 | 4.558e-06 |
| 139 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 10 | 505 | 1.391e-07 | 4.656e-06 |
| 140 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 8 | 272 | 1.472e-07 | 4.893e-06 |
| 141 | ENDOCYTOSIS | 10 | 509 | 1.496e-07 | 4.938e-06 |
| 142 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 10 | 514 | 1.638e-07 | 5.368e-06 |
| 143 | ENDOTHELIAL CELL MIGRATION | 5 | 57 | 1.76e-07 | 5.726e-06 |
| 144 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 8 | 282 | 1.94e-07 | 6.226e-06 |
| 145 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 13 | 1004 | 1.939e-07 | 6.226e-06 |
| 146 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 15 | 1395 | 1.957e-07 | 6.236e-06 |
| 147 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 8 | 289 | 2.339e-07 | 7.404e-06 |
| 148 | RESPONSE TO PEPTIDE | 9 | 404 | 2.386e-07 | 7.501e-06 |
| 149 | RESPONSE TO ABIOTIC STIMULUS | 13 | 1024 | 2.433e-07 | 7.597e-06 |
| 150 | CELL DEVELOPMENT | 15 | 1426 | 2.602e-07 | 8.072e-06 |
| 151 | POSITIVE REGULATION OF CHEMOTAXIS | 6 | 120 | 2.768e-07 | 8.528e-06 |
| 152 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 8 | 296 | 2.807e-07 | 8.592e-06 |
| 153 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 6 | 121 | 2.907e-07 | 8.841e-06 |
| 154 | VESICLE MEDIATED TRANSPORT | 14 | 1239 | 3.096e-07 | 9.355e-06 |
| 155 | CELLULAR RESPONSE TO HORMONE STIMULUS | 10 | 552 | 3.163e-07 | 9.494e-06 |
| 156 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 16 | 1672 | 3.301e-07 | 9.846e-06 |
| 157 | INOSITOL LIPID MEDIATED SIGNALING | 6 | 124 | 3.361e-07 | 9.96e-06 |
| 158 | REGULATION OF PROTEIN TARGETING | 8 | 307 | 3.703e-07 | 1.09e-05 |
| 159 | LIPID MODIFICATION | 7 | 210 | 4.119e-07 | 1.205e-05 |
| 160 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 6 | 129 | 4.245e-07 | 1.235e-05 |
| 161 | REGULATION OF CELL JUNCTION ASSEMBLY | 5 | 68 | 4.302e-07 | 1.236e-05 |
| 162 | RESPONSE TO HORMONE | 12 | 893 | 4.292e-07 | 1.236e-05 |
| 163 | REGULATION OF CELL PROLIFERATION | 15 | 1496 | 4.825e-07 | 1.377e-05 |
| 164 | RESPONSE TO GROWTH HORMONE | 4 | 30 | 5.871e-07 | 1.666e-05 |
| 165 | CELLULAR GLUCOSE HOMEOSTASIS | 5 | 75 | 7.04e-07 | 1.985e-05 |
| 166 | REGULATION OF PROTEIN LOCALIZATION | 12 | 950 | 8.281e-07 | 2.321e-05 |
| 167 | REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 5 | 78 | 8.568e-07 | 2.387e-05 |
| 168 | CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 146 | 8.794e-07 | 2.436e-05 |
| 169 | RESPONSE TO GROWTH FACTOR | 9 | 475 | 9.241e-07 | 2.544e-05 |
| 170 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 13 | 1152 | 9.302e-07 | 2.546e-05 |
| 171 | INSULIN RECEPTOR SIGNALING PATHWAY | 5 | 80 | 9.724e-07 | 2.646e-05 |
| 172 | TISSUE MIGRATION | 5 | 84 | 1.24e-06 | 3.356e-05 |
| 173 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 12 | 1008 | 1.545e-06 | 4.155e-05 |
| 174 | POSITIVE REGULATION OF CELL ADHESION | 8 | 376 | 1.701e-06 | 4.548e-05 |
| 175 | POSITIVE REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 3 | 11 | 1.734e-06 | 4.562e-05 |
| 176 | WNT SIGNALING PATHWAY CALCIUM MODULATING PATHWAY | 4 | 39 | 1.736e-06 | 4.562e-05 |
| 177 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 9 | 513 | 1.745e-06 | 4.562e-05 |
| 178 | POSITIVE REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 3 | 11 | 1.734e-06 | 4.562e-05 |
| 179 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 8 | 381 | 1.876e-06 | 4.877e-05 |
| 180 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 4 | 42 | 2.351e-06 | 6.077e-05 |
| 181 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 8 | 404 | 2.899e-06 | 7.452e-05 |
| 182 | REGULATION OF GLUCOSE TRANSPORT | 5 | 100 | 2.945e-06 | 7.53e-05 |
| 183 | REGULATION OF CHEMOTAXIS | 6 | 180 | 2.976e-06 | 7.566e-05 |
| 184 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 9 | 552 | 3.181e-06 | 7.958e-05 |
| 185 | ANATOMICAL STRUCTURE HOMEOSTASIS | 7 | 285 | 3.165e-06 | 7.958e-05 |
| 186 | REGULATION OF CELL MORPHOGENESIS | 9 | 552 | 3.181e-06 | 7.958e-05 |
| 187 | REGULATION OF PROTEIN IMPORT | 6 | 183 | 3.274e-06 | 8.146e-05 |
| 188 | VASCULAR ENDOTHELIAL GROWTH FACTOR SIGNALING PATHWAY | 3 | 14 | 3.808e-06 | 9.424e-05 |
| 189 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 5 | 106 | 3.925e-06 | 9.663e-05 |
| 190 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 11 | 926 | 4.712e-06 | 0.0001154 |
| 191 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 13 | 1340 | 4.999e-06 | 0.0001218 |
| 192 | REGULATION OF ENDOCYTOSIS | 6 | 199 | 5.306e-06 | 0.0001286 |
| 193 | REGULATION OF EARLY ENDOSOME TO LATE ENDOSOME TRANSPORT | 3 | 16 | 5.84e-06 | 0.0001408 |
| 194 | REGULATION OF HOMEOSTATIC PROCESS | 8 | 447 | 6.103e-06 | 0.0001464 |
| 195 | RESPONSE TO INSULIN | 6 | 205 | 6.292e-06 | 0.0001494 |
| 196 | NEURON PROJECTION GUIDANCE | 6 | 205 | 6.292e-06 | 0.0001494 |
| 197 | REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 3 | 17 | 7.08e-06 | 0.0001647 |
| 198 | NEGATIVE REGULATION OF ANOIKIS | 3 | 17 | 7.08e-06 | 0.0001647 |
| 199 | PHOSPHATIDYLGLYCEROL ACYL CHAIN REMODELING | 3 | 17 | 7.08e-06 | 0.0001647 |
| 200 | REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 3 | 17 | 7.08e-06 | 0.0001647 |
| 201 | SINGLE ORGANISM CELL ADHESION | 8 | 459 | 7.406e-06 | 0.0001715 |
| 202 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 6 | 218 | 8.942e-06 | 0.000206 |
| 203 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 6 | 220 | 9.42e-06 | 0.0002159 |
| 204 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 7 | 337 | 9.486e-06 | 0.0002164 |
| 205 | CELL DEATH | 11 | 1001 | 9.879e-06 | 0.0002231 |
| 206 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 5 | 128 | 9.878e-06 | 0.0002231 |
| 207 | REGULATION OF GLUCOSE IMPORT | 4 | 60 | 9.945e-06 | 0.0002235 |
| 208 | POSITIVE REGULATION OF CELL PROLIFERATION | 10 | 814 | 1.031e-05 | 0.0002307 |
| 209 | REGULATION OF CELL ACTIVATION | 8 | 484 | 1.089e-05 | 0.0002425 |
| 210 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 6 | 232 | 1.274e-05 | 0.0002824 |
| 211 | PEPTIDYL AMINO ACID MODIFICATION | 10 | 841 | 1.37e-05 | 0.0003006 |
| 212 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 3 | 21 | 1.376e-05 | 0.0003006 |
| 213 | BONE RESORPTION | 3 | 21 | 1.376e-05 | 0.0003006 |
| 214 | CELLULAR RESPONSE TO UV | 4 | 66 | 1.455e-05 | 0.0003165 |
| 215 | NON CANONICAL WNT SIGNALING PATHWAY | 5 | 140 | 1.526e-05 | 0.0003287 |
| 216 | REGULATION OF SYNAPTIC PLASTICITY | 5 | 140 | 1.526e-05 | 0.0003287 |
| 217 | CELL PROLIFERATION | 9 | 672 | 1.551e-05 | 0.0003326 |
| 218 | CELLULAR HOMEOSTASIS | 9 | 676 | 1.626e-05 | 0.000347 |
| 219 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 7 | 368 | 1.676e-05 | 0.0003562 |
| 220 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 7 | 370 | 1.736e-05 | 0.0003671 |
| 221 | PHOSPHATIDYLETHANOLAMINE ACYL CHAIN REMODELING | 3 | 23 | 1.827e-05 | 0.0003846 |
| 222 | REGULATION OF POSITIVE CHEMOTAXIS | 3 | 24 | 2.084e-05 | 0.0004291 |
| 223 | POSITIVE REGULATION OF CELL JUNCTION ASSEMBLY | 3 | 24 | 2.084e-05 | 0.0004291 |
| 224 | REGULATION OF CELL CELL ADHESION | 7 | 380 | 2.061e-05 | 0.0004291 |
| 225 | REGULATION OF ANOIKIS | 3 | 24 | 2.084e-05 | 0.0004291 |
| 226 | POSITIVE REGULATION OF LAMELLIPODIUM ORGANIZATION | 3 | 24 | 2.084e-05 | 0.0004291 |
| 227 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 14 | 1784 | 2.254e-05 | 0.000462 |
| 228 | CELLULAR RESPONSE TO CARBOHYDRATE STIMULUS | 4 | 74 | 2.293e-05 | 0.000468 |
| 229 | THYROID GLAND DEVELOPMENT | 3 | 25 | 2.365e-05 | 0.0004805 |
| 230 | AMEBOIDAL TYPE CELL MIGRATION | 5 | 154 | 2.417e-05 | 0.0004889 |
| 231 | CELLULAR COMPONENT MORPHOGENESIS | 10 | 900 | 2.459e-05 | 0.0004953 |
| 232 | CELL PROJECTION ORGANIZATION | 10 | 902 | 2.506e-05 | 0.0005026 |
| 233 | NEURON PROJECTION DEVELOPMENT | 8 | 545 | 2.56e-05 | 0.0005113 |
| 234 | GLAND DEVELOPMENT | 7 | 395 | 2.641e-05 | 0.0005251 |
| 235 | CELLULAR RESPONSE TO STRESS | 13 | 1565 | 2.662e-05 | 0.0005262 |
| 236 | PHOSPHATIDYLCHOLINE ACYL CHAIN REMODELING | 3 | 26 | 2.669e-05 | 0.0005262 |
| 237 | LYMPHOCYTE COSTIMULATION | 4 | 78 | 2.824e-05 | 0.0005545 |
| 238 | NEURON PROJECTION MORPHOGENESIS | 7 | 402 | 2.954e-05 | 0.0005775 |
| 239 | REGULATION OF CELL PROJECTION ORGANIZATION | 8 | 558 | 3.029e-05 | 0.0005897 |
| 240 | REGULATION OF CYTOKINE PRODUCTION | 8 | 563 | 3.228e-05 | 0.0006258 |
| 241 | REPRODUCTIVE SYSTEM DEVELOPMENT | 7 | 408 | 3.247e-05 | 0.0006268 |
| 242 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 6 | 278 | 3.531e-05 | 0.0006761 |
| 243 | CELLULAR CHEMICAL HOMEOSTASIS | 8 | 570 | 3.524e-05 | 0.0006761 |
| 244 | LEUKOCYTE ACTIVATION | 7 | 414 | 3.563e-05 | 0.0006794 |
| 245 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 8 | 573 | 3.658e-05 | 0.0006938 |
| 246 | RESPONSE TO CARBOHYDRATE | 5 | 168 | 3.668e-05 | 0.0006938 |
| 247 | REGULATION OF VACUOLAR TRANSPORT | 3 | 29 | 3.733e-05 | 0.0007033 |
| 248 | GLUCOSE HOMEOSTASIS | 5 | 170 | 3.882e-05 | 0.0007254 |
| 249 | CARBOHYDRATE HOMEOSTASIS | 5 | 170 | 3.882e-05 | 0.0007254 |
| 250 | POSITIVE REGULATION OF ERK1 AND ERK2 CASCADE | 5 | 172 | 4.105e-05 | 0.000764 |
| 251 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 3 | 30 | 4.142e-05 | 0.0007678 |
| 252 | TISSUE REMODELING | 4 | 87 | 4.343e-05 | 0.0008019 |
| 253 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 9 | 767 | 4.386e-05 | 0.0008067 |
| 254 | NEUROGENESIS | 12 | 1402 | 4.404e-05 | 0.0008067 |
| 255 | HOMEOSTASIS OF NUMBER OF CELLS | 5 | 175 | 4.458e-05 | 0.0008134 |
| 256 | REGULATION OF ORGANELLE ORGANIZATION | 11 | 1178 | 4.485e-05 | 0.0008151 |
| 257 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 4 | 88 | 4.542e-05 | 0.0008192 |
| 258 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 4 | 88 | 4.542e-05 | 0.0008192 |
| 259 | PHOSPHATIDYLGLYCEROL METABOLIC PROCESS | 3 | 31 | 4.578e-05 | 0.0008193 |
| 260 | HOMEOSTASIS OF NUMBER OF CELLS WITHIN A TISSUE | 3 | 31 | 4.578e-05 | 0.0008193 |
| 261 | LEUKOCYTE DIFFERENTIATION | 6 | 292 | 4.643e-05 | 0.0008278 |
| 262 | POSITIVE REGULATION OF LIPID KINASE ACTIVITY | 3 | 32 | 5.044e-05 | 0.0008957 |
| 263 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 12 | 1423 | 5.093e-05 | 0.000901 |
| 264 | CELLULAR RESPONSE TO LIGHT STIMULUS | 4 | 91 | 5.181e-05 | 0.0009105 |
| 265 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 8 | 602 | 5.185e-05 | 0.0009105 |
| 266 | MODULATION OF SYNAPTIC TRANSMISSION | 6 | 301 | 5.496e-05 | 0.0009614 |
| 267 | RESPONSE TO FLUID SHEAR STRESS | 3 | 34 | 6.066e-05 | 0.001057 |
| 268 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 6 | 307 | 6.131e-05 | 0.001064 |
| 269 | BONE REMODELING | 3 | 35 | 6.624e-05 | 0.001146 |
| 270 | POSITIVE REGULATION OF CELL SUBSTRATE ADHESION | 4 | 99 | 7.202e-05 | 0.001241 |
| 271 | CELL PART MORPHOGENESIS | 8 | 633 | 7.375e-05 | 0.001266 |
| 272 | HEART DEVELOPMENT | 7 | 466 | 7.518e-05 | 0.001286 |
| 273 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 9 | 823 | 7.562e-05 | 0.001289 |
| 274 | REGULATION OF LAMELLIPODIUM ORGANIZATION | 3 | 37 | 7.839e-05 | 0.001317 |
| 275 | RESPIRATORY SYSTEM DEVELOPMENT | 5 | 197 | 7.819e-05 | 0.001317 |
| 276 | BIOLOGICAL ADHESION | 10 | 1032 | 7.831e-05 | 0.001317 |
| 277 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 6 | 321 | 7.843e-05 | 0.001317 |
| 278 | ASTROCYTE DIFFERENTIATION | 3 | 39 | 9.192e-05 | 0.001532 |
| 279 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 3 | 39 | 9.192e-05 | 0.001532 |
| 280 | ACTIVATION OF INNATE IMMUNE RESPONSE | 5 | 204 | 9.218e-05 | 0.001532 |
| 281 | TISSUE DEVELOPMENT | 12 | 1518 | 9.522e-05 | 0.001577 |
| 282 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 3 | 40 | 9.921e-05 | 0.001637 |
| 283 | SYSTEM PROCESS | 13 | 1785 | 0.0001045 | 0.001718 |
| 284 | LYMPHOCYTE ACTIVATION | 6 | 342 | 0.0001111 | 0.00182 |
| 285 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 5 | 213 | 0.0001129 | 0.001844 |
| 286 | POSITIVE REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 3 | 42 | 0.0001149 | 0.00187 |
| 287 | CHEMICAL HOMEOSTASIS | 9 | 874 | 0.0001197 | 0.001934 |
| 288 | NEURON DIFFERENTIATION | 9 | 874 | 0.0001197 | 0.001934 |
| 289 | POSITIVE REGULATION OF HOMEOSTATIC PROCESS | 5 | 216 | 0.0001206 | 0.001941 |
| 290 | ALCOHOL METABOLIC PROCESS | 6 | 348 | 0.0001221 | 0.00196 |
| 291 | REGULATION OF SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 3 | 43 | 0.0001233 | 0.001972 |
| 292 | WNT SIGNALING PATHWAY | 6 | 351 | 0.000128 | 0.00204 |
| 293 | NEURON DEVELOPMENT | 8 | 687 | 0.0001302 | 0.00206 |
| 294 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 6 | 352 | 0.00013 | 0.00206 |
| 295 | REGULATION OF CELLULAR RESPONSE TO STRESS | 8 | 691 | 0.0001355 | 0.002137 |
| 296 | MAMMARY GLAND DEVELOPMENT | 4 | 117 | 0.0001377 | 0.002165 |
| 297 | LUNG MORPHOGENESIS | 3 | 45 | 0.0001414 | 0.002215 |
| 298 | EMBRYO DEVELOPMENT | 9 | 894 | 0.0001421 | 0.002215 |
| 299 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 4 | 118 | 0.0001423 | 0.002215 |
| 300 | PEPTIDYL THREONINE MODIFICATION | 3 | 46 | 0.000151 | 0.002342 |
| 301 | CYTOKINE PRODUCTION | 4 | 120 | 0.0001518 | 0.002347 |
| 302 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 6 | 363 | 0.0001537 | 0.002368 |
| 303 | POSITIVE REGULATION OF DEFENSE RESPONSE | 6 | 364 | 0.000156 | 0.002396 |
| 304 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 6 | 365 | 0.0001584 | 0.002424 |
| 305 | THYMUS DEVELOPMENT | 3 | 47 | 0.000161 | 0.002457 |
| 306 | HEAD DEVELOPMENT | 8 | 709 | 0.0001617 | 0.002458 |
| 307 | RESPONSE TO UV | 4 | 126 | 0.0001832 | 0.002777 |
| 308 | MUSCLE CELL DIFFERENTIATION | 5 | 237 | 0.0001859 | 0.002809 |
| 309 | REGULATION OF ERK1 AND ERK2 CASCADE | 5 | 238 | 0.0001896 | 0.002855 |
| 310 | FACE DEVELOPMENT | 3 | 50 | 0.0001937 | 0.002908 |
| 311 | MUSCLE CELL DEVELOPMENT | 4 | 128 | 0.0001946 | 0.002912 |
| 312 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 4 | 131 | 0.0002127 | 0.003172 |
| 313 | TUBE DEVELOPMENT | 7 | 552 | 0.0002145 | 0.003189 |
| 314 | EPITHELIUM DEVELOPMENT | 9 | 945 | 0.0002155 | 0.003193 |
| 315 | B CELL ACTIVATION | 4 | 132 | 0.000219 | 0.003229 |
| 316 | REGULATION OF NEURON DIFFERENTIATION | 7 | 554 | 0.0002193 | 0.003229 |
| 317 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 5 | 246 | 0.000221 | 0.003245 |
| 318 | LIPID CATABOLIC PROCESS | 5 | 247 | 0.0002252 | 0.003296 |
| 319 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 3 | 53 | 0.0002305 | 0.003362 |
| 320 | ESTABLISHMENT OF LOCALIZATION IN CELL | 12 | 1676 | 0.0002431 | 0.003534 |
| 321 | GLIAL CELL DIFFERENTIATION | 4 | 136 | 0.0002455 | 0.003559 |
| 322 | CELLULAR RESPONSE TO RADIATION | 4 | 137 | 0.0002525 | 0.003637 |
| 323 | ACTIVATION OF MAPK ACTIVITY | 4 | 137 | 0.0002525 | 0.003637 |
| 324 | PLACENTA DEVELOPMENT | 4 | 138 | 0.0002596 | 0.003728 |
| 325 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 6 | 406 | 0.0002812 | 0.004026 |
| 326 | POSITIVE REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 3 | 57 | 0.0002861 | 0.004083 |
| 327 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 5 | 263 | 0.0003009 | 0.004282 |
| 328 | VASCULOGENESIS | 3 | 59 | 0.0003168 | 0.004495 |
| 329 | REGULATION OF GOLGI ORGANIZATION | 2 | 12 | 0.0003221 | 0.004527 |
| 330 | CEREBRAL CORTEX GABAERGIC INTERNEURON DIFFERENTIATION | 2 | 12 | 0.0003221 | 0.004527 |
| 331 | TRACHEA MORPHOGENESIS | 2 | 12 | 0.0003221 | 0.004527 |
| 332 | POSITIVE REGULATION OF LYASE ACTIVITY | 3 | 61 | 0.0003497 | 0.004886 |
| 333 | POSITIVE REGULATION OF CYCLASE ACTIVITY | 3 | 61 | 0.0003497 | 0.004886 |
| 334 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 4 | 152 | 0.000375 | 0.005224 |
| 335 | LIPOXYGENASE PATHWAY | 2 | 13 | 0.0003801 | 0.005263 |
| 336 | LYMPH VESSEL MORPHOGENESIS | 2 | 13 | 0.0003801 | 0.005263 |
| 337 | CELL CELL ADHESION | 7 | 608 | 0.0003854 | 0.005289 |
| 338 | REGULATION OF MUSCLE ADAPTATION | 3 | 63 | 0.0003846 | 0.005289 |
| 339 | RESPONSE TO OSMOTIC STRESS | 3 | 63 | 0.0003846 | 0.005289 |
| 340 | RESPONSE TO LIGHT STIMULUS | 5 | 280 | 0.000401 | 0.005488 |
| 341 | PHOSPHATIDYLCHOLINE METABOLIC PROCESS | 3 | 64 | 0.0004029 | 0.005493 |
| 342 | REGULATION OF CYCLIC NUCLEOTIDE METABOLIC PROCESS | 4 | 155 | 0.0004038 | 0.005493 |
| 343 | REGULATION OF RESPIRATORY BURST | 2 | 14 | 0.0004428 | 0.005989 |
| 344 | POSITIVE REGULATION OF SPROUTING ANGIOGENESIS | 2 | 14 | 0.0004428 | 0.005989 |
| 345 | APOPTOTIC SIGNALING PATHWAY | 5 | 289 | 0.0004633 | 0.006248 |
| 346 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 4 | 162 | 0.0004771 | 0.006417 |
| 347 | PROTEIN LOCALIZATION | 12 | 1805 | 0.0004814 | 0.006439 |
| 348 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 3 | 68 | 0.0004816 | 0.006439 |
| 349 | VASCULAR PROCESS IN CIRCULATORY SYSTEM | 4 | 163 | 0.0004883 | 0.006511 |
| 350 | CYTOSKELETON ORGANIZATION | 8 | 838 | 0.0004993 | 0.006637 |
| 351 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 2 | 15 | 0.0005102 | 0.006687 |
| 352 | NEGATIVE REGULATION OF DENDRITE MORPHOGENESIS | 2 | 15 | 0.0005102 | 0.006687 |
| 353 | GABAERGIC NEURON DIFFERENTIATION | 2 | 15 | 0.0005102 | 0.006687 |
| 354 | REGULATION OF HYDROGEN PEROXIDE METABOLIC PROCESS | 2 | 15 | 0.0005102 | 0.006687 |
| 355 | NEUROTROPHIN TRK RECEPTOR SIGNALING PATHWAY | 2 | 15 | 0.0005102 | 0.006687 |
| 356 | CENTRAL NERVOUS SYSTEM NEURON DEVELOPMENT | 3 | 70 | 0.0005244 | 0.006854 |
| 357 | NEGATIVE REGULATION OF TRANSPORT | 6 | 458 | 0.0005333 | 0.006951 |
| 358 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 4 | 167 | 0.000535 | 0.006954 |
| 359 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 3 | 71 | 0.0005467 | 0.007085 |
| 360 | AMMONIUM ION METABOLIC PROCESS | 4 | 169 | 0.0005596 | 0.007232 |
| 361 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 9 | 1079 | 0.0005714 | 0.007364 |
| 362 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 5 | 303 | 0.0005743 | 0.007382 |
| 363 | POSITIVE REGULATION OF LAMELLIPODIUM ASSEMBLY | 2 | 16 | 0.0005822 | 0.007422 |
| 364 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 16 | 0.0005822 | 0.007422 |
| 365 | PHOSPHATIDYLINOSITOL ACYL CHAIN REMODELING | 2 | 16 | 0.0005822 | 0.007422 |
| 366 | STRIATED MUSCLE CELL DIFFERENTIATION | 4 | 173 | 0.0006109 | 0.007767 |
| 367 | GLIOGENESIS | 4 | 175 | 0.0006379 | 0.008087 |
| 368 | IN UTERO EMBRYONIC DEVELOPMENT | 5 | 311 | 0.0006462 | 0.008126 |
| 369 | RESPONSE TO OXYGEN LEVELS | 5 | 311 | 0.0006462 | 0.008126 |
| 370 | POSITIVE REGULATION OF CELL ACTIVATION | 5 | 311 | 0.0006462 | 0.008126 |
| 371 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 8 | 872 | 0.0006491 | 0.008141 |
| 372 | RESPONSE TO CORTICOSTEROID | 4 | 176 | 0.0006516 | 0.008151 |
| 373 | PHOSPHATIDYLSERINE ACYL CHAIN REMODELING | 2 | 17 | 0.0006589 | 0.008176 |
| 374 | REGULATION OF CHROMATIN BINDING | 2 | 17 | 0.0006589 | 0.008176 |
| 375 | REGULATION OF PRI MIRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 2 | 17 | 0.0006589 | 0.008176 |
| 376 | RESPONSE TO INORGANIC SUBSTANCE | 6 | 479 | 0.0006748 | 0.00835 |
| 377 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 6 | 482 | 0.0006971 | 0.008604 |
| 378 | RESPONSE TO BIOTIC STIMULUS | 8 | 886 | 0.0007206 | 0.00887 |
| 379 | REGULATION OF MITOCHONDRIAL DEPOLARIZATION | 2 | 18 | 0.0007402 | 0.009016 |
| 380 | REGULATION OF CELL MATURATION | 2 | 18 | 0.0007402 | 0.009016 |
| 381 | OVULATION | 2 | 18 | 0.0007402 | 0.009016 |
| 382 | MAST CELL MEDIATED IMMUNITY | 2 | 18 | 0.0007402 | 0.009016 |
| 383 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 4 | 185 | 0.0007852 | 0.009539 |
| 384 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 3 | 81 | 0.0008033 | 0.009684 |
| 385 | PEPTIDYL TYROSINE MODIFICATION | 4 | 186 | 0.0008012 | 0.009684 |
| 386 | POSITIVE REGULATION OF LEUKOCYTE CHEMOTAXIS | 3 | 81 | 0.0008033 | 0.009684 |
| 387 | CELLULAR RESPONSE TO FLUID SHEAR STRESS | 2 | 19 | 0.0008261 | 0.009907 |
| 388 | REGULATION OF CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 2 | 19 | 0.0008261 | 0.009907 |
| 389 | NEGATIVE REGULATION OF PROTEOLYSIS | 5 | 329 | 0.0008323 | 0.009955 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | KINASE ACTIVITY | 20 | 842 | 2.873e-16 | 2.669e-13 |
| 2 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 20 | 992 | 6.553e-15 | 3.044e-12 |
| 3 | KINASE BINDING | 15 | 606 | 2.088e-12 | 6.466e-10 |
| 4 | RIBONUCLEOTIDE BINDING | 22 | 1860 | 8.984e-12 | 2.086e-09 |
| 5 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 70 | 1.931e-10 | 3.145e-08 |
| 6 | ENZYME BINDING | 20 | 1737 | 2.031e-10 | 3.145e-08 |
| 7 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 6 | 43 | 5.255e-10 | 6.974e-08 |
| 8 | PROTEIN KINASE ACTIVITY | 13 | 640 | 9.303e-10 | 1.08e-07 |
| 9 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 6 | 51 | 1.532e-09 | 1.581e-07 |
| 10 | KINASE REGULATOR ACTIVITY | 8 | 186 | 7.73e-09 | 7.182e-07 |
| 11 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 10 | 445 | 4.274e-08 | 3.609e-06 |
| 12 | ADENYL NUCLEOTIDE BINDING | 16 | 1514 | 8.433e-08 | 6.528e-06 |
| 13 | INSULIN RECEPTOR SUBSTRATE BINDING | 3 | 11 | 1.734e-06 | 0.0001239 |
| 14 | CALCIUM DEPENDENT PROTEIN KINASE ACTIVITY | 3 | 12 | 2.309e-06 | 0.0001341 |
| 15 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 6 | 172 | 2.287e-06 | 0.0001341 |
| 16 | PHOSPHOLIPASE ACTIVITY | 5 | 94 | 2.169e-06 | 0.0001341 |
| 17 | PROTEIN KINASE C BINDING | 4 | 50 | 4.774e-06 | 0.0002609 |
| 18 | PROTEIN KINASE C ACTIVITY | 3 | 16 | 5.84e-06 | 0.0002855 |
| 19 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 3 | 16 | 5.84e-06 | 0.0002855 |
| 20 | LIPASE ACTIVITY | 5 | 117 | 6.372e-06 | 0.000296 |
| 21 | MOLECULAR FUNCTION REGULATOR | 12 | 1353 | 3.103e-05 | 0.001373 |
| 22 | HYDROLASE ACTIVITY ACTING ON ESTER BONDS | 9 | 739 | 3.282e-05 | 0.001386 |
| 23 | PHOSPHOLIPASE A2 ACTIVITY | 3 | 31 | 4.578e-05 | 0.001849 |
| 24 | RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY | 4 | 92 | 5.407e-05 | 0.002093 |
| 25 | PHOSPHORIC ESTER HYDROLASE ACTIVITY | 6 | 368 | 0.0001656 | 0.006152 |
| 26 | ENZYME REGULATOR ACTIVITY | 9 | 959 | 0.0002405 | 0.008383 |
| 27 | PROTEIN TYROSINE KINASE BINDING | 3 | 54 | 0.0002437 | 0.008383 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 7 | 20 | 1.357e-14 | 7.924e-12 |
| 2 | EXTRINSIC COMPONENT OF MEMBRANE | 11 | 252 | 7.106e-12 | 2.075e-09 |
| 3 | CELL LEADING EDGE | 9 | 350 | 7.067e-08 | 1.376e-05 |
| 4 | LAMELLIPODIUM | 7 | 172 | 1.061e-07 | 1.549e-05 |
| 5 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 7 | 237 | 9.288e-07 | 0.0001085 |
| 6 | CELL SUBSTRATE JUNCTION | 8 | 398 | 2.595e-06 | 0.0002526 |
| 7 | CELL JUNCTION | 12 | 1151 | 6.1e-06 | 0.0005089 |
| 8 | ANCHORING JUNCTION | 8 | 489 | 1.174e-05 | 0.0008568 |
| 9 | ACTIN FILAMENT | 4 | 70 | 1.839e-05 | 0.001194 |
| 10 | MEMBRANE MICRODOMAIN | 6 | 288 | 4.3e-05 | 0.002283 |
| 11 | PLASMA MEMBRANE RAFT | 4 | 86 | 4.15e-05 | 0.002283 |
| 12 | ACTIN CYTOSKELETON | 7 | 444 | 5.548e-05 | 0.0027 |
| 13 | CATALYTIC COMPLEX | 10 | 1038 | 8.219e-05 | 0.003692 |
| 14 | CELL PROJECTION | 13 | 1786 | 0.0001051 | 0.004383 |
| 15 | PHOSPHATASE COMPLEX | 3 | 48 | 0.0001715 | 0.00626 |
| 16 | MEMBRANE REGION | 10 | 1134 | 0.0001706 | 0.00626 |
| 17 | EXTRINSIC COMPONENT OF PLASMA MEMBRANE | 4 | 136 | 0.0002455 | 0.008434 |
| 18 | CYTOSKELETON | 13 | 1967 | 0.0002771 | 0.008992 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
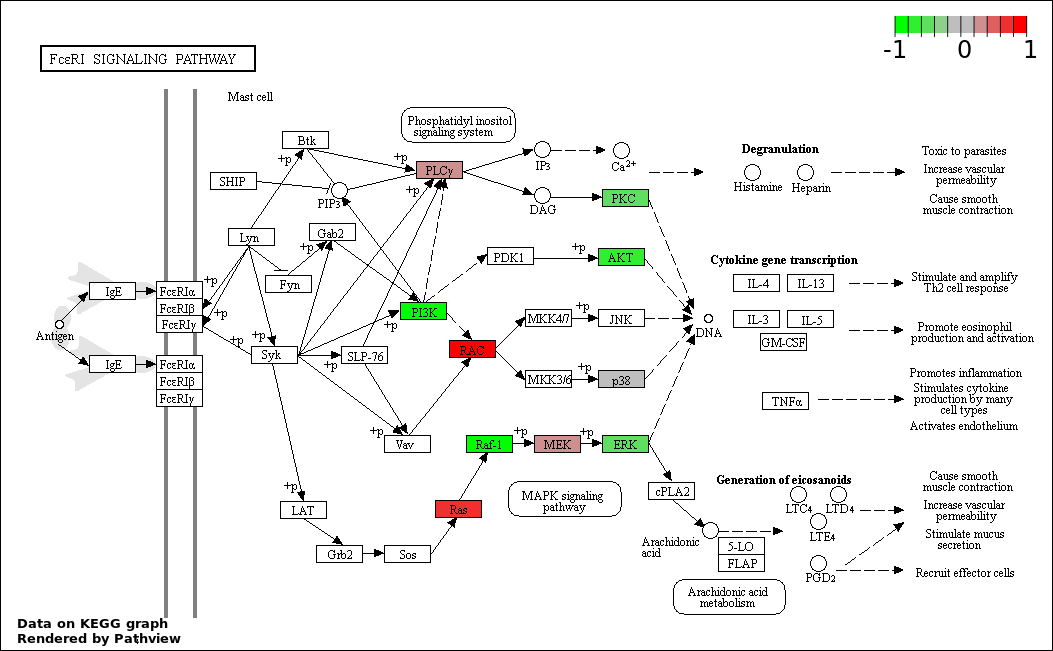
| 1 | hsa04370_VEGF_signaling_pathway | 45 | 76 | 6.848e-117 | 1.233e-114 | |
| 2 | hsa04662_B_cell_receptor_signaling_pathway | 27 | 75 | 2.494e-56 | 2.244e-54 | |
| 3 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 28 | 136 | 1.058e-50 | 6.349e-49 | |
| 4 | hsa04664_Fc_epsilon_RI_signaling_pathway | 25 | 79 | 3.528e-50 | 1.588e-48 | |
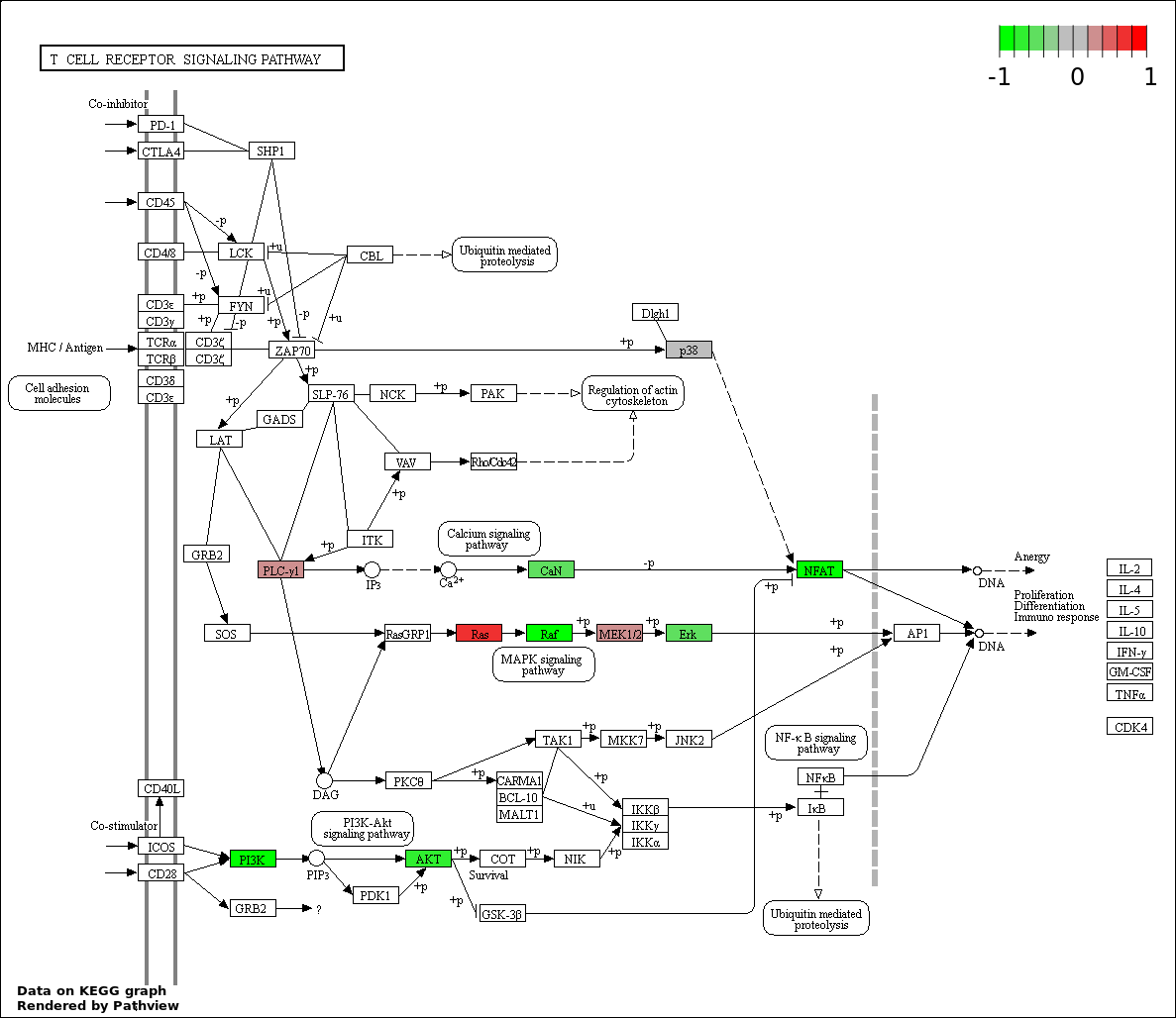
| 5 | hsa04660_T_cell_receptor_signaling_pathway | 24 | 108 | 8.372e-44 | 3.014e-42 | |
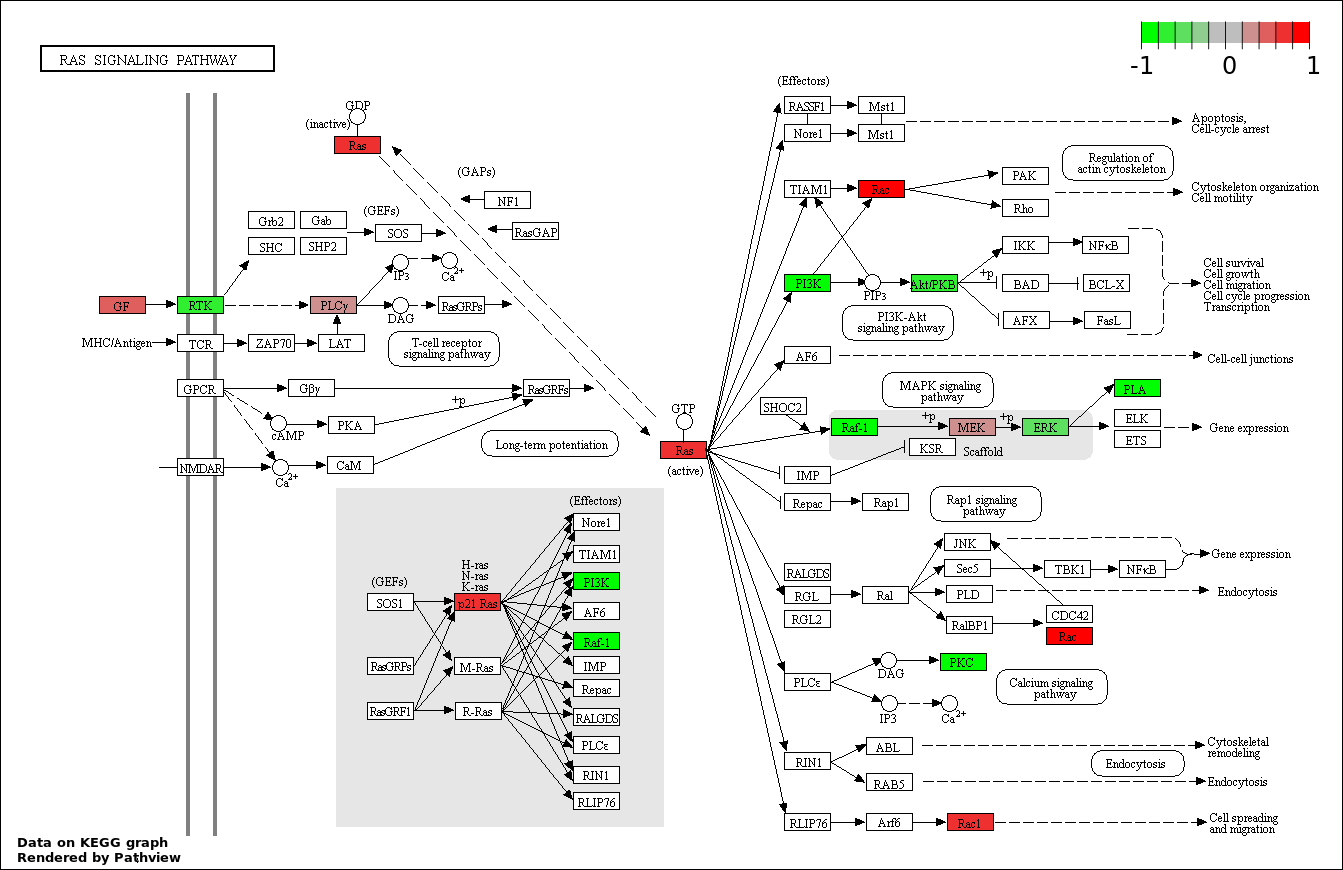
| 6 | hsa04014_Ras_signaling_pathway | 26 | 236 | 3.616e-39 | 1.085e-37 | |
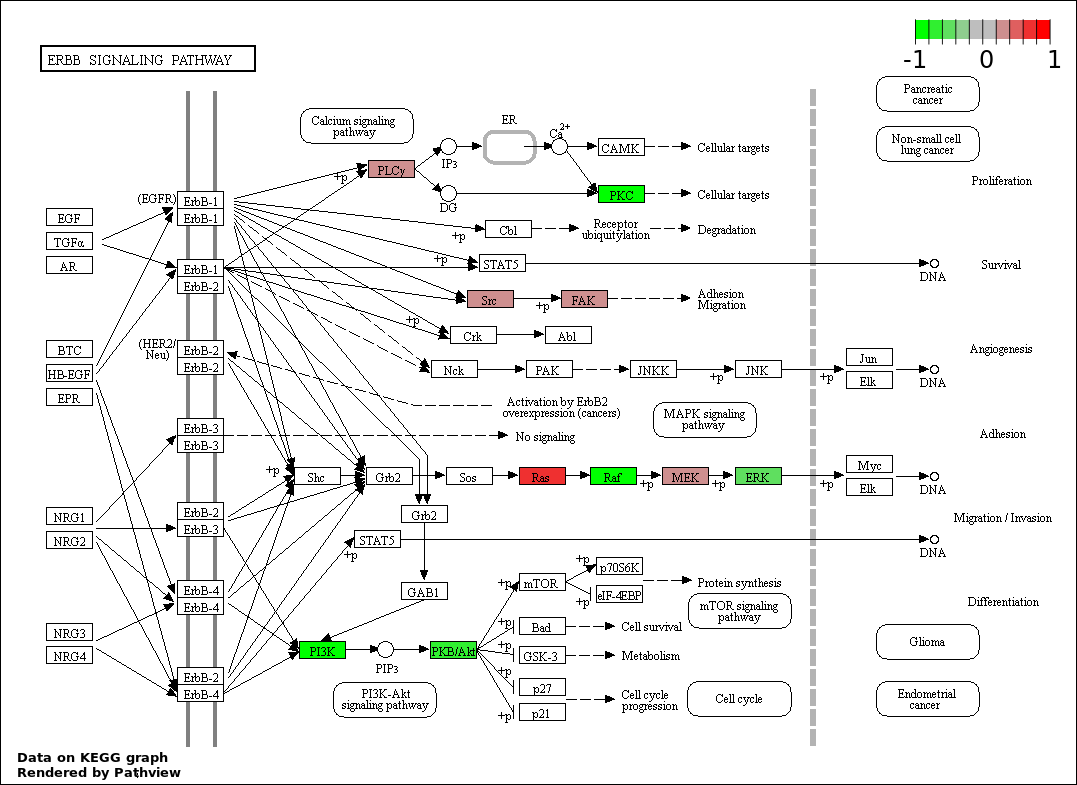
| 7 | hsa04012_ErbB_signaling_pathway | 21 | 87 | 6.527e-39 | 1.678e-37 | |
| 8 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 20 | 95 | 1.162e-35 | 2.614e-34 | |
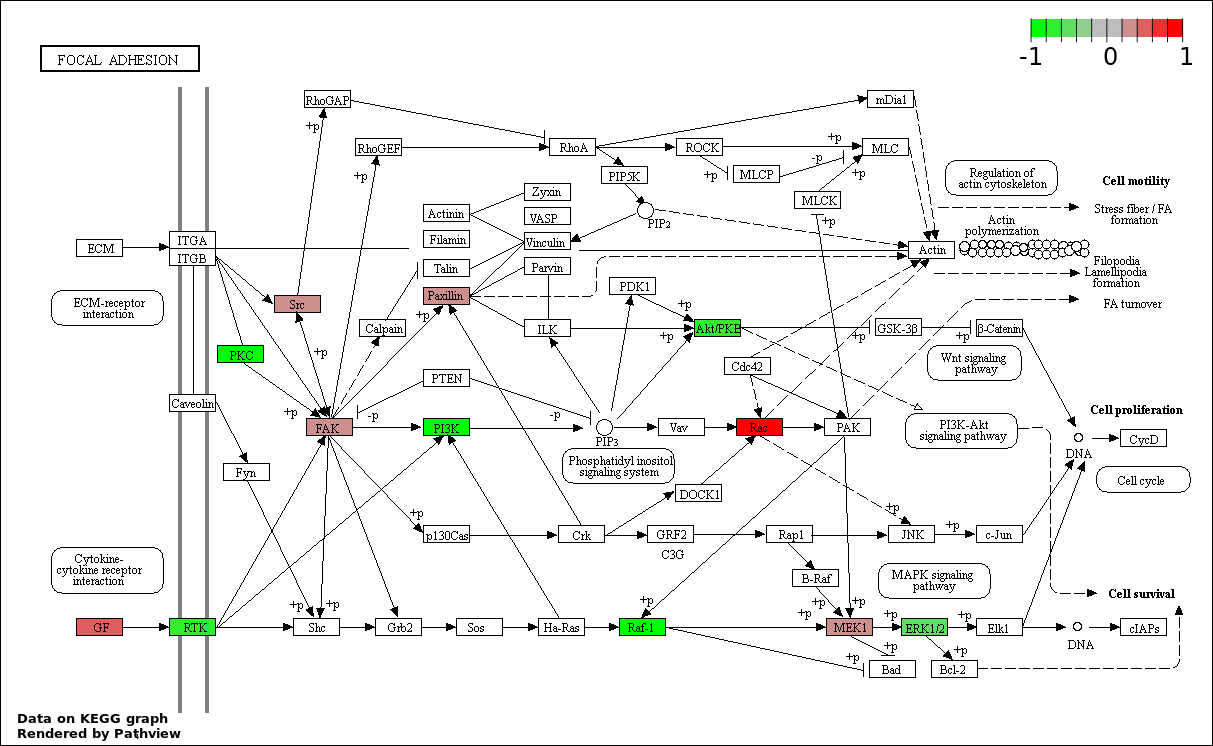
| 9 | hsa04510_Focal_adhesion | 23 | 200 | 9.275e-35 | 1.855e-33 | |
| 10 | hsa04010_MAPK_signaling_pathway | 23 | 268 | 1.02e-31 | 1.836e-30 | |
| 11 | hsa04722_Neurotrophin_signaling_pathway | 18 | 127 | 1.197e-28 | 1.958e-27 | |
| 12 | hsa04380_Osteoclast_differentiation | 18 | 128 | 1.391e-28 | 2.086e-27 | |
| 13 | hsa04062_Chemokine_signaling_pathway | 19 | 189 | 2.666e-27 | 3.692e-26 | |
| 14 | hsa04670_Leukocyte_transendothelial_migration | 17 | 117 | 3.156e-27 | 4.058e-26 | |
| 15 | hsa04151_PI3K_AKT_signaling_pathway | 21 | 351 | 1.907e-25 | 2.289e-24 | |
| 16 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 12 | 42 | 3.565e-23 | 4.01e-22 | |
| 17 | hsa04914_Progesterone.mediated_oocyte_maturation | 14 | 87 | 4.34e-23 | 4.595e-22 | |
| 18 | hsa04810_Regulation_of_actin_cytoskeleton | 17 | 214 | 1.406e-22 | 1.406e-21 | |
| 19 | hsa04360_Axon_guidance | 15 | 130 | 1.989e-22 | 1.884e-21 | |
| 20 | hsa04210_Apoptosis | 13 | 89 | 6.994e-21 | 6.295e-20 | |
| 21 | hsa04910_Insulin_signaling_pathway | 14 | 138 | 3.925e-20 | 3.364e-19 | |
| 22 | hsa04620_Toll.like_receptor_signaling_pathway | 13 | 102 | 4.563e-20 | 3.734e-19 | |
| 23 | hsa04150_mTOR_signaling_pathway | 11 | 52 | 1.124e-19 | 8.797e-19 | |
| 24 | hsa04070_Phosphatidylinositol_signaling_system | 12 | 78 | 1.325e-19 | 9.938e-19 | |
| 25 | hsa04310_Wnt_signaling_pathway | 14 | 151 | 1.443e-19 | 1.039e-18 | |
| 26 | hsa04973_Carbohydrate_digestion_and_absorption | 10 | 44 | 2.663e-18 | 1.844e-17 | |
| 27 | hsa04912_GnRH_signaling_pathway | 12 | 101 | 3.511e-18 | 2.341e-17 | |
| 28 | hsa04720_Long.term_potentiation | 11 | 70 | 3.915e-18 | 2.43e-17 | |
| 29 | hsa04730_Long.term_depression | 11 | 70 | 3.915e-18 | 2.43e-17 | |
| 30 | hsa04540_Gap_junction | 9 | 90 | 3.896e-13 | 2.338e-12 | |
| 31 | hsa04270_Vascular_smooth_muscle_contraction | 9 | 116 | 4.031e-12 | 2.341e-11 | |
| 32 | hsa04020_Calcium_signaling_pathway | 10 | 177 | 5.571e-12 | 3.134e-11 | |
| 33 | hsa04630_Jak.STAT_signaling_pathway | 9 | 155 | 5.574e-11 | 3.04e-10 | |
| 34 | hsa04916_Melanogenesis | 8 | 101 | 5.894e-11 | 3.12e-10 | |
| 35 | hsa04530_Tight_junction | 8 | 133 | 5.42e-10 | 2.788e-09 | |
| 36 | hsa04972_Pancreatic_secretion | 7 | 101 | 2.631e-09 | 1.315e-08 | |
| 37 | hsa00562_Inositol_phosphate_metabolism | 5 | 57 | 1.76e-07 | 8.561e-07 | |
| 38 | hsa04520_Adherens_junction | 5 | 73 | 6.147e-07 | 2.912e-06 | |
| 39 | hsa04114_Oocyte_meiosis | 5 | 114 | 5.611e-06 | 2.59e-05 | |
| 40 | hsa00590_Arachidonic_acid_metabolism | 4 | 59 | 9.297e-06 | 4.184e-05 | |
| 41 | hsa00592_alpha.Linolenic_acid_metabolism | 3 | 20 | 1.181e-05 | 5.186e-05 | |
| 42 | hsa04320_Dorso.ventral_axis_formation | 3 | 25 | 2.365e-05 | 0.0001013 | |
| 43 | hsa00591_Linoleic_acid_metabolism | 3 | 30 | 4.142e-05 | 0.0001734 | |
| 44 | hsa00565_Ether_lipid_metabolism | 3 | 36 | 7.215e-05 | 0.0002952 | |
| 45 | hsa04975_Fat_digestion_and_absorption | 3 | 46 | 0.000151 | 0.000604 | |
| 46 | hsa04971_Gastric_acid_secretion | 3 | 74 | 0.0006171 | 0.002415 | |
| 47 | hsa00564_Glycerophospholipid_metabolism | 3 | 80 | 0.0007748 | 0.002967 | |
| 48 | hsa04970_Salivary_secretion | 3 | 89 | 0.001056 | 0.00396 | |
| 49 | hsa04621_NOD.like_receptor_signaling_pathway | 2 | 59 | 0.007807 | 0.02868 | |
| 50 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.01026 | 0.03695 | |
| 51 | hsa04144_Endocytosis | 2 | 203 | 0.0764 | 0.2696 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RFPL1S |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p | 11 | AKT3 | Sponge network | -0.223 | 0.70704 | -0.749 | 0.06936 | 0.466 |
| 2 | CASC2 | hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-193b-5p;hsa-miR-218-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-654-3p;hsa-miR-944 | 12 | PRKCA | Sponge network | -0.561 | 0.05962 | -0.525 | 0.15685 | 0.436 |
| 3 | PCA3 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-452-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-616-5p | 12 | PRKCB | Sponge network | -2.778 | 8.0E-5 | -1.403 | 0.00367 | 0.387 |
| 4 | EMX2OS |
hsa-miR-107;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-32-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577 | 12 | AKT3 | Sponge network | -1.088 | 0.10042 | -0.749 | 0.06936 | 0.354 |
| 5 | MEG3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-616-5p | 11 | AKT3 | Sponge network | -1.645 | 0.00049 | -0.749 | 0.06936 | 0.312 |
| 6 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 28 | PIK3R1 | Sponge network | -2.778 | 8.0E-5 | -1.094 | 2.0E-5 | 0.309 |
| 7 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b | 11 | NFAT5 | Sponge network | -1.728 | 0.00016 | -0.044 | 0.82158 | 0.278 |
| 8 | PCA3 |
hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-2355-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-944 | 11 | PRKCA | Sponge network | -2.778 | 8.0E-5 | -0.525 | 0.15685 | 0.278 |
| 9 | MEG3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-652-3p | 12 | NFAT5 | Sponge network | -1.645 | 0.00049 | -0.044 | 0.82158 | 0.265 |